

You can:
| Name | G-protein coupled receptor 84 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GPR84 |
| Synonym | GPCR4 GPR84 Inflammation-related G-protein coupled receptor EX33 |
| Disease | N/A |
| Length | 396 |
| Amino acid sequence | MWNSSDANFSCYHESVLGYRYVAVSWGVVVAVTGTVGNVLTLLALAIQPKLRTRFNLLIANLTLADLLYCTLLQPFSVDTYLHLHWRTGATFCRVFGLLLFASNSVSILTLCLIALGRYLLIAHPKLFPQVFSAKGIVLALVSTWVVGVASFAPLWPIYILVPVVCTCSFDRIRGRPYTTILMGIYFVLGLSSVGIFYCLIHRQVKRAAQALDQYKLRQASIHSNHVARTDEAMPGRFQELDSRLASGGPSEGISSEPVSAATTQTLEGDSSEVGDQINSKRAKQMAEKSPPEASAKAQPIKGARRAPDSSSEFGKVTRMCFAVFLCFALSYIPFLLLNILDARVQAPRVVHMLAANLTWLNGCINPVLYAAMNRQFRQAYGSILKRGPRSFHRLH |
| UniProt | Q9NQS5 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9NQS5 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9NQS5. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3714079 |
| IUPHAR | 120 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 521474 | 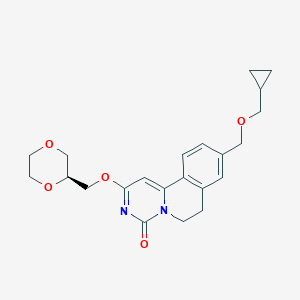 CHEMBL3715817 CHEMBL3715817 | C22H26N2O5 | 398.459 | 5 / 0 | 1.5 | Yes |
| 521521 | 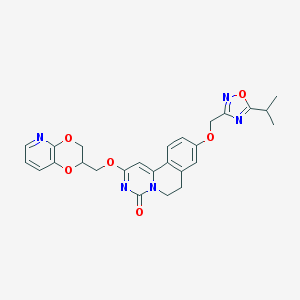 CHEMBL3716930 CHEMBL3716930 | C26H25N5O6 | 503.515 | 9 / 0 | 3.2 | No |
| 521618 | 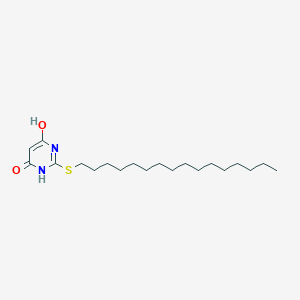 CHEMBL3828578 CHEMBL3828578 | C20H36N2O2S | 368.58 | 4 / 2 | 7.8 | No |
| 521665 | 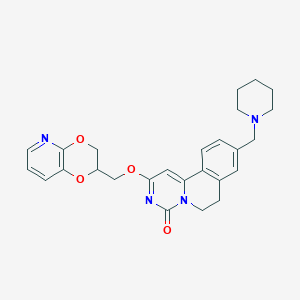 CHEMBL3719012 CHEMBL3719012 | C26H28N4O4 | 460.534 | 6 / 0 | 3.0 | Yes |
| 521670 | 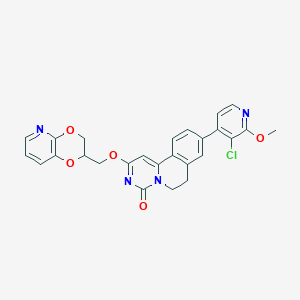 CHEMBL3718281 CHEMBL3718281 | C26H21ClN4O5 | 504.927 | 7 / 0 | 3.8 | No |
| 521676 | 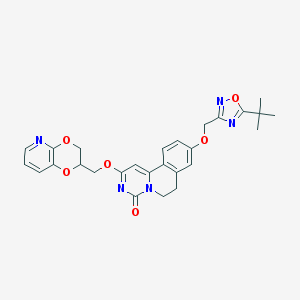 CHEMBL3715645 CHEMBL3715645 | C27H27N5O6 | 517.542 | 9 / 0 | 3.7 | No |
| 521679 | 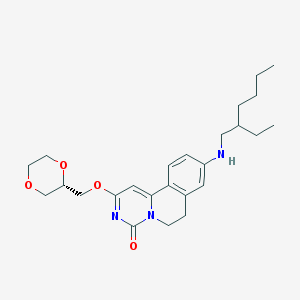 CHEMBL3715870 CHEMBL3715870 | C25H35N3O4 | 441.572 | 5 / 1 | 4.2 | Yes |
| 521834 | 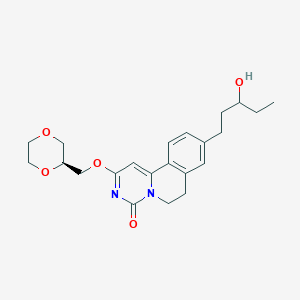 CHEMBL3715265 CHEMBL3715265 | C22H28N2O5 | 400.475 | 5 / 1 | 2.0 | Yes |
| 521849 | 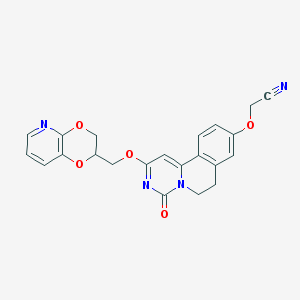 CHEMBL3715646 CHEMBL3715646 | C22H18N4O5 | 418.409 | 7 / 0 | 2.0 | Yes |
| 521976 | 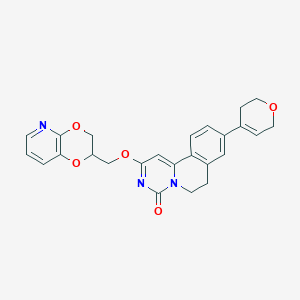 CHEMBL3717958 CHEMBL3717958 | C25H23N3O5 | 445.475 | 6 / 0 | 2.3 | Yes |
| 521980 | 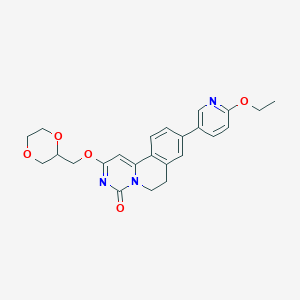 CHEMBL3718999 CHEMBL3718999 | C24H25N3O5 | 435.48 | 6 / 0 | 2.3 | Yes |
| 522055 | 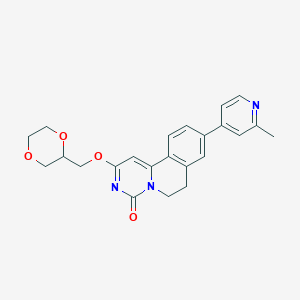 CHEMBL3719139 CHEMBL3719139 | C23H23N3O4 | 405.454 | 5 / 0 | 2.0 | Yes |
| 522077 | 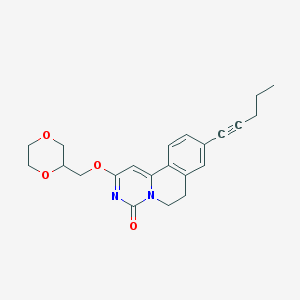 CHEMBL3715514 CHEMBL3715514 | C22H24N2O4 | 380.444 | 4 / 0 | 2.8 | Yes |
| 522098 | 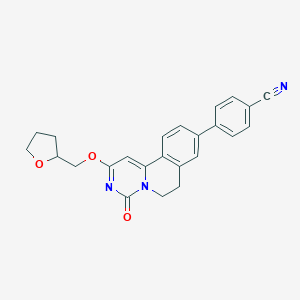 CHEMBL3717904 CHEMBL3717904 | C24H21N3O3 | 399.45 | 4 / 0 | 3.3 | Yes |
| 522099 | 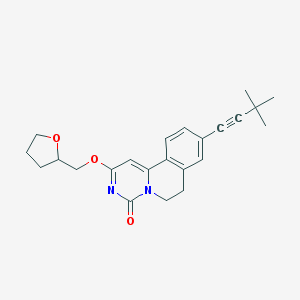 CHEMBL3718208 CHEMBL3718208 | C23H26N2O3 | 378.472 | 3 / 0 | 4.0 | Yes |
| 522249 | 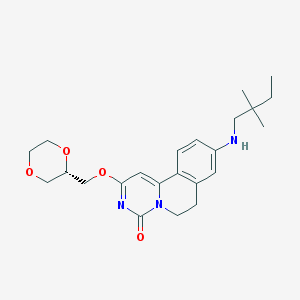 CHEMBL3716561 CHEMBL3716561 | C23H31N3O4 | 413.518 | 5 / 1 | 3.1 | Yes |
| 522371 |  CHEMBL3718718 CHEMBL3718718 | C27H23N3O5 | 469.497 | 6 / 0 | 3.9 | Yes |
| 522396 |  CHEMBL3714917 CHEMBL3714917 | C22H20N2O5 | 392.411 | 5 / 0 | 3.0 | Yes |
| 522515 |  CHEMBL3717267 CHEMBL3717267 | C26H22N4O5 | 470.485 | 7 / 0 | 3.1 | Yes |
| 522516 |  CHEMBL3717323 CHEMBL3717323 | C27H30N4O6 | 506.559 | 8 / 0 | 1.6 | No |
| 522541 |  CHEMBL3716619 CHEMBL3716619 | C24H32N2O5 | 428.529 | 5 / 0 | 3.5 | Yes |
| 522578 |  CHEMBL3827283 CHEMBL3827283 | C12H12N2O2S | 248.3 | 4 / 2 | 2.0 | Yes |
| 522653 |  CHEMBL3828033 CHEMBL3828033 | C10H14N2O2S | 226.294 | 4 / 2 | 1.8 | Yes |
| 522740 |  CHEMBL3714952 CHEMBL3714952 | C24H23FN2O4 | 422.456 | 5 / 0 | 3.5 | Yes |
| 522753 | 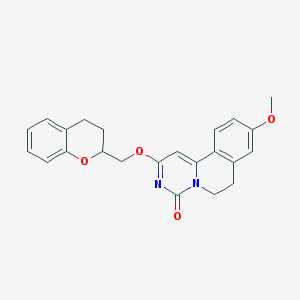 CHEMBL3715962 CHEMBL3715962 | C23H22N2O4 | 390.439 | 4 / 0 | 3.6 | Yes |
| 522759 | 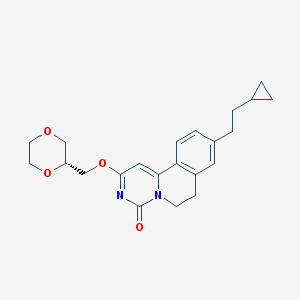 CHEMBL3715907 CHEMBL3715907 | C22H26N2O4 | 382.46 | 4 / 0 | 3.0 | Yes |
| 522760 | 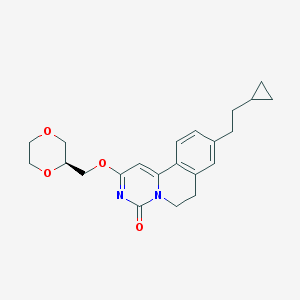 CHEMBL3718720 CHEMBL3718720 | C22H26N2O4 | 382.46 | 4 / 0 | 3.0 | Yes |
| 522804 | 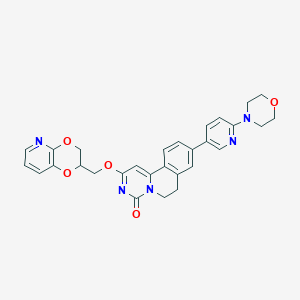 CHEMBL3718169 CHEMBL3718169 | C29H27N5O5 | 525.565 | 8 / 0 | 2.9 | No |
| 522826 | 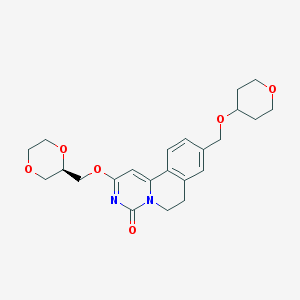 CHEMBL3718828 CHEMBL3718828 | C23H28N2O6 | 428.485 | 6 / 0 | 1.1 | Yes |
| 522849 | 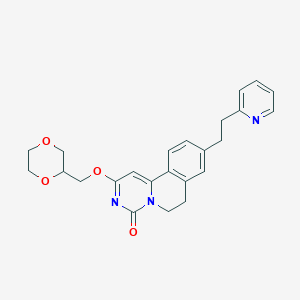 CHEMBL3716446 CHEMBL3716446 | C24H25N3O4 | 419.481 | 5 / 0 | 2.3 | Yes |
| 522853 | 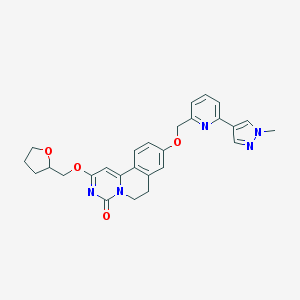 CHEMBL3718451 CHEMBL3718451 | C27H27N5O4 | 485.544 | 6 / 0 | 2.3 | Yes |
| 522943 | 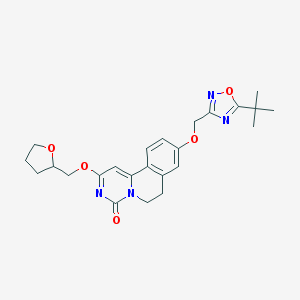 CHEMBL3719047 CHEMBL3719047 | C24H28N4O5 | 452.511 | 7 / 0 | 3.4 | Yes |
| 522955 | 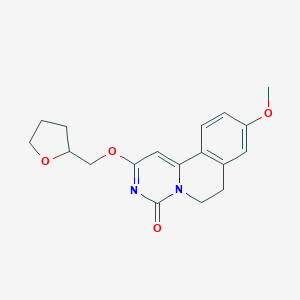 CHEMBL3717000 CHEMBL3717000 | C18H20N2O4 | 328.368 | 4 / 0 | 1.9 | Yes |
| 522969 | 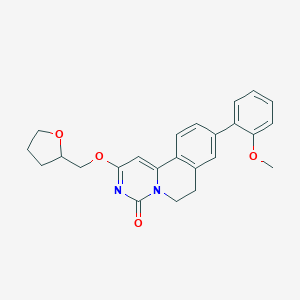 CHEMBL3715368 CHEMBL3715368 | C24H24N2O4 | 404.466 | 4 / 0 | 3.5 | Yes |
| 523002 | 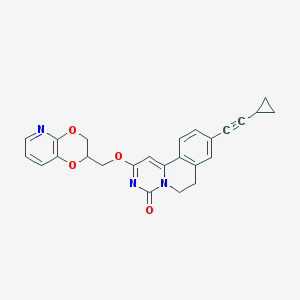 CHEMBL3718614 CHEMBL3718614 | C25H21N3O4 | 427.46 | 5 / 0 | 3.6 | Yes |
| 523184 | 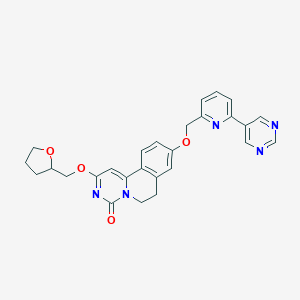 CHEMBL3714897 CHEMBL3714897 | C27H25N5O4 | 483.528 | 7 / 0 | 2.3 | Yes |
| 523230 | 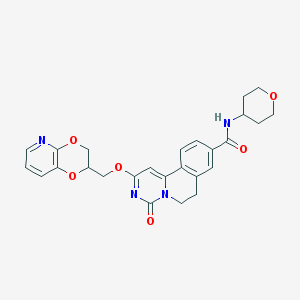 CHEMBL3717979 CHEMBL3717979 | C26H26N4O6 | 490.516 | 7 / 1 | 2.0 | Yes |
| 523254 | 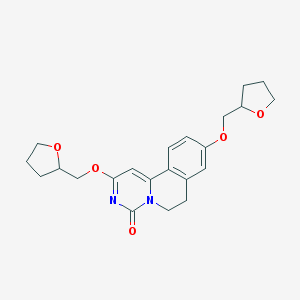 CHEMBL3718665 CHEMBL3718665 | C22H26N2O5 | 398.459 | 5 / 0 | 2.3 | Yes |
| 523257 | 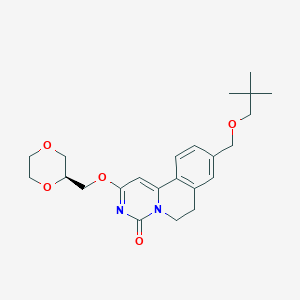 CHEMBL3714785 CHEMBL3714785 | C23H30N2O5 | 414.502 | 5 / 0 | 2.5 | Yes |
| 523274 | 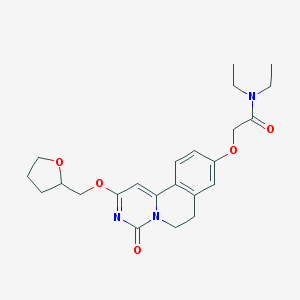 CHEMBL3718686 CHEMBL3718686 | C23H29N3O5 | 427.501 | 5 / 0 | 2.2 | Yes |
| 523299 | 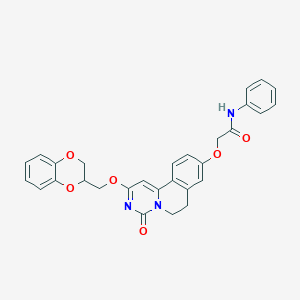 CHEMBL3716399 CHEMBL3716399 | C29H25N3O6 | 511.534 | 6 / 1 | 3.9 | No |
| 523309 | 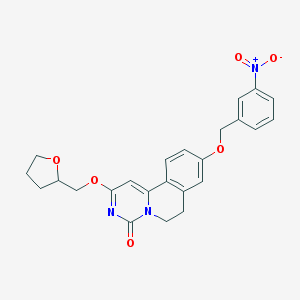 CHEMBL3718388 CHEMBL3718388 | C24H23N3O6 | 449.463 | 6 / 0 | 3.2 | Yes |
| 523317 | 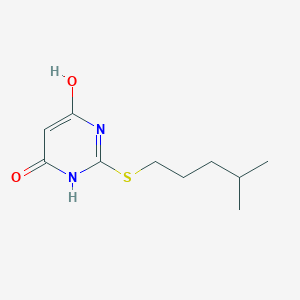 CHEMBL3828230 CHEMBL3828230 | C10H16N2O2S | 228.31 | 4 / 2 | 2.1 | Yes |
| 523326 | 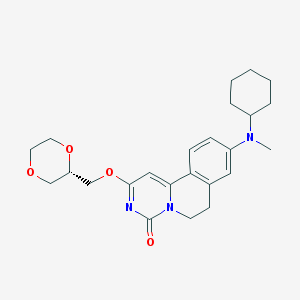 CHEMBL3718852 CHEMBL3718852 | C24H31N3O4 | 425.529 | 5 / 0 | 3.0 | Yes |
| 523330 | 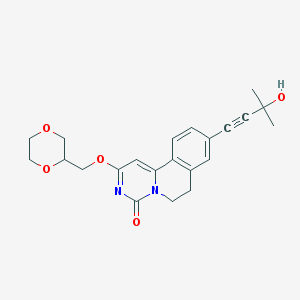 CHEMBL3719155 CHEMBL3719155 | C22H24N2O5 | 396.443 | 5 / 1 | 1.2 | Yes |
| 523349 | 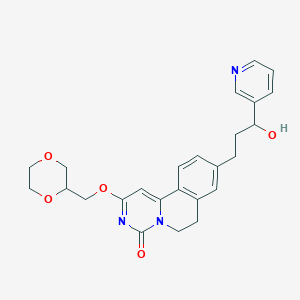 CHEMBL3715055 CHEMBL3715055 | C25H27N3O5 | 449.507 | 6 / 1 | 1.5 | Yes |
| 523452 | 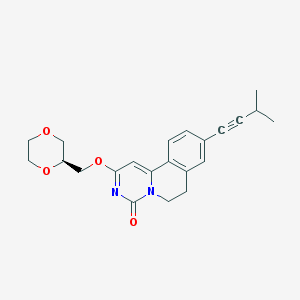 CHEMBL3715064 CHEMBL3715064 | C22H24N2O4 | 380.444 | 4 / 0 | 2.6 | Yes |
| 523453 | 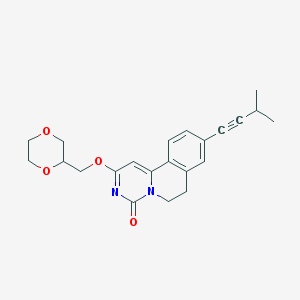 CHEMBL3714904 CHEMBL3714904 | C22H24N2O4 | 380.444 | 4 / 0 | 2.6 | Yes |
| 523477 | 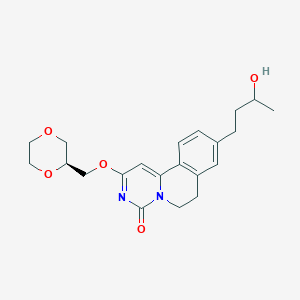 CHEMBL3718174 CHEMBL3718174 | C21H26N2O5 | 386.448 | 5 / 1 | 1.4 | Yes |
| 523514 | 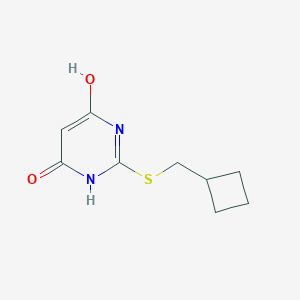 CHEMBL3827914 CHEMBL3827914 | C9H12N2O2S | 212.267 | 4 / 2 | 1.4 | Yes |
| 523551 | 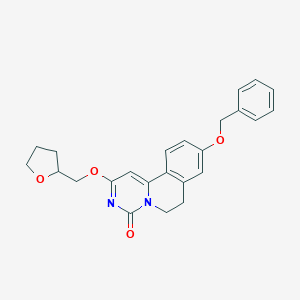 CHEMBL3718668 CHEMBL3718668 | C24H24N2O4 | 404.466 | 4 / 0 | 3.4 | Yes |
| 523578 | 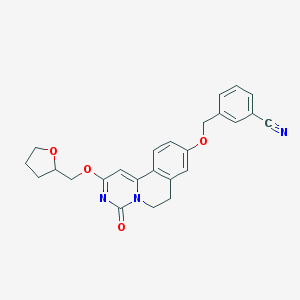 CHEMBL3716270 CHEMBL3716270 | C25H23N3O4 | 429.476 | 5 / 0 | 3.1 | Yes |
| 523754 | 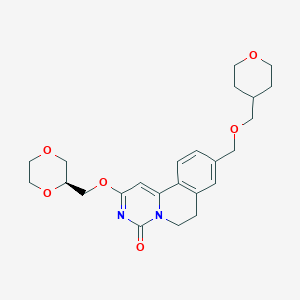 CHEMBL3715918 CHEMBL3715918 | C24H30N2O6 | 442.512 | 6 / 0 | 1.3 | Yes |
| 523791 | 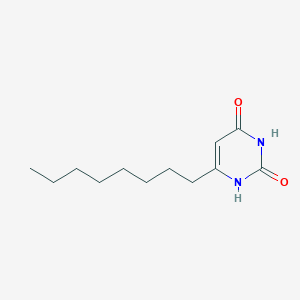 CHEMBL3827933 CHEMBL3827933 | C12H20N2O2 | 224.304 | 2 / 2 | 2.7 | Yes |
| 523805 | 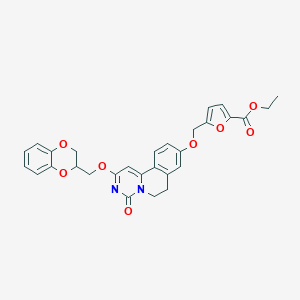 CHEMBL3715990 CHEMBL3715990 | C29H26N2O8 | 530.533 | 8 / 0 | 4.1 | No |
| 523867 | 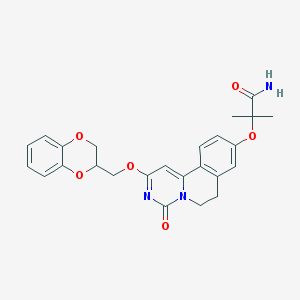 CHEMBL3717695 CHEMBL3717695 | C25H25N3O6 | 463.49 | 6 / 1 | 2.5 | Yes |
| 523869 | 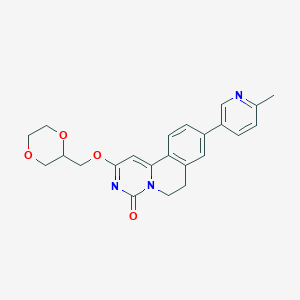 CHEMBL3717653 CHEMBL3717653 | C23H23N3O4 | 405.454 | 5 / 0 | 2.0 | Yes |
| 523885 | 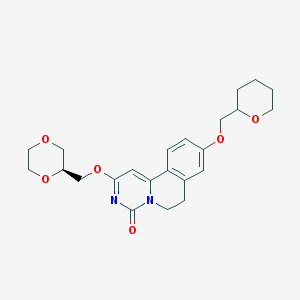 CHEMBL3716889 CHEMBL3716889 | C23H28N2O6 | 428.485 | 6 / 0 | 1.8 | Yes |
| 523917 | 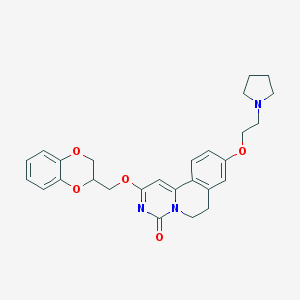 CHEMBL3716334 CHEMBL3716334 | C27H29N3O5 | 475.545 | 6 / 0 | 3.5 | Yes |
| 523928 | 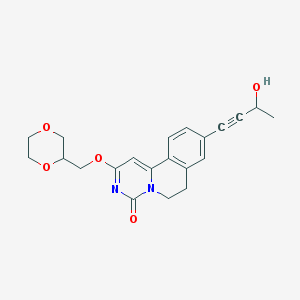 CHEMBL3717415 CHEMBL3717415 | C21H22N2O5 | 382.416 | 5 / 1 | 1.0 | Yes |
| 523961 |  CHEMBL3718113 CHEMBL3718113 | C23H23N3O5 | 421.453 | 6 / 0 | 1.9 | Yes |
| 523997 |  CHEMBL3716761 CHEMBL3716761 | C26H26N2O6 | 462.502 | 6 / 0 | 3.4 | Yes |
| 524003 |  CHEMBL3716024 CHEMBL3716024 | C23H26N2O5 | 410.47 | 5 / 1 | 1.9 | Yes |
| 524011 |  CHEMBL3828642 CHEMBL3828642 | C9H10N2O2S | 210.251 | 4 / 2 | 1.0 | Yes |
| 524022 | 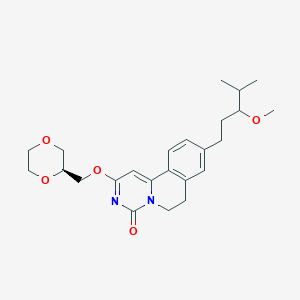 CHEMBL3716528 CHEMBL3716528 | C24H32N2O5 | 428.529 | 5 / 0 | 2.9 | Yes |
| 524039 | 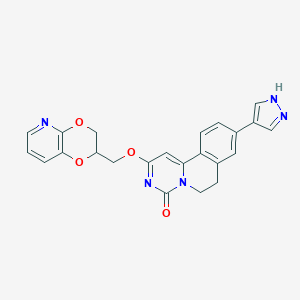 CHEMBL3718482 CHEMBL3718482 | C23H19N5O4 | 429.436 | 6 / 1 | 2.2 | Yes |
| 524059 | 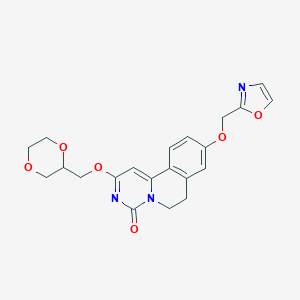 CHEMBL3718012 CHEMBL3718012 | C21H21N3O6 | 411.414 | 7 / 0 | 1.0 | Yes |
| 524061 | 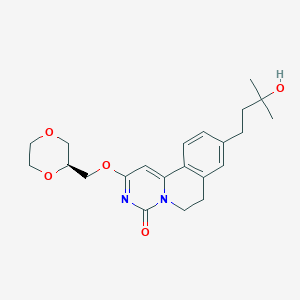 CHEMBL3718393 CHEMBL3718393 | C22H28N2O5 | 400.475 | 5 / 1 | 1.6 | Yes |
| 524113 |  CHEMBL3719143 CHEMBL3719143 | C24H30N2O5 | 426.513 | 5 / 1 | 2.1 | Yes |
| 524114 |  CHEMBL3714986 CHEMBL3714986 | C24H30N2O5 | 426.513 | 5 / 1 | 2.1 | Yes |
| 524125 |  CHEMBL3717795 CHEMBL3717795 | C23H26N2O4 | 394.471 | 4 / 0 | 3.4 | Yes |
| 553711 |  3-Hydroxydecanoic acid 3-Hydroxydecanoic acid | C10H20O3 | 188.267 | 3 / 2 | 2.5 | Yes |
| 524160 | 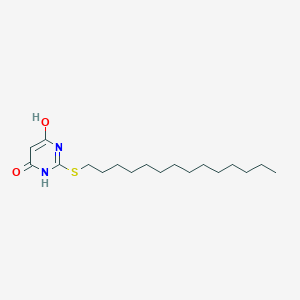 CHEMBL3827657 CHEMBL3827657 | C18H32N2O2S | 340.526 | 4 / 2 | 6.7 | No |
| 524181 | 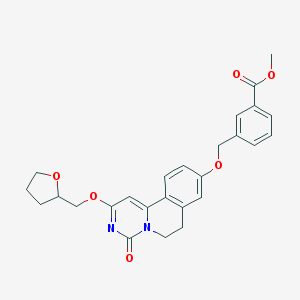 CHEMBL3716311 CHEMBL3716311 | C26H26N2O6 | 462.502 | 6 / 0 | 3.3 | Yes |
| 524263 | 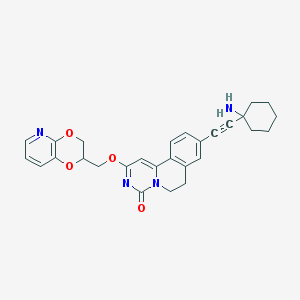 CHEMBL3715550 CHEMBL3715550 | C28H28N4O4 | 484.556 | 6 / 1 | 3.1 | Yes |
| 553733 | 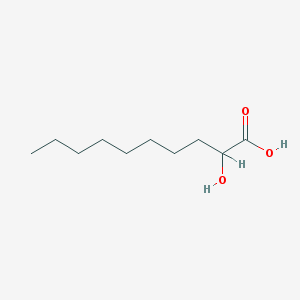 2-HYDROXYDECANOIC ACID 2-HYDROXYDECANOIC ACID | C10H20O3 | 188.267 | 3 / 2 | 3.1 | Yes |
| 553734 | 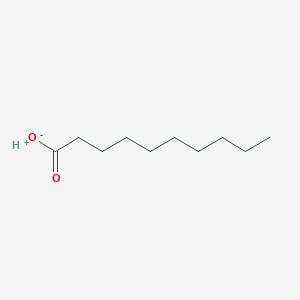 SCHEMBL20553507 SCHEMBL20553507 | C10H20O2 | 172.268 | 2 / 1 | N/A | N/A |
| 524383 | 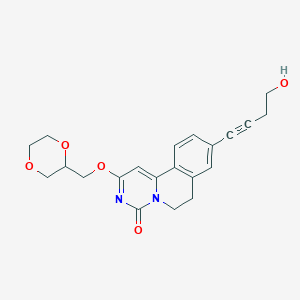 CHEMBL3717936 CHEMBL3717936 | C21H22N2O5 | 382.416 | 5 / 1 | 1.0 | Yes |
| 524410 | 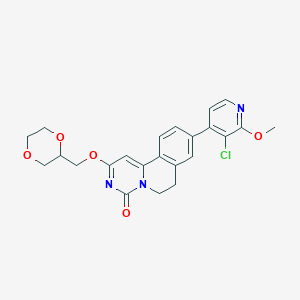 CHEMBL3719358 CHEMBL3719358 | C23H22ClN3O5 | 455.895 | 6 / 0 | 2.6 | Yes |
| 524441 | 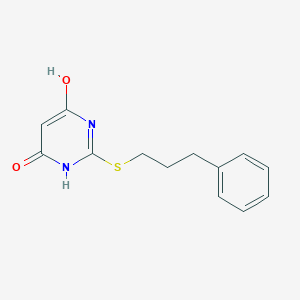 CHEMBL3828415 CHEMBL3828415 | C13H14N2O2S | 262.327 | 4 / 2 | 2.4 | Yes |
| 524486 |  CHEMBL3715192 CHEMBL3715192 | C25H28N2O4 | 420.509 | 4 / 0 | 4.0 | Yes |
| 524509 |  CHEMBL3715639 CHEMBL3715639 | C26H19N5O4 | 465.469 | 7 / 0 | 2.9 | Yes |
| 524565 |  CHEMBL3717805 CHEMBL3717805 | C26H27N3O6 | 477.517 | 7 / 0 | 3.0 | Yes |
| 524645 |  CHEMBL3716166 CHEMBL3716166 | C27H28N2O7 | 492.528 | 7 / 0 | 3.9 | Yes |
| 524662 | 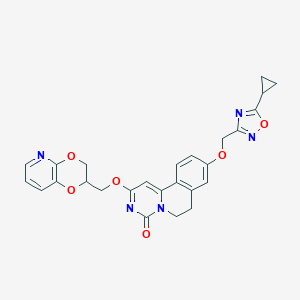 CHEMBL3717081 CHEMBL3717081 | C26H23N5O6 | 501.499 | 9 / 0 | 2.6 | No |
| 524719 | 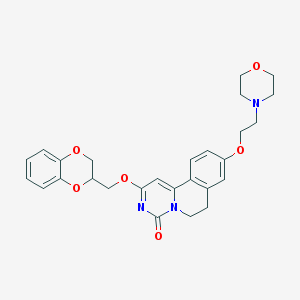 CHEMBL3718258 CHEMBL3718258 | C27H29N3O6 | 491.544 | 7 / 0 | 2.6 | Yes |
| 524763 | 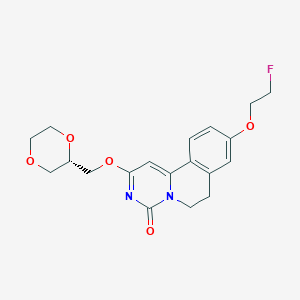 CHEMBL3719125 CHEMBL3719125 | C19H21FN2O5 | 376.384 | 6 / 0 | 1.4 | Yes |
| 524765 | 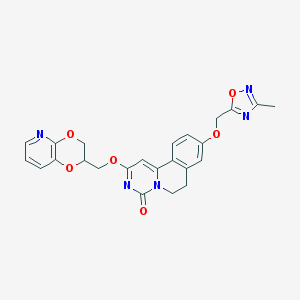 CHEMBL3718310 CHEMBL3718310 | C24H21N5O6 | 475.461 | 9 / 0 | 2.4 | Yes |
| 524783 | 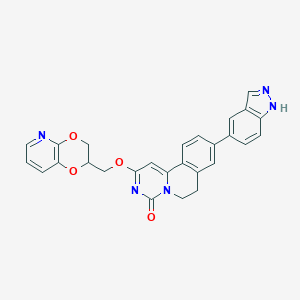 CHEMBL3717155 CHEMBL3717155 | C27H21N5O4 | 479.496 | 6 / 1 | 3.5 | Yes |
| 524817 | 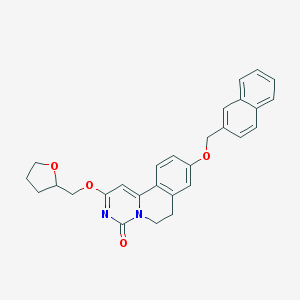 CHEMBL3717594 CHEMBL3717594 | C28H26N2O4 | 454.526 | 4 / 0 | 4.7 | Yes |
| 524848 | 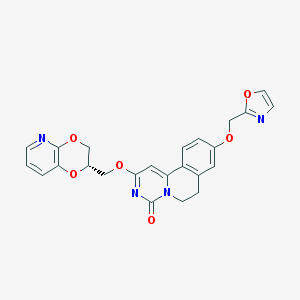 CHEMBL3714891 CHEMBL3714891 | C24H20N4O6 | 460.446 | 8 / 0 | 2.2 | Yes |
| 524849 | 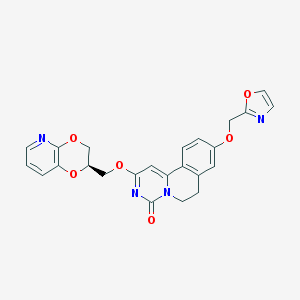 CHEMBL3717676 CHEMBL3717676 | C24H20N4O6 | 460.446 | 8 / 0 | 2.2 | Yes |
| 524850 | 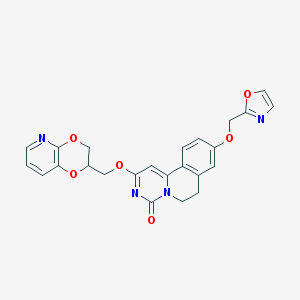 CHEMBL3718963 CHEMBL3718963 | C24H20N4O6 | 460.446 | 8 / 0 | 2.2 | Yes |
| 524866 | 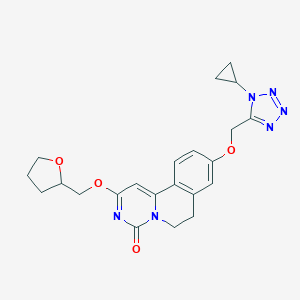 CHEMBL3715120 CHEMBL3715120 | C22H24N6O4 | 436.472 | 7 / 0 | 1.5 | Yes |
| 524882 | 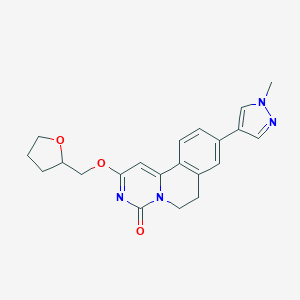 CHEMBL3717536 CHEMBL3717536 | C21H22N4O3 | 378.432 | 4 / 0 | 1.9 | Yes |
| 524916 | 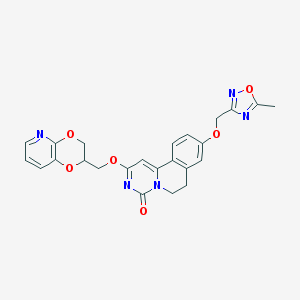 CHEMBL3718050 CHEMBL3718050 | C24H21N5O6 | 475.461 | 9 / 0 | 2.4 | Yes |
| 524933 |  CHEMBL3715009 CHEMBL3715009 | C27H25N3O5 | 471.513 | 5 / 1 | 2.4 | Yes |
| 524942 |  CHEMBL3827935 CHEMBL3827935 | C15H18N2O2S | 290.381 | 4 / 2 | 3.3 | Yes |
| 524988 |  CHEMBL3715086 CHEMBL3715086 | C26H26N2O6 | 462.502 | 6 / 0 | 3.3 | Yes |
| 524989 |  CHEMBL3715772 CHEMBL3715772 | C25H26N2O4 | 418.493 | 4 / 0 | 3.9 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417