

You can:
| Name | Free fatty acid receptor 1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Ffar1 |
| Synonym | FFA1 receptor FFA1R G protein-coupled receptor 40 G-protein coupled receptor 40 GPR40 |
| Disease | N/A for non-human GPCRs |
| Length | 300 |
| Amino acid sequence | MDLPPQLSFALYVSAFALGFPLNLLAIRGAVSHAKLRLTPSLVYTLHLACSDLLLAITLPLKAVEALASGVWPLPLPFCPVFALAHFAPLYAGGGFLAALSAGRYLGAAFPFGYQAIRRPCYSWGVCVAIWALVLCHLGLALGLEAPRGWVDNTTSSLGINIPVNGSPVCLEAWDPDSARPARLSFSILLFFLPLVITAFCYVGCLRALVHSGLSHKRKLRAAWVAGGALLTLLLCLGPYNASNVASFINPDLEGSWRKLGLITGAWSVVLNPLVTGYLGTGPGQGTICVTRTPRGTIQK |
| UniProt | Q8K3T4 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL1795180 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 886 | 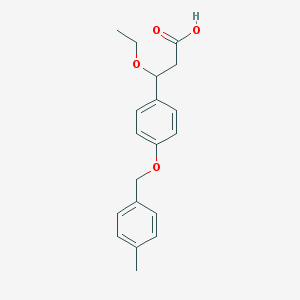 CHEMBL3286417 CHEMBL3286417 | C19H22O4 | 314.381 | 4 / 1 | 3.3 | Yes |
| 2403 | 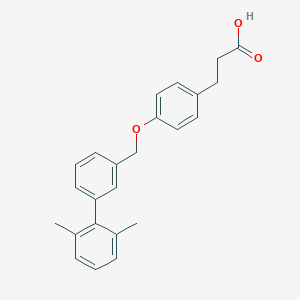 CHEMBL1688486 CHEMBL1688486 | C24H24O3 | 360.453 | 3 / 1 | 5.5 | No |
| 4159 | 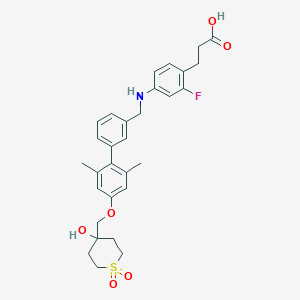 CHEMBL2048627 CHEMBL2048627 | C30H34FNO6S | 555.661 | 8 / 3 | 4.6 | No |
| 557420 | 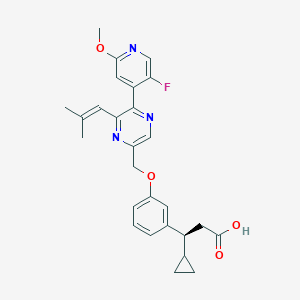 SCHEMBL17419035 SCHEMBL17419035 | C27H28FN3O4 | 477.536 | 8 / 1 | 4.6 | Yes |
