

You can:
| Name | Neuropeptide FF receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | NPFFR1 |
| Synonym | GnIH-R G protein-coupled receptor 147 OT7T022 NPFF1R1 NPFF1 receptor [ Show all ] |
| Disease | N/A |
| Length | 430 |
| Amino acid sequence | MEGEPSQPPNSSWPLSQNGTNTEATPATNLTFSSYYQHTSPVAAMFIVAYALIFLLCMVGNTLVCFIVLKNRHMHTVTNMFILNLAVSDLLVGIFCMPTTLVDNLITGWPFDNATCKMSGLVQGMSVSASVFTLVAIAVERFRCIVHPFREKLTLRKALVTIAVIWALALLIMCPSAVTLTVTREEHHFMVDARNRSYPLYSCWEAWPEKGMRRVYTTVLFSHIYLAPLALIVVMYARIARKLCQAPGPAPGGEEAADPRASRRRARVVHMLVMVALFFTLSWLPLWALLLLIDYGQLSAPQLHLVTVYAFPFAHWLAFFNSSANPIIYGYFNENFRRGFQAAFRARLCPRPSGSHKEAYSERPGGLLHRRVFVVVRPSDSGLPSESGPSSGAPRPGRLPLRNGRVAHHGLPREGPGCSHLPLTIPAWDI |
| UniProt | Q9GZQ6 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9GZQ6 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9GZQ6. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL5951 |
| IUPHAR | 300 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 441756 | 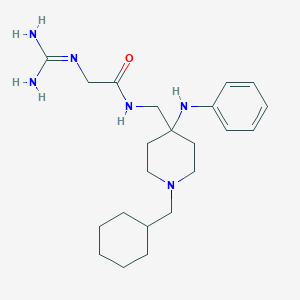 CHEMBL3361427 CHEMBL3361427 | C22H36N6O | 400.571 | 4 / 4 | 2.7 | Yes |
| 441837 | 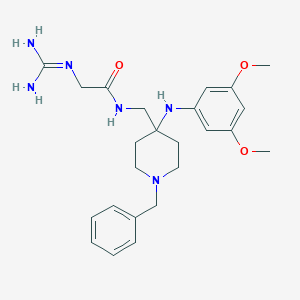 CHEMBL3361419 CHEMBL3361419 | C24H34N6O3 | 454.575 | 6 / 4 | 1.7 | Yes |
| 441916 | 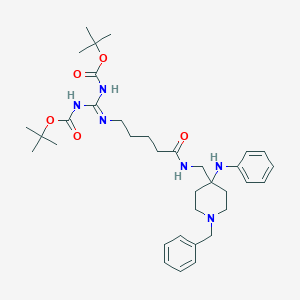 CHEMBL3361412 CHEMBL3361412 | C35H52N6O5 | 636.838 | 8 / 4 | 6.5 | No |
| 442050 | 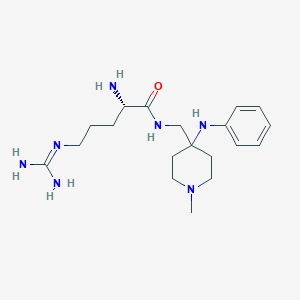 CHEMBL3360831 CHEMBL3360831 | C19H33N7O | 375.521 | 5 / 5 | -0.1 | Yes |
| 16897 | 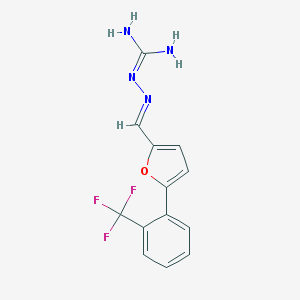 CHEMBL574565 CHEMBL574565 | C13H11F3N4O | 296.253 | 6 / 2 | 2.4 | Yes |
