

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MEGEPSQPPNSSWPLSQNGTNTEATPATNLTFSSYYQHTSPVAAMFIVAYALIFLLCMVGNTLVCFIVLKNRHMHTVTNMFILNLAVSDLLVGIFCMPTTLVDNLITGWPFDNATCKMSGLVQGMSVSASVFTLVAIAVERFRCIVHPFREKLTLRKALVTIAVIWALALLIMCPSAVTLTVTREEHHFMVDARNRSYPLYSCWEAWPEKGMRRVYTTVLFSHIYLAPLALIVVMYARIARKLCQAPGPAPGGEEAADPRASRRRARVVHMLVMVALFFTLSWLPLWALLLLIDYGQLSAPQLHLVTVYAFPFAHWLAFFNSSANPIIYGYFNENFRRGFQAAFRARLCPRPSGSHKEAYSERPGGLLHRRVFVVVRPSDSGLPSESGPSSGAPRPGRLPLRNGRVAHHGLPREGPGCSHLPLTIPAWDI | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9899999998676677888777778887877654446760899999999999999997777762523571477773768999999999999999987399999994883783689988999999999999999999999999998736886401756874525999999999999999918873046544466667777505540088678999998999999999999999999999999993078999862026678988742169899999999999998999999999997465665332699999999999999999999999995899999999999614258998887655666678986204666500578888877778888988888666789863478889889997728877758999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MEGEPSQPPNSSWPLSQNGTNTEATPATNLTFSSYYQHTSPVAAMFIVAYALIFLLCMVGNTLVCFIVLKNRHMHTVTNMFILNLAVSDLLVGIFCMPTTLVDNLITGWPFDNATCKMSGLVQGMSVSASVFTLVAIAVERFRCIVHPFREKLTLRKALVTIAVIWALALLIMCPSAVTLTVTREEHHFMVDARNRSYPLYSCWEAWPEKGMRRVYTTVLFSHIYLAPLALIVVMYARIARKLCQAPGPAPGGEEAADPRASRRRARVVHMLVMVALFFTLSWLPLWALLLLIDYGQLSAPQLHLVTVYAFPFAHWLAFFNSSANPIIYGYFNENFRRGFQAAFRARLCPRPSGSHKEAYSERPGGLLHRRVFVVVRPSDSGLPSESGPSSGAPRPGRLPLRNGRVAHHGLPREGPGCSHLPLTIPAWDI | |
| 8444344331211313323133433333323243223222000000010112002102301200000000134020000000100010010000000100000000320001200200000000000000000000001220100000013322331000000001110000010000002023343422233334421100010201263022000000022313310210020000000101324433544443344433333100000000000000000120000000100022224321000000101000000100000000000014310410240021111234444444433434443343433444334444343444444444454414344341333344462542331203021276 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MEGEPSQPPNSSWPLSQNGTNTEATPATNLTFSSYYQHTSPVAAMFIVAYALIFLLCMVGNTLVCFIVLKNRHMHTVTNMFILNLAVSDLLVGIFCMPTTLVDNLITGWPFDNATCKMSGLVQGMSVSASVFTLVAIAVERFRCIVHPFREKLTLRKALVTIAVIWALALLIMCPSAVTLTVTREEHHFMVDARNRSYPLYSCWEAWPEKGMRRVYTTVLFSHIYLAPLALIVVMYARIARKLCQAPGPAPGGEEAADPRASRRRARVVHMLVMVALFFTLSWLPLWALLLLIDYGQLSAPQLHLVTVYAFPFAHWLAFFNSSANPIIYGYFNENFRRGFQAAFRARLCPRPSGSHKEAYSERPGGLLHRRVFVVVRPSDSGLPSESGPSSGAPRPGRLPLRNGRVAHHGLPREGPGCSHLPLTIPAWDI | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.24 | 0.21 | 0.69 | 2.68 | Download | ------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG------------AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD--PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4xeeA | 0.19 | 0.23 | 0.88 | 3.74 | Download | ----------------------------GPNSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA----RKSTVHYHLGSLALSDLLILLLAMPVELYNFIWHPWAFGDAGCRGYYFLRDACTYATALNVASLSVERYLAICHPFKAKMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTP-------IVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMVP-----------GRVQALRHGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTLFDFYHYFYMLTNALFYASSAINPILYNLVSANFRQVFLSTLNIFEMLRIDEGLRLKIYKDTEGYYTIGIGHLLTKSPSLNAAKSELDKAIGRNTNGVITKDEAENQDVDAAVRGILRNAKLKPVYDS | |||||||||||||||||||
| 3 | 4n6hA | 0.24 | 0.23 | 0.69 | 3.73 | Download | -----------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDP--LVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP----------------------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 4n6hA | 0.24 | 0.21 | 0.69 | 3.49 | Download | ------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDP--LVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.26 | 0.23 | 0.67 | 1.51 | Download | --------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED----------VDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIKTPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA--A------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------------------------------------------------ | |||||||||||||||||||
| 6 | 4rnb | 0.36 | 0.27 | 0.68 | 1.15 | Download | --------------------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNHHMRTVTNYFIVNLSLADVLVTITCLPATLVVDITETWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPS----TAKRARNSIVIIWIVSCIIMIPQAIVMECSTVFK---------TTLFTVCDERWGGEIYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSDPNMSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFAHD-RETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC------------------------------------------------------------------------------------ | |||||||||||||||||||
| 7 | 4n6hA | 0.24 | 0.21 | 0.69 | 3.06 | Download | --------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI--DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 3uon | 0.22 | 0.23 | 0.66 | 1.68 | Download | ------------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTV---------EDGECYIQFFSN---AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGAATWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC------IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 4ea3A | 0.28 | 0.20 | 0.65 | 5.12 | Download | --------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHP-----TSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEEI----------ECLVEIPTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS------REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPS---SETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR----------------------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5glhA | 0.23 | 0.18 | 0.66 | 2.90 | Download | -----------------------------PPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYII-----YKNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWW--------TAVEIVLIWVVSVVLAVPEAIGFDIITMDYK--------GSYLRICLLHPVQQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKN------------DHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPLLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL-------------------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
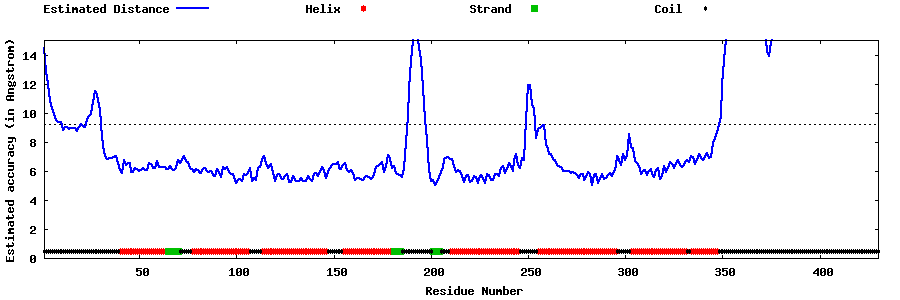
| Generated 3D models | Estimated local accuracy of models | ||
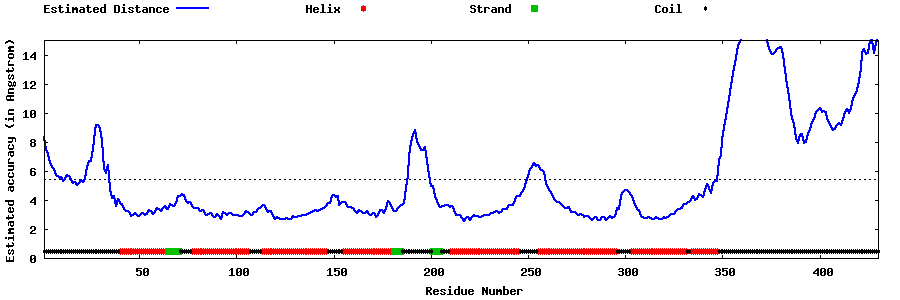 | |||
|
|
|||
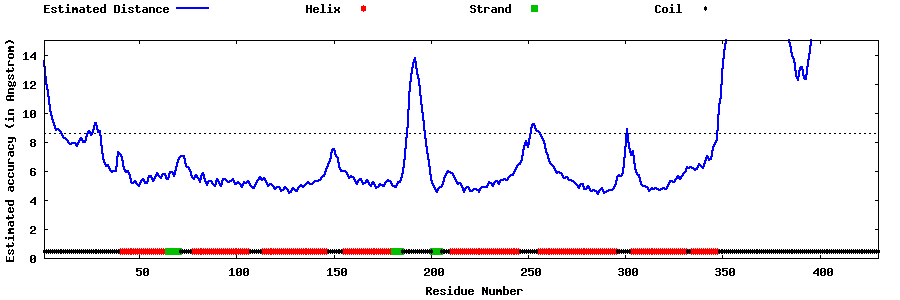 | |||
|
| |||
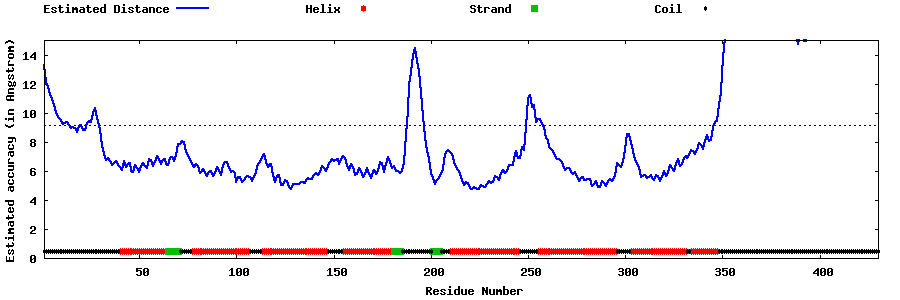 | |||
|
| |||
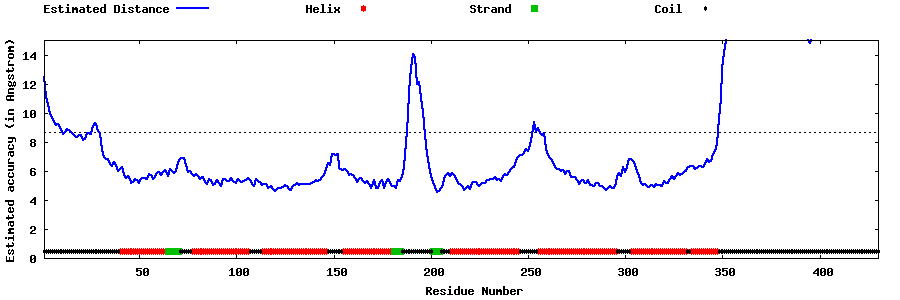 | |||
|
| |||
 | |||
|
| |||