| 6799 | 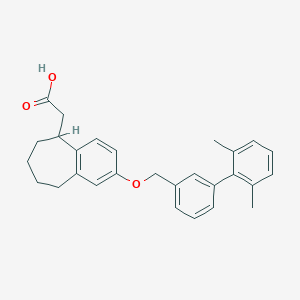 CHEMBL2022245 CHEMBL2022245 | C28H30O3 | 414.545 | 3 / 1 | 7.0 | No |
| 11850 | 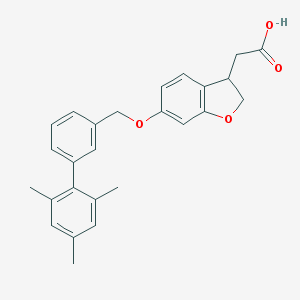 CHEMBL2022258 CHEMBL2022258 | C26H26O4 | 402.49 | 4 / 1 | 5.4 | No |
| 12027 | 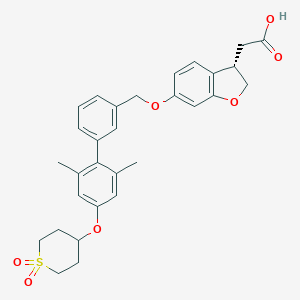 CHEMBL2047155 CHEMBL2047155 | C30H32O7S | 536.639 | 7 / 1 | 4.9 | No |
| 12029 | 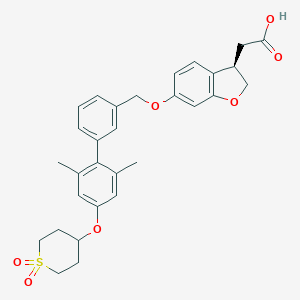 CHEMBL2047156 CHEMBL2047156 | C30H32O7S | 536.639 | 7 / 1 | 4.9 | No |
| 12031 | 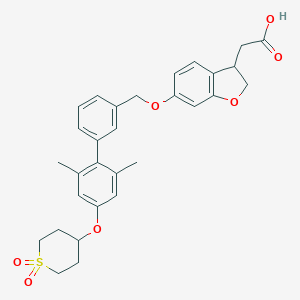 CHEMBL2047154 CHEMBL2047154 | C30H32O7S | 536.639 | 7 / 1 | 4.9 | No |
| 521844 | 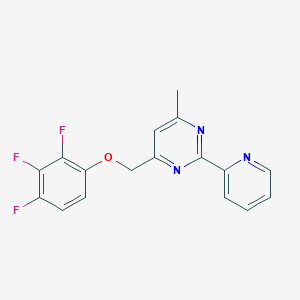 CHEMBL3739712 CHEMBL3739712 | C17H12F3N3O | 331.298 | 7 / 0 | 3.0 | Yes |
| 15560 | 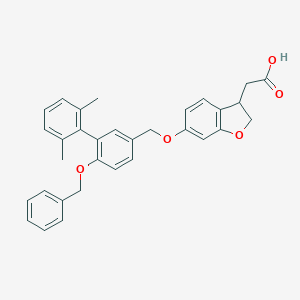 CHEMBL2022256 CHEMBL2022256 | C32H30O5 | 494.587 | 5 / 1 | 6.5 | No |
| 557885 | 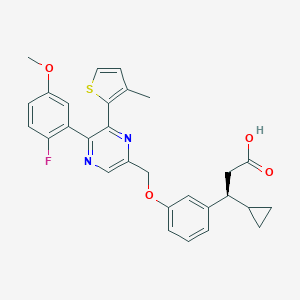 SCHEMBL17418599 SCHEMBL17418599 | C29H27FN2O4S | 518.603 | 8 / 1 | 5.4 | No |
| 19903 | 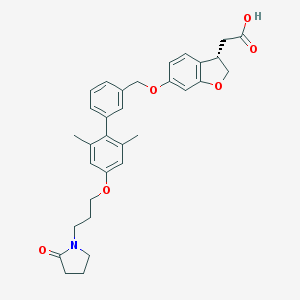 CHEMBL2047160 CHEMBL2047160 | C32H35NO6 | 529.633 | 6 / 1 | 4.9 | No |
| 557905 | 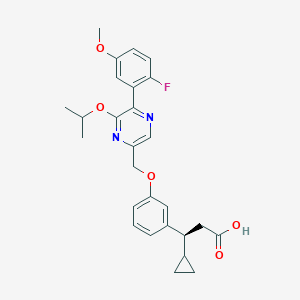 SCHEMBL17418781 SCHEMBL17418781 | C27H29FN2O5 | 480.536 | 8 / 1 | 4.8 | Yes |
| 23995 | 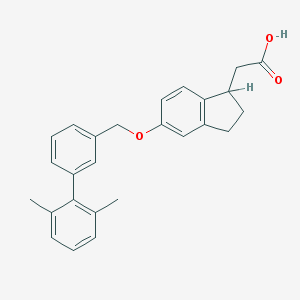 CHEMBL2022243 CHEMBL2022243 | C26H26O3 | 386.491 | 3 / 1 | 5.9 | No |
| 33187 | 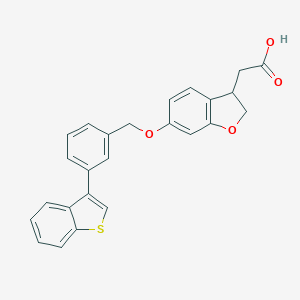 CHEMBL2022253 CHEMBL2022253 | C25H20O4S | 416.491 | 5 / 1 | 5.3 | No |
| 558296 | 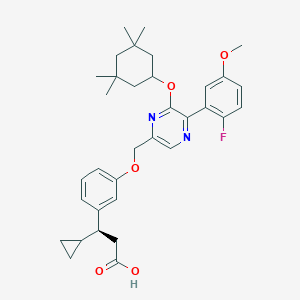 SCHEMBL17418575 SCHEMBL17418575 | C34H41FN2O5 | 576.709 | 8 / 1 | 7.5 | No |
| 35485 | 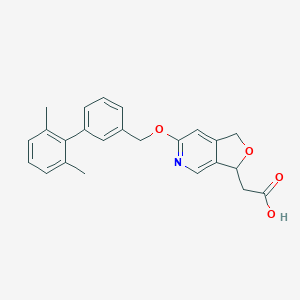 CHEMBL3288360 CHEMBL3288360 | C24H23NO4 | 389.451 | 5 / 1 | 3.8 | Yes |
| 36230 | 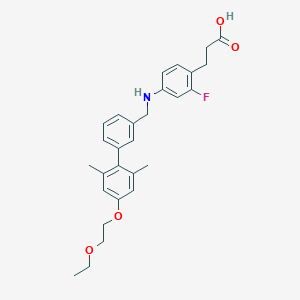 CHEMBL2048628 CHEMBL2048628 | C28H32FNO4 | 465.565 | 6 / 2 | 5.8 | No |
| 36311 | 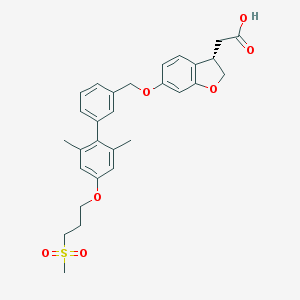 TAK-875 TAK-875 | C29H32O7S | 524.628 | 7 / 1 | 4.7 | No |
| 36314 | 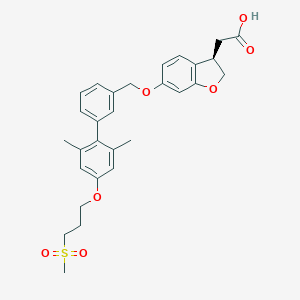 CHEMBL2179472 CHEMBL2179472 | C29H32O7S | 524.628 | 7 / 1 | 4.7 | No |
| 36368 | 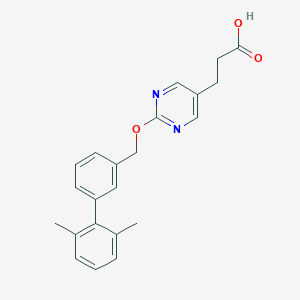 CHEMBL3288350 CHEMBL3288350 | C22H22N2O3 | 362.429 | 5 / 1 | 4.1 | Yes |
| 558402 | 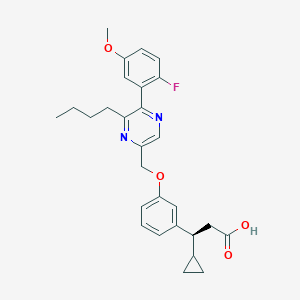 SCHEMBL17419038 SCHEMBL17419038 | C28H31FN2O4 | 478.564 | 7 / 1 | 5.4 | No |
| 558574 | 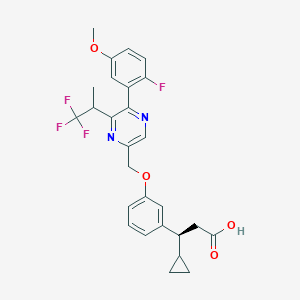 SCHEMBL17418846 SCHEMBL17418846 | C27H26F4N2O4 | 518.509 | 10 / 1 | 5.4 | No |
| 548410 | 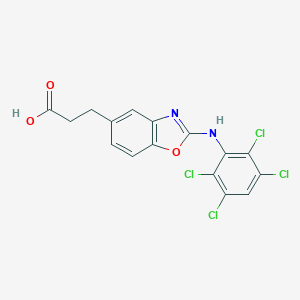 CHEMBL3954105 CHEMBL3954105 | C16H10Cl4N2O3 | 420.067 | 5 / 2 | 5.9 | No |
| 558716 | 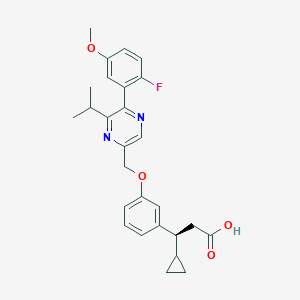 SCHEMBL17418617 SCHEMBL17418617 | C27H29FN2O4 | 464.537 | 7 / 1 | 4.8 | Yes |
| 48796 | 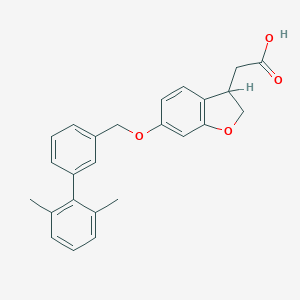 CHEMBL2022247 CHEMBL2022247 | C25H24O4 | 388.463 | 4 / 1 | 5.0 | Yes |
| 49706 | 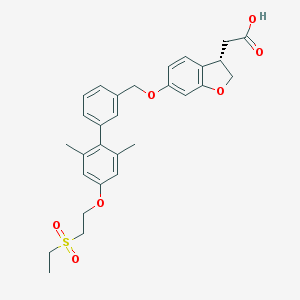 CHEMBL2047158 CHEMBL2047158 | C29H32O7S | 524.628 | 7 / 1 | 4.8 | No |
| 59171 | 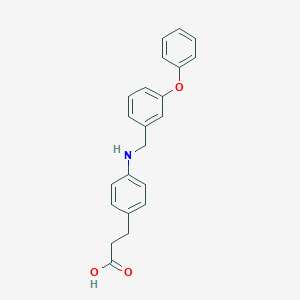 GW9508 GW9508 | C22H21NO3 | 347.414 | 4 / 2 | 4.7 | Yes |
| 65545 | 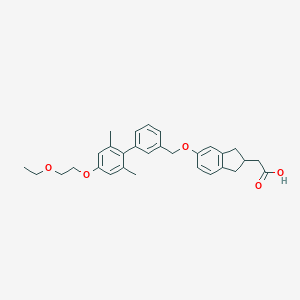 CHEMBL2022579 CHEMBL2022579 | C30H34O5 | 474.597 | 5 / 1 | 6.0 | No |
| 68052 | 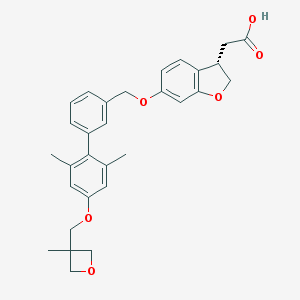 CHEMBL2047152 CHEMBL2047152 | C30H32O6 | 488.58 | 6 / 1 | 5.3 | No |
| 68054 | 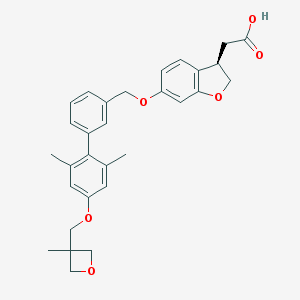 CHEMBL2047153 CHEMBL2047153 | C30H32O6 | 488.58 | 6 / 1 | 5.3 | No |
| 68056 | 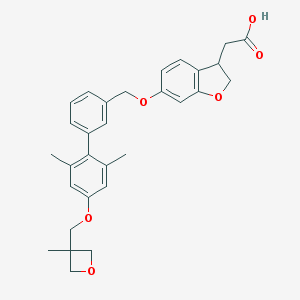 CHEMBL2047151 CHEMBL2047151 | C30H32O6 | 488.58 | 6 / 1 | 5.3 | No |
| 77640 | 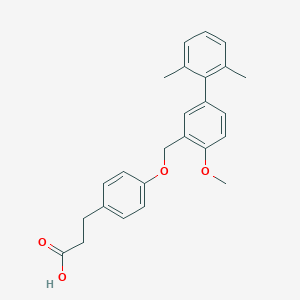 CHEMBL2048611 CHEMBL2048611 | C25H26O4 | 390.479 | 4 / 1 | 5.5 | No |
| 559766 | 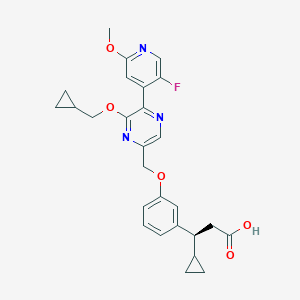 SCHEMBL17418795 SCHEMBL17418795 | C27H28FN3O5 | 493.535 | 9 / 1 | 4.0 | Yes |
| 80263 | 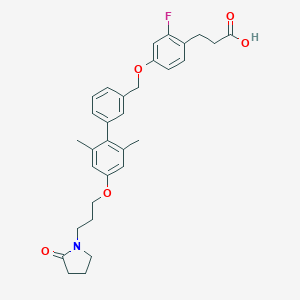 CHEMBL2048621 CHEMBL2048621 | C31H34FNO5 | 519.613 | 6 / 1 | 5.5 | No |
| 559875 | 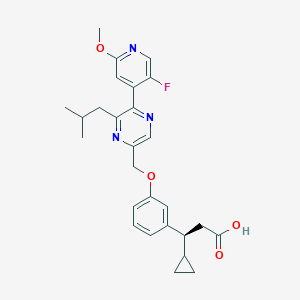 SCHEMBL17418997 SCHEMBL17418997 | C27H30FN3O4 | 479.552 | 8 / 1 | 4.6 | Yes |
| 83243 | 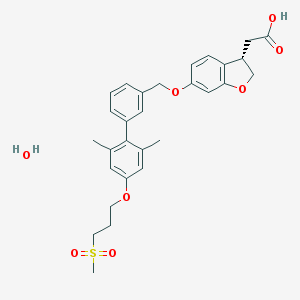 CHEMBL2047159 CHEMBL2047159 | C29H34O8S | 542.643 | 8 / 2 | N/A | No |
| 445106 | 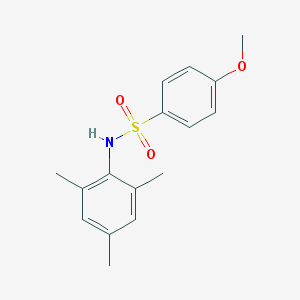 GSK137647A GSK137647A | C16H19NO3S | 305.392 | 4 / 1 | 3.5 | Yes |
| 560049 | 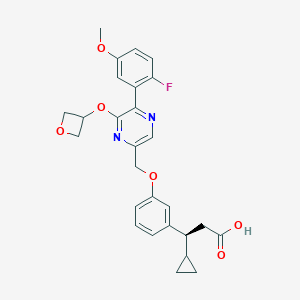 SCHEMBL17418823 SCHEMBL17418823 | C27H27FN2O6 | 494.519 | 9 / 1 | 3.7 | Yes |
| 89562 | 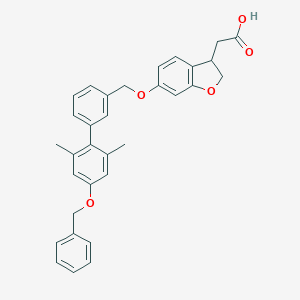 CHEMBL2022576 CHEMBL2022576 | C32H30O5 | 494.587 | 5 / 1 | 6.5 | No |
| 91181 | 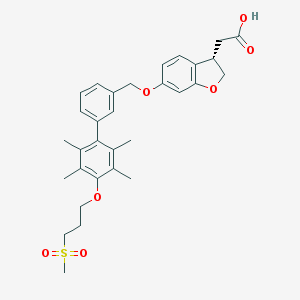 CHEMBL2047166 CHEMBL2047166 | C31H36O7S | 552.682 | 7 / 1 | 5.5 | No |
| 93325 | 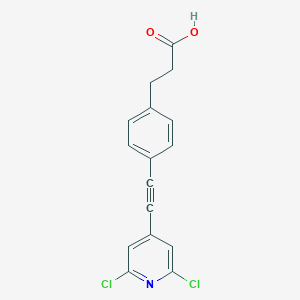 CHEMBL1829170 CHEMBL1829170 | C16H11Cl2NO2 | 320.169 | 3 / 1 | 4.6 | Yes |
| 94569 | 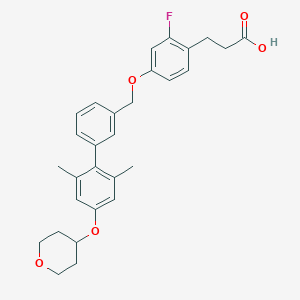 CHEMBL2048619 CHEMBL2048619 | C29H31FO5 | 478.56 | 6 / 1 | 6.0 | No |
| 524256 | 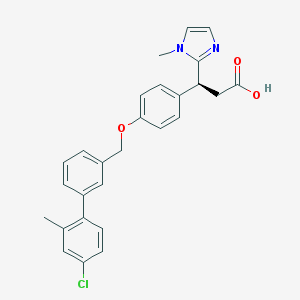 CHEMBL3287569 CHEMBL3287569 | C27H25ClN2O3 | 460.958 | 4 / 1 | 5.3 | No |
| 101615 | 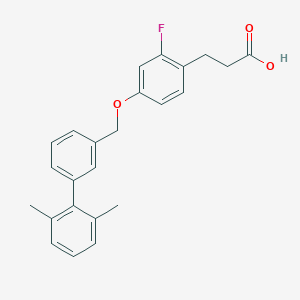 CHEMBL1688473 CHEMBL1688473 | C24H23FO3 | 378.443 | 4 / 1 | 5.6 | No |
| 103066 | 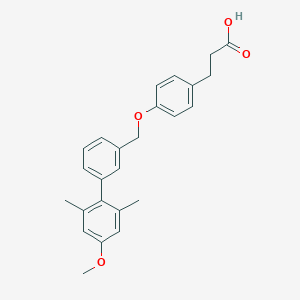 CHEMBL2048614 CHEMBL2048614 | C25H26O4 | 390.479 | 4 / 1 | 5.5 | No |
| 106960 | 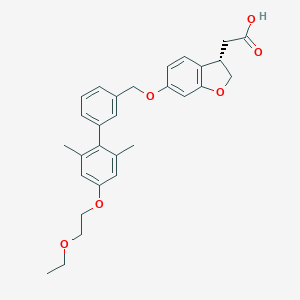 CHEMBL2047149 CHEMBL2047149 | C29H32O6 | 476.569 | 6 / 1 | 5.2 | No |
| 106962 | 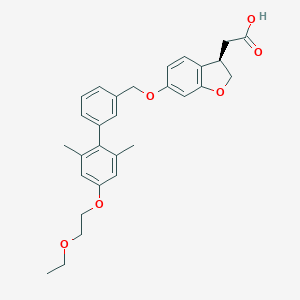 CHEMBL2047150 CHEMBL2047150 | C29H32O6 | 476.569 | 6 / 1 | 5.2 | No |
| 106964 | 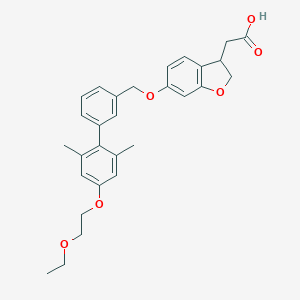 CHEMBL2022577 CHEMBL2022577 | C29H32O6 | 476.569 | 6 / 1 | 5.2 | No |
| 560769 | 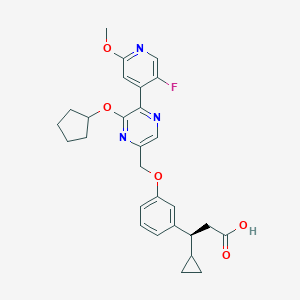 SCHEMBL17418598 SCHEMBL17418598 | C28H30FN3O5 | 507.562 | 9 / 1 | 4.5 | No |
| 115414 | 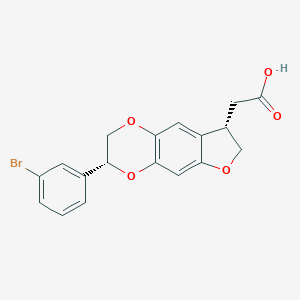 CHEMBL2381282 CHEMBL2381282 | C18H15BrO5 | 391.217 | 5 / 1 | 3.2 | Yes |
| 120275 | 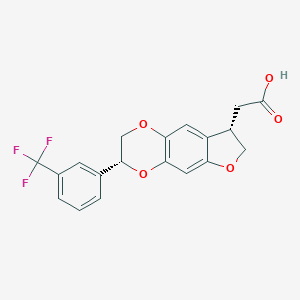 CHEMBL2381289 CHEMBL2381289 | C19H15F3O5 | 380.319 | 8 / 1 | 3.3 | Yes |
| 121519 | 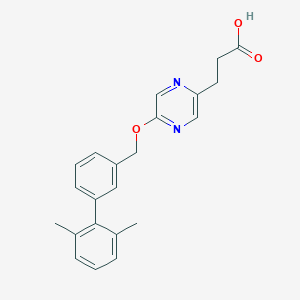 CHEMBL3288348 CHEMBL3288348 | C22H22N2O3 | 362.429 | 5 / 1 | 3.7 | Yes |
| 561148 | 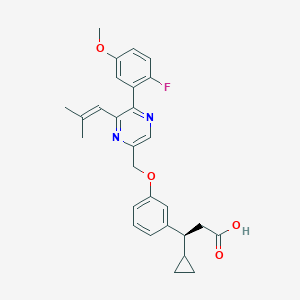 SCHEMBL17418897 SCHEMBL17418897 | C28H29FN2O4 | 476.548 | 7 / 1 | 5.3 | No |
| 124144 | 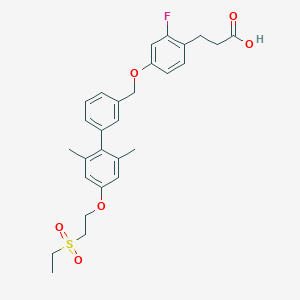 CHEMBL2048622 CHEMBL2048622 | C28H31FO6S | 514.608 | 7 / 1 | 5.3 | No |
| 561222 | 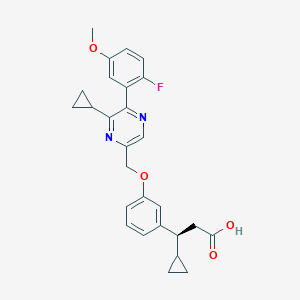 SCHEMBL17418658 SCHEMBL17418658 | C27H27FN2O4 | 462.521 | 7 / 1 | 4.3 | Yes |
| 131743 | 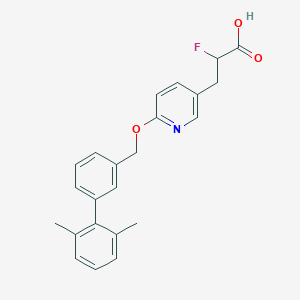 CHEMBL3288354 CHEMBL3288354 | C23H22FNO3 | 379.431 | 5 / 1 | 5.2 | No |
| 561409 | 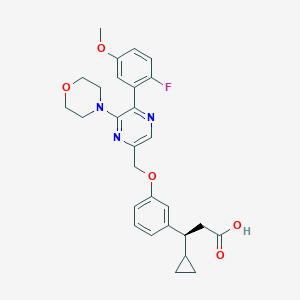 SCHEMBL17418790 SCHEMBL17418790 | C28H30FN3O5 | 507.562 | 9 / 1 | 3.8 | No |
| 132943 | 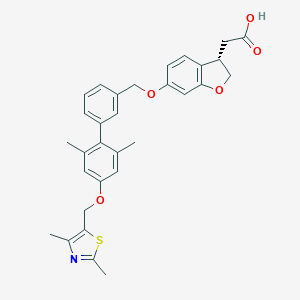 CHEMBL2047162 CHEMBