

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| METSSPRPPRPSSNPGLSLDARLGVDTRLWAKVLFTALYALIWALGAAGNALSAHVVLKARAGRAGRLRHHVLSLALAGLLLLLVGVPVELYSFVWFHYPWVFGDLGCRGYYFVHELCAYATVLSVAGLSAERCLAVCQPLRARSLLTPRRTRWLVALSWAASLGLALPMAVIMGQKHELETADGEPEPASRVCTVLVSRTALQVFIQVNVLVSFVLPLALTAFLNGVTVSHLLALCSQVPSTSTPGSSTPSRLELLSEEGLLSFIVWKKTFIQGGQVSLVRHKDVRRIRSLQRSVQVLRAIVVMYVICWLPYHARRLMYCYVPDDAWTDPLYNFYHYFYMVTNTLFYVSSAVTPLLYNAVSSSFRKLFLEAVSSLCGEHHPMKRLPPKPQSPTLMDTASGFGDPPETRT | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSSCCCCCCCCCCCCSSSCCCCCCHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99999999999999998877666778536999999999999999999999998798740688888650999999999999999998169999999806997686324436999999999999999999998868741142745313569999999999999999999999998022541134677788887173365783112025159999999999999999999999999998755102356655564422223434444555444455677775444307788888987899999999999999830999999999984534456405789999999999999999999999999418999999999974643799855678888988775668889999999999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| METSSPRPPRPSSNPGLSLDARLGVDTRLWAKVLFTALYALIWALGAAGNALSAHVVLKARAGRAGRLRHHVLSLALAGLLLLLVGVPVELYSFVWFHYPWVFGDLGCRGYYFVHELCAYATVLSVAGLSAERCLAVCQPLRARSLLTPRRTRWLVALSWAASLGLALPMAVIMGQKHELETADGEPEPASRVCTVLVSRTALQVFIQVNVLVSFVLPLALTAFLNGVTVSHLLALCSQVPSTSTPGSSTPSRLELLSEEGLLSFIVWKKTFIQGGQVSLVRHKDVRRIRSLQRSVQVLRAIVVMYVICWLPYHARRLMYCYVPDDAWTDPLYNFYHYFYMVTNTLFYVSSAVTPLLYNAVSSSFRKLFLEAVSSLCGEHHPMKRLPPKPQSPTLMDTASGFGDPPETRT | |
| 74443442343333332333343425230001001011012002202311200000001123023110000000001000000000000000010133320000100010000000000000000000000000100010020323113300000000000100000110000011233232343342221100010124321200000000210120010000000100020122134233443444333433433444433434434444364444344444434333342200100000000100110100000001000342324431110000010101110000000214320001464140012001000333444443454443333332333444454468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSSCCCCCCCCCCCCSSSCCCCCCHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC METSSPRPPRPSSNPGLSLDARLGVDTRLWAKVLFTALYALIWALGAAGNALSAHVVLKARAGRAGRLRHHVLSLALAGLLLLLVGVPVELYSFVWFHYPWVFGDLGCRGYYFVHELCAYATVLSVAGLSAERCLAVCQPLRARSLLTPRRTRWLVALSWAASLGLALPMAVIMGQKHELETADGEPEPASRVCTVLVSRTALQVFIQVNVLVSFVLPLALTAFLNGVTVSHLLALCSQVPSTSTPGSSTPSRLELLSEEGLLSFIVWKKTFIQGGQVSLVRHKDVRRIRSLQRSVQVLRAIVVMYVICWLPYHARRLMYCYVPDDAWTDPLYNFYHYFYMVTNTLFYVSSAVTPLLYNAVSSSFRKLFLEAVSSLCGEHHPMKRLPPKPQSPTLMDTASGFGDPPETRT | |||||||||||||||||||||||||
| 1 | 5vblB | 0.18 | 0.21 | 0.86 | 2.42 | Download | ----------------------CEYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSREKRRSADIFIASLAVADLTFVVTL-PLWATYTYRD-YDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTNKVQCYMDYSMVATVSSEAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKYTCTVCGYIYNPEDGDPDNGVNPGTDFKDIPDDWVC---PLCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWPCDFDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.20 | 0.24 | 0.86 | 4.27 | Download | ----------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGP-NILIASLALGDLLHIVIAIPINVYKLLA--EDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDY-----KGSYLRICLLHPVQKYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIFEMLRIDEGGGSGGDEAEKLFNQDVDAAVRGILRNAKLRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNRLLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC----------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.18 | 0.20 | 0.82 | 3.02 | Download | --------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKS-MTDIYLLNLAISDLFFLLTVPFWAHYAAA----QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLH---------YTCSSHFPQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKYTCTVCGYIYNPEDGDPDNGVNPGTDFKDIPVCPLCGVGKDQFEEVEEEK--KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.19 | 0.81 | 1.56 | Download | -------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQ-YATNYFLMSLAVADLLVGLFVMPIALLTIM-FEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK-GIETN---------PNNITCVLTKE--RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLADNAMRAAALDKKDGFDIANEGKEAAEQLKTNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC--N---QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-------------------------------- | |||||||||||||||||||
| 5 | 3uon | 0.15 | 0.20 | 0.80 | 1.19 | Download | ----------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVN-NYFLFSLACADLIIGVFSMNLYTLYTV--IGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVR-----TVEDGECYIQFFS---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKEGAAIGRNTNGVITKDVRGILRNAKKSTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCI--------PNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------ | |||||||||||||||||||
| 6 | 5vblB | 0.18 | 0.21 | 0.84 | 2.96 | Download | ------------------------YTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYD--WPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGD-------LENTNKVQCYMDSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKYTCTVCGYIYNPEDGDPDNGVNPGTDFKDIPDDWV---CPLCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWPCDFDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.17 | 0.18 | 0.74 | 1.70 | Download | -------------------------QVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPA-NMFIINLAFSDFTFSLVGFPLMTISCF--LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEG--------VLCNCSFDYRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLN------------------------------AKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVT-------PYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPW----------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.44 | 0.34 | 0.74 | 4.58 | Download | -------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQS-TVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNL---SGDGTHPGGLVCTPIVDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQ----------------------------------------------PGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTTFLFDFYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------------------------------------- | |||||||||||||||||||
| 9 | 5vblB | 0.18 | 0.21 | 0.86 | 2.62 | Download | ----------------------CEYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRD--YDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTNKVQCYMDYSMVATVSSEAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKYTCTVCGYIYNPEDGDPDNGVNPGTDFKDIPDDWVCP---LCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWPCDFDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR------------------------------- | |||||||||||||||||||
| 10 | 3zpqA | 0.21 | 0.20 | 0.70 | 2.35 | Download | ----------------------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTL-TNLFITSLACADLVVGLLVVPFGATLVVR--GTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDE-DPQALKCYQDPGCCDFVTN----RAYAIASSIISFYIPLLIMIFVALRVYREAKEQ------------------------------------------------SRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP--------DWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLLA------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
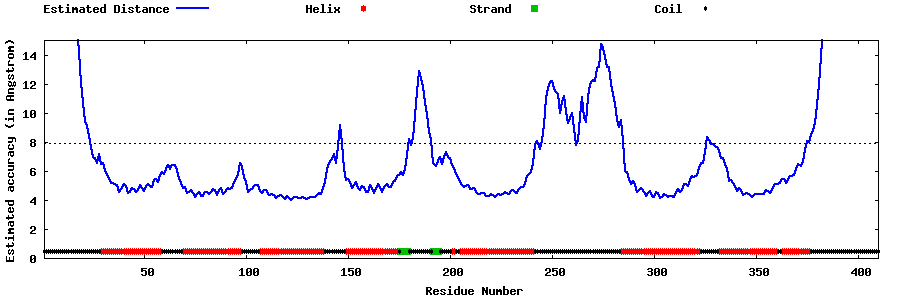 | |||
|
|
|||
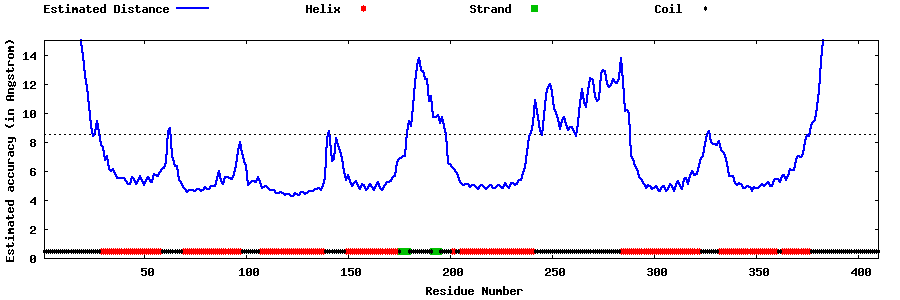 | |||
|
| |||
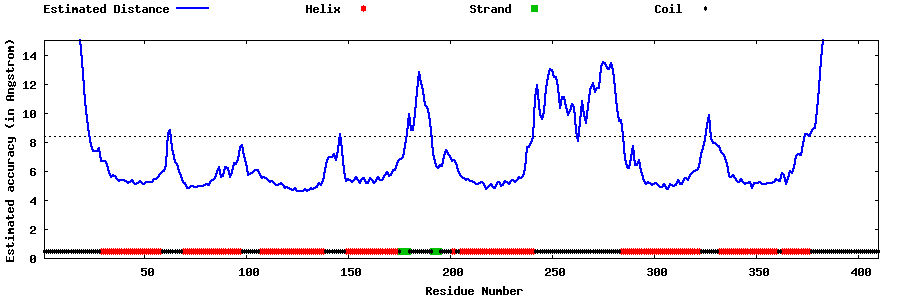 | |||
|
| |||
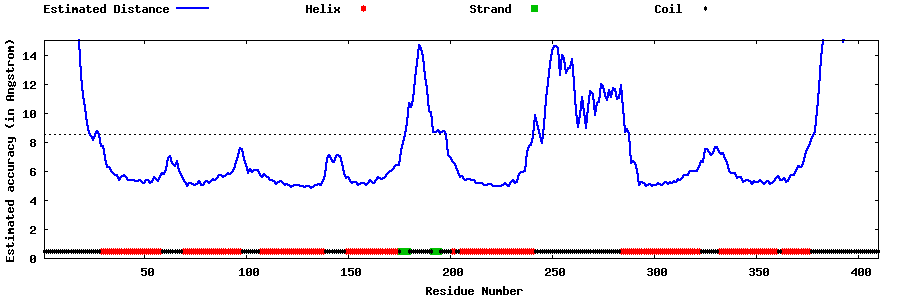 | |||
|
| |||
 | |||
|
| |||