| 16899 | 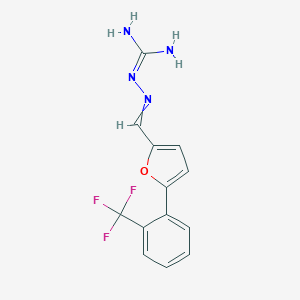 CID 69536911 CID 69536911 | C13H11F3N4O | 296.253 | 6 / 2 | 2.4 | Yes |
| 17311 | 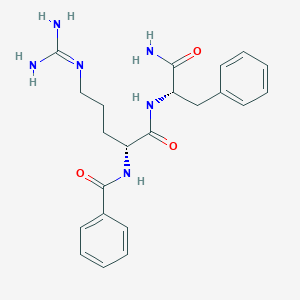 CHEMBL2208340 CHEMBL2208340 | C22H28N6O3 | 424.505 | 4 / 5 | 0.5 | Yes |
| 17313 | 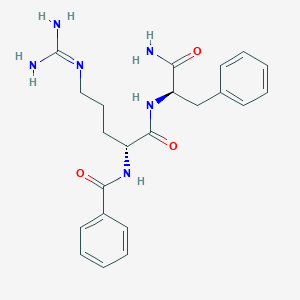 CHEMBL2204019 CHEMBL2204019 | C22H28N6O3 | 424.505 | 4 / 5 | 0.5 | Yes |
| 17315 |  CHEMBL2208339 CHEMBL2208339 | C22H28N6O3 | 424.505 | 4 / 5 | 0.5 | Yes |
| 21424 |  CHEMBL572825 CHEMBL572825 | C15H18N4O | 270.336 | 3 / 2 | 2.7 | Yes |
| 442546 |  CHEMBL3361418 CHEMBL3361418 | C26H32N6O | 444.583 | 4 / 4 | 3.0 | Yes |
| 24521 |  CHEMBL2208309 CHEMBL2208309 | C19H28N6O5 | 420.47 | 6 / 6 | -1.7 | No |
| 442790 | 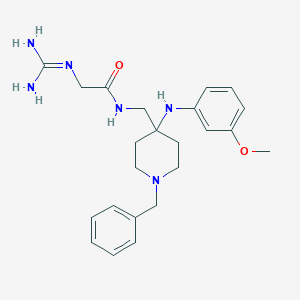 CHEMBL3361420 CHEMBL3361420 | C23H32N6O2 | 424.549 | 5 / 4 | 1.7 | Yes |
| 443046 | 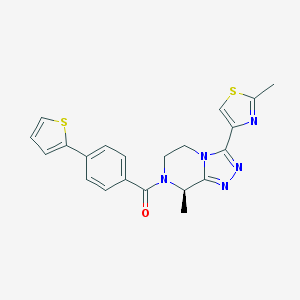 CHEMBL3422010 CHEMBL3422010 | C21H19N5OS2 | 421.537 | 6 / 0 | 3.0 | Yes |
| 35318 | 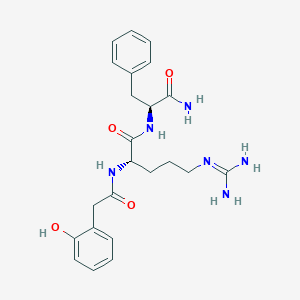 CHEMBL2208074 CHEMBL2208074 | C23H30N6O4 | 454.531 | 5 / 6 | 0.3 | No |
| 443160 | 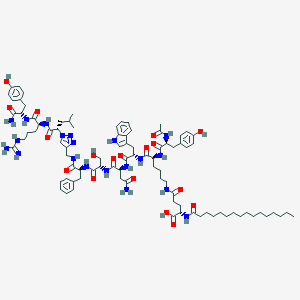 CHEMBL3422516 CHEMBL3422516 | C89H128N20O18 | 1766.12 | 21 / 21 | 6.5 | No |
| 443243 |  CHEMBL3422407 CHEMBL3422407 | C65H85N17O16 | 1360.5 | 17 / 20 | -1.9 | No |
| 40230 |  CHEMBL2208297 CHEMBL2208297 | C30H33N7O3 | 539.64 | 4 / 6 | 2.8 | No |
| 46849 |  CHEMBL573738 CHEMBL573738 | C13H14N4O | 242.282 | 3 / 2 | 1.9 | Yes |
| 49371 |  CHEMBL388586 CHEMBL388586 | C41H51FN10O6 | 798.921 | 8 / 9 | 2.4 | No |
| 58067 |  CHEMBL2208295 CHEMBL2208295 | C26H30N6O3 | 474.565 | 4 / 5 | 1.9 | Yes |
| 444071 |  CHEMBL3422018 CHEMBL3422018 | C20H18N6OS2 | 422.525 | 7 / 0 | 2.8 | Yes |
| 523375 |  CHEMBL508974 CHEMBL508974 | C32H38N6O6 | 602.692 | 7 / 6 | 3.4 | No |
| 63702 |  CHEMBL2208311 CHEMBL2208311 | C25H29N7O3 | 475.553 | 5 / 5 | 1.0 | Yes |
| 444353 | 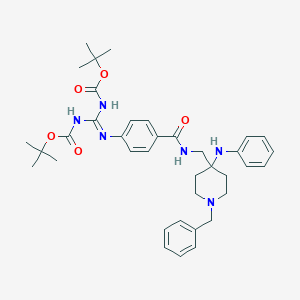 CHEMBL3361414 CHEMBL3361414 | C37H48N6O5 | 656.828 | 8 / 4 | 7.5 | No |
| 444652 | 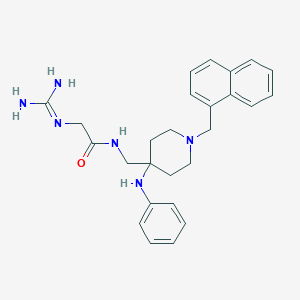 CHEMBL3361429 CHEMBL3361429 | C26H32N6O | 444.583 | 4 / 4 | 3.0 | Yes |
| 74582 | 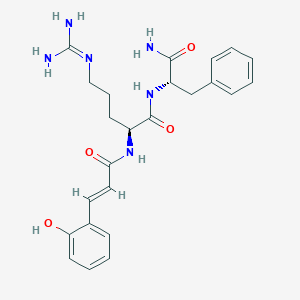 CHEMBL2208076 CHEMBL2208076 | C24H30N6O4 | 466.542 | 5 / 6 | 0.8 | No |
| 76025 | 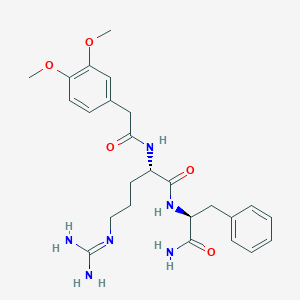 CHEMBL2208070 CHEMBL2208070 | C25H34N6O5 | 498.584 | 6 / 5 | 0.6 | Yes |
| 445506 | 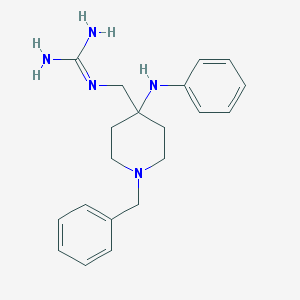 CHEMBL3360836 CHEMBL3360836 | C20H27N5 | 337.471 | 3 / 3 | 2.3 | Yes |
| 97180 | 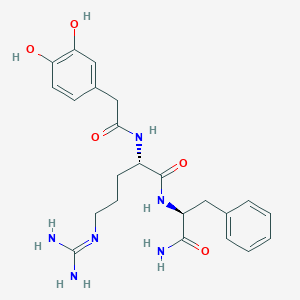 CHEMBL2208075 CHEMBL2208075 | C23H30N6O5 | 470.53 | 6 / 7 | -0.1 | No |
| 445632 | 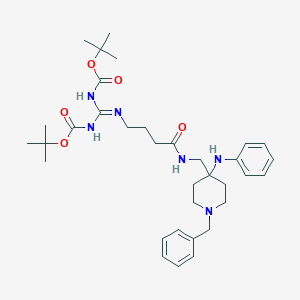 CHEMBL3352907 CHEMBL3352907 | C34H50N6O5 | 622.811 | 8 / 4 | 6.1 | No |
| 111285 | 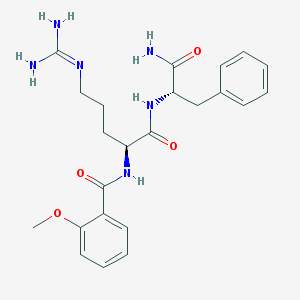 CHEMBL2208333 CHEMBL2208333 | C23H30N6O4 | 454.531 | 5 / 5 | 0.7 | Yes |
| 116679 | 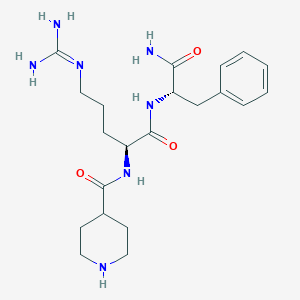 CHEMBL2208308 CHEMBL2208308 | C21H33N7O3 | 431.541 | 5 / 6 | -1.1 | No |
| 117109 | 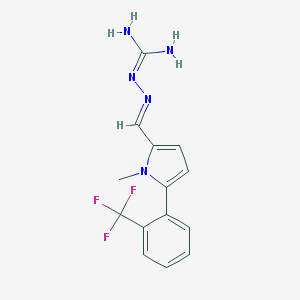 CHEMBL573519 CHEMBL573519 | C14H14F3N5 | 309.296 | 5 / 2 | 2.1 | Yes |
| 117983 | 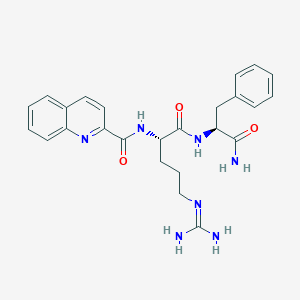 CHEMBL2208312 CHEMBL2208312 | C25H29N7O3 | 475.553 | 5 / 5 | 1.3 | Yes |
| 122295 | 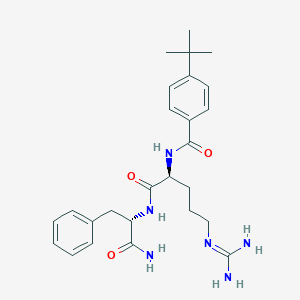 CHEMBL2208080 CHEMBL2208080 | C26H36N6O3 | 480.613 | 4 / 5 | 2.4 | Yes |
| 137596 | 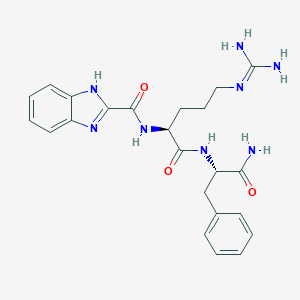 CHEMBL2208314 CHEMBL2208314 | C23H28N8O3 | 464.53 | 5 / 6 | 0.6 | No |
| 447158 | 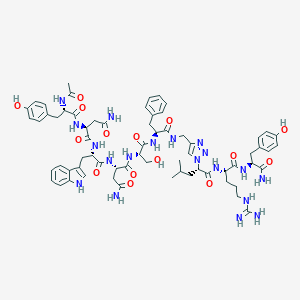 CHEMBL3422408 CHEMBL3422408 | C66H85N19O15 | 1384.52 | 18 / 19 | -1.6 | No |
| 447249 | 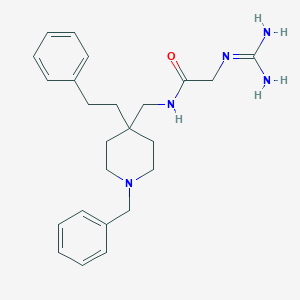 CHEMBL3361424 CHEMBL3361424 | C24H33N5O | 407.562 | 3 / 3 | 2.6 | Yes |
| 141684 | 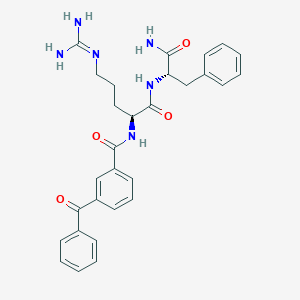 CHEMBL2208078 CHEMBL2208078 | C29H32N6O4 | 528.613 | 5 / 5 | 2.0 | No |
| 447588 | 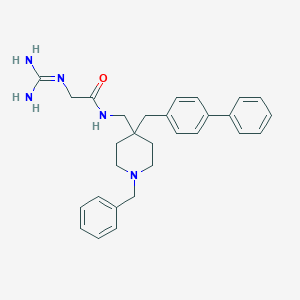 CHEMBL3361436 CHEMBL3361436 | C29H35N5O | 469.633 | 3 / 3 | 3.9 | Yes |
| 150045 | 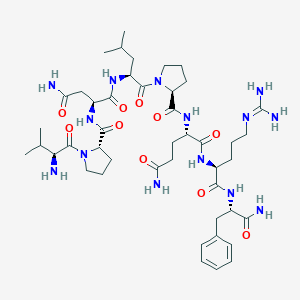 311309-27-0 311309-27-0 | C45H72N14O10 | 969.159 | 12 / 11 | -2.5 | No |
| 151718 | 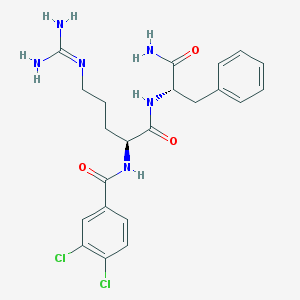 CHEMBL2208331 CHEMBL2208331 | C22H26Cl2N6O3 | 493.389 | 4 / 5 | 1.9 | Yes |
| 447750 | 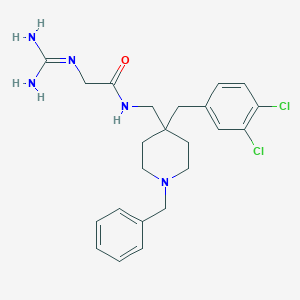 CHEMBL3361434 CHEMBL3361434 | C23H29Cl2N5O | 462.419 | 3 / 3 | 3.5 | Yes |
| 447866 | 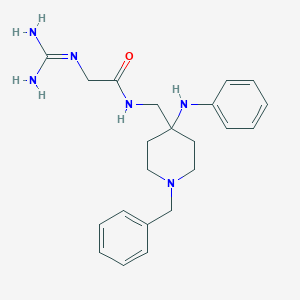 CHEMBL3361416 CHEMBL3361416 | C22H30N6O | 394.523 | 4 / 4 | 1.8 | Yes |
| 447879 | 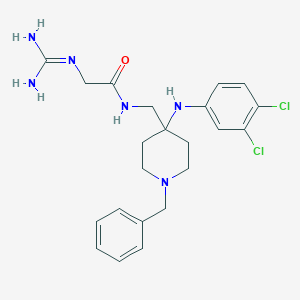 CHEMBL3361421 CHEMBL3361421 | C22H28Cl2N6O | 463.407 | 4 / 4 | 3.0 | Yes |
| 157689 | 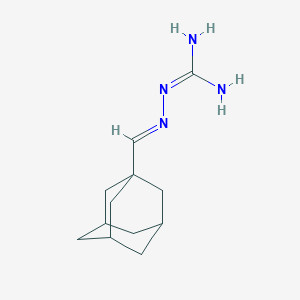 CHEMBL2165922 CHEMBL2165922 | C12H20N4 | 220.32 | 2 / 2 | 1.4 | Yes |
| 448000 | 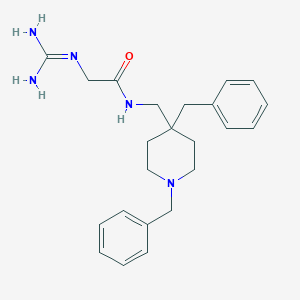 CHEMBL3361423 CHEMBL3361423 | C23H31N5O | 393.535 | 3 / 3 | 2.3 | Yes |
| 161517 | 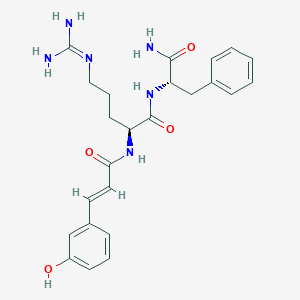 CHEMBL2208059 CHEMBL2208059 | C24H30N6O4 | 466.542 | 5 / 6 | 0.8 | No |
| 164510 | 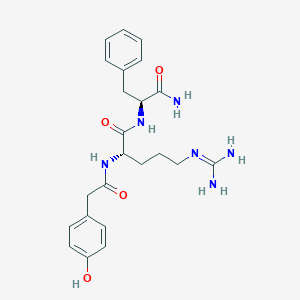 CHEMBL2208073 CHEMBL2208073 | C23H30N6O4 | 454.531 | 5 / 6 | 0.3 | No |
| 170739 | 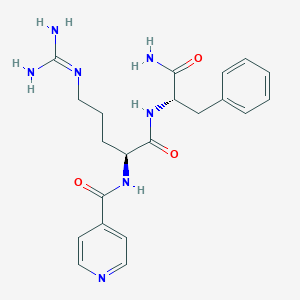 CHEMBL2208328 CHEMBL2208328 | C21H27N7O3 | 425.493 | 5 / 5 | -0.6 | Yes |
| 448721 | 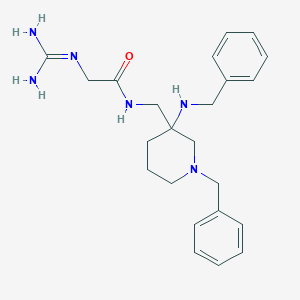 CHEMBL3361417 CHEMBL3361417 | C23H32N6O | 408.55 | 4 / 4 | 1.2 | Yes |
| 448746 | 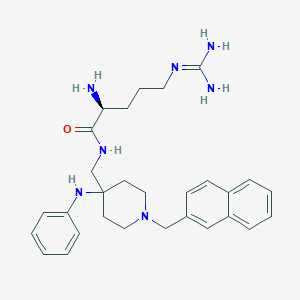 CHEMBL3360834 CHEMBL3360834 | C29H39N7O | 501.679 | 5 / 5 | 2.7 | No |
| 178861 | 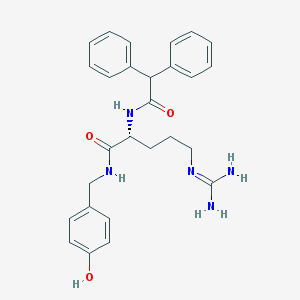 BIBP3226 BIBP3226 | C27H31N5O3 | 473.577 | 4 / 5 | 2.5 | Yes |
| 448762 | 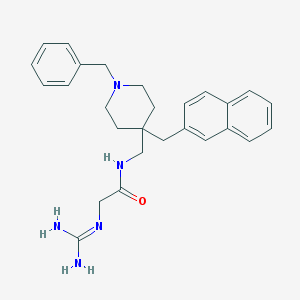 CHEMBL3361435 CHEMBL3361435 | C27H33N5O | 443.595 | 3 / 3 | 3.5 | Yes |
| 183317 | 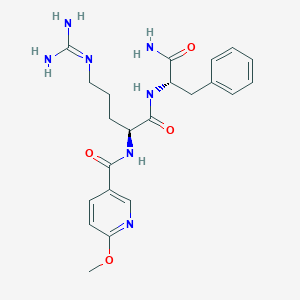 CHEMBL2208081 CHEMBL2208081 | C22H29N7O4 | 455.519 | 6 / 5 | -0.1 | Yes |
| 448963 | 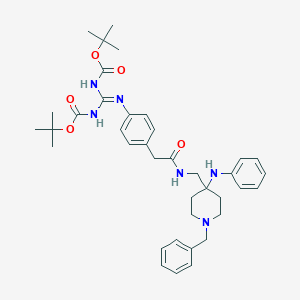 CHEMBL3361415 CHEMBL3361415 | C38H50N6O5 | 670.855 | 8 / 4 | 7.4 | No |
| 192771 | 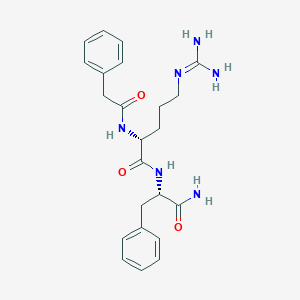 CHEMBL2208316 CHEMBL2208316 | C23H30N6O3 | 438.532 | 4 / 5 | 0.4 | Yes |
| 192772 | 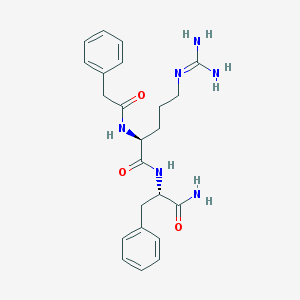 SCHEMBL7535795 SCHEMBL7535795 | C23H30N6O3 | 438.532 | 4 / 5 | 0.4 | Yes |
| 192775 | 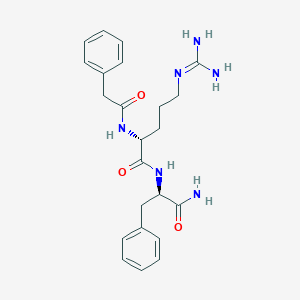 CHEMBL2208317 CHEMBL2208317 | C23H30N6O3 | 438.532 | 4 / 5 | 0.4 | Yes |
| 198384 | 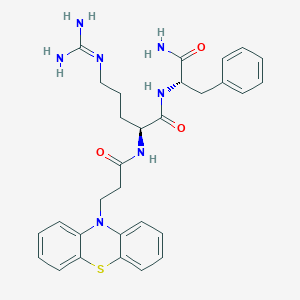 CHEMBL2208301 CHEMBL2208301 | C30H35N7O3S | 573.716 | 6 / 5 | 2.5 | No |
| 199222 | 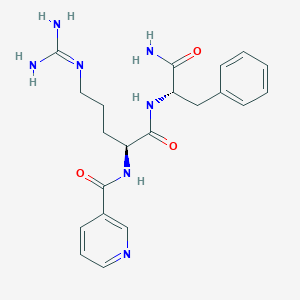 CHEMBL2208327 CHEMBL2208327 | C21H27N7O3 | 425.493 | 5 / 5 | -0.4 | Yes |
| 449713 | 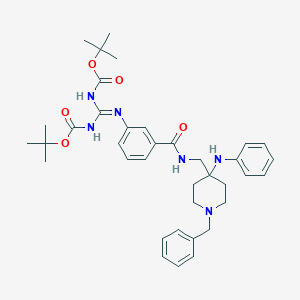 CHEMBL3361413 CHEMBL3361413 | C37H48N6O5 | 656.828 | 8 / 4 | 7.5 | No |
| 208506 | 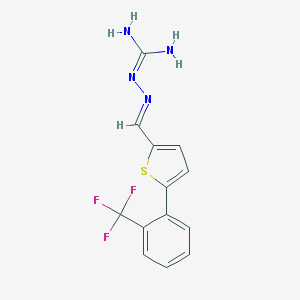 CHEMBL573518 CHEMBL573518 | C13H11F3N4S | 312.314 | 6 / 2 | 3.0 | Yes |
| 223184 | 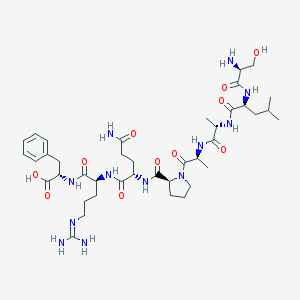 CHEMBL1672379 CHEMBL1672379 | C40H64N12O11 | 889.025 | 13 / 12 | -5.0 | No |
| 227935 | 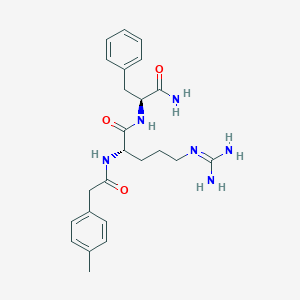 CHEMBL2208067 CHEMBL2208067 | C24H32N6O3 | 452.559 | 4 / 5 | 0.8 | Yes |
| 237559 | 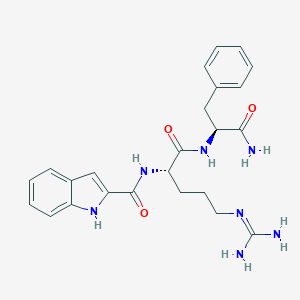 SCHEMBL7560327 SCHEMBL7560327 | C24H29N7O3 | 463.542 | 4 / 6 | 1.1 | No |
| 244311 | 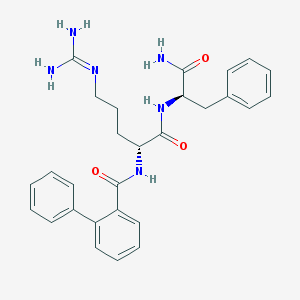 CHEMBL2208320 CHEMBL2208320 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 244314 | 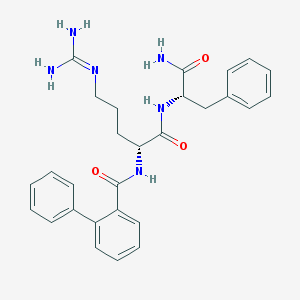 CHEMBL2208338 CHEMBL2208338 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 244315 | 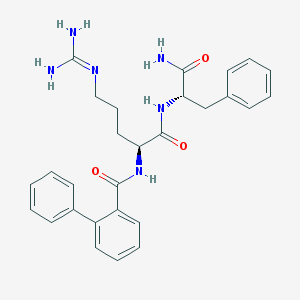 CHEMBL2208337 CHEMBL2208337 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 451356 | 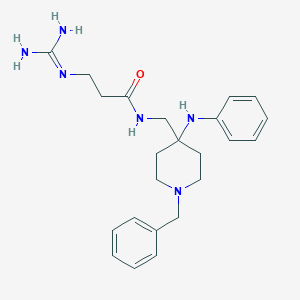 CHEMBL3361410 CHEMBL3361410 | C23H32N6O | 408.55 | 4 / 4 | 1.5 | Yes |
| 249090 | 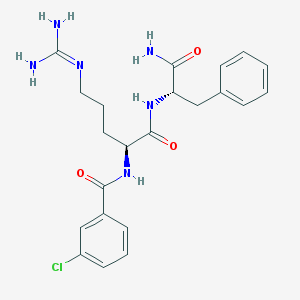 SCHEMBL7528463 SCHEMBL7528463 | C22H27ClN6O3 | 458.947 | 4 / 5 | 1.3 | Yes |
| 252536 | 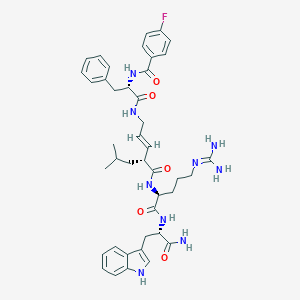 Peptide analogue, 19 Peptide analogue, 19 | C42H52FN9O5 | 781.934 | 7 / 8 | 3.4 | No |
| 256197 | 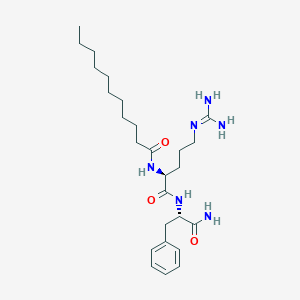 CHEMBL2208065 CHEMBL2208065 | C26H44N6O3 | 488.677 | 4 / 5 | 3.4 | Yes |
| 257769 | 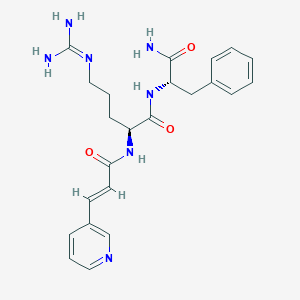 CHEMBL2208062 CHEMBL2208062 | C23H29N7O3 | 451.531 | 5 / 5 | 0.0 | Yes |
| 265081 | 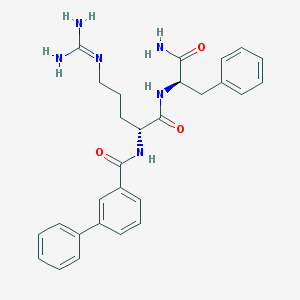 CHEMBL2208323 CHEMBL2208323 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 265082 |  CHEMBL2208322 CHEMBL2208322 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 265084 |  CHEMBL2208321 CHEMBL2208321 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 452480 |  CHEMBL3361428 CHEMBL3361428 | C23H32N6O | 408.55 | 4 / 4 | 2.2 | Yes |
| 277783 |  CHEMBL2208298 CHEMBL2208298 | C23H28N8O3 | 464.53 | 5 / 6 | 0.2 | No |
| 278621 | 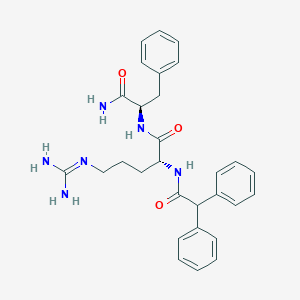 CHEMBL2208299 CHEMBL2208299 | C29H34N6O3 | 514.63 | 4 / 5 | 2.3 | No |
| 278624 | 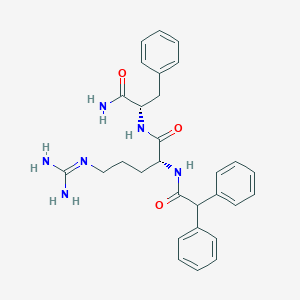 CHEMBL2208319 CHEMBL2208319 | C29H34N6O3 | 514.63 | 4 / 5 | 2.3 | No |
| 278626 | 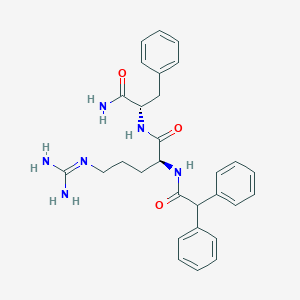 CHEMBL2208318 CHEMBL2208318 | C29H34N6O3 | 514.63 | 4 / 5 | 2.3 | No |
| 279334 | 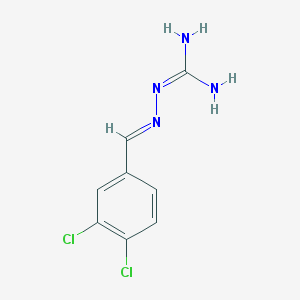 CHEMBL572531 CHEMBL572531 | C8H8Cl2N4 | 231.08 | 2 / 2 | 1.9 | Yes |
| 287582 | 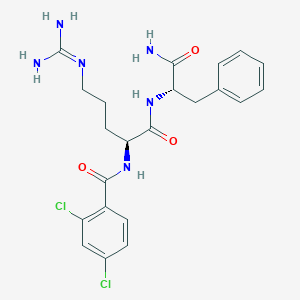 CHEMBL2208332 CHEMBL2208332 | C22H26Cl2N6O3 | 493.389 | 4 / 5 | 1.9 | Yes |
| 453142 | 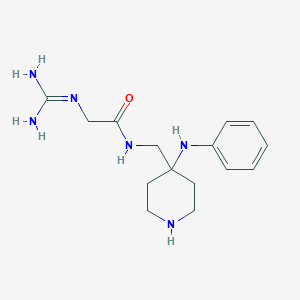 CHEMBL3361426 CHEMBL3361426 | C15H24N6O | 304.398 | 4 / 5 | -0.2 | Yes |
| 453156 | 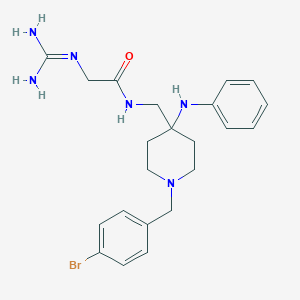 CHEMBL3361431 CHEMBL3361431 | C22H29BrN6O | 473.419 | 4 / 4 | 2.5 | Yes |
| 294346 | 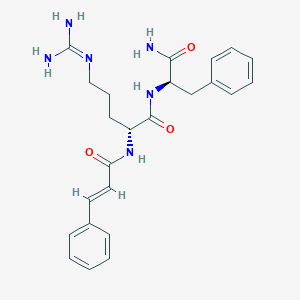 CHEMBL2208304 CHEMBL2208304 | C24H30N6O3 | 450.543 | 4 / 5 | 1.1 | Yes |
| 294348 | 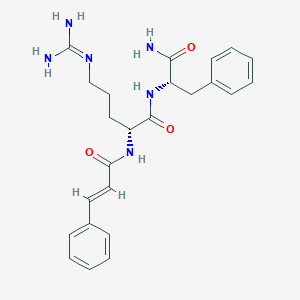 CHEMBL2208303 CHEMBL2208303 | C24H30N6O3 | 450.543 | 4 / 5 | 1.1 | Yes |
| 294351 | 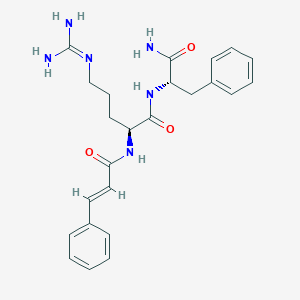 CHEMBL2208302 CHEMBL2208302 | C24H30N6O3 | 450.543 | 4 / 5 | 1.1 | Yes |
| 294771 | 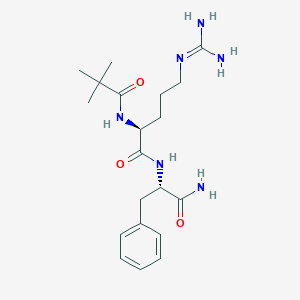 CHEMBL2208066 CHEMBL2208066 | C20H32N6O3 | 404.515 | 4 / 5 | 0.2 | Yes |
| 295017 | 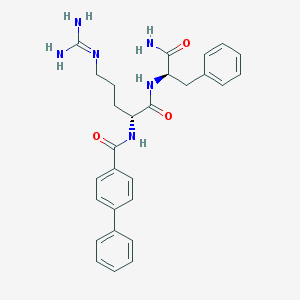 CHEMBL2208326 CHEMBL2208326 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 295018 | 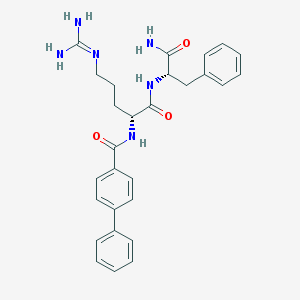 CHEMBL2208325 CHEMBL2208325 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 295020 | 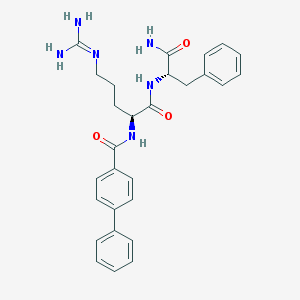 CHEMBL2208324 CHEMBL2208324 | C28H32N6O3 | 500.603 | 4 / 5 | 2.3 | No |
| 453413 | 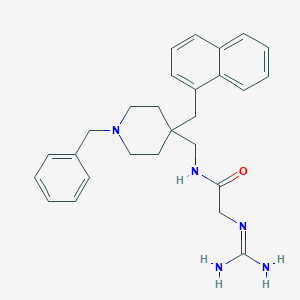 CHEMBL3361433 CHEMBL3361433 | C27H33N5O | 443.595 | 3 / 3 | 3.5 | Yes |
| 297199 | 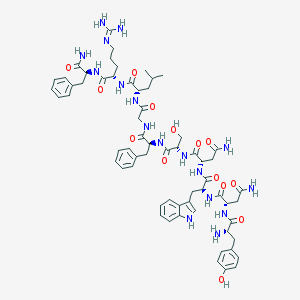 Kisspeptin-10 Kisspeptin-10 | C63H83N17O14 | 1302.46 | 16 / 18 | -1.9 | No |
| 453978 | 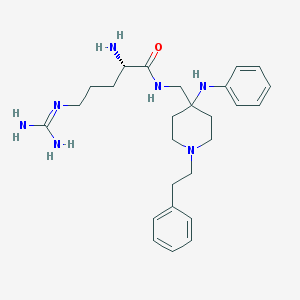 CHEMBL3360833 CHEMBL3360833 | C26H39N7O | 465.646 | 5 / 5 | 1.9 | Yes |
| 454040 | 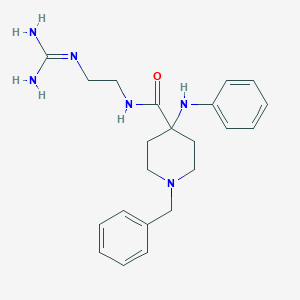 CHEMBL3361425 CHEMBL3361425 | C22H30N6O | 394.523 | 4 / 4 | 1.6 | Yes |
| 316096 | 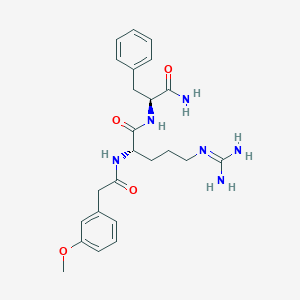 CHEMBL2208068 CHEMBL2208068 | C24H32N6O4 | 468.558 | 5 / 5 | 0.6 | Yes |
| 530603 | 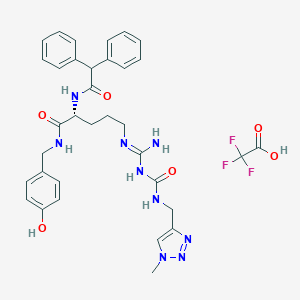 CHEMBL3746851 CHEMBL3746851 | C34H38F3N9O6 | 725.73 | 12 / 7 | N/A | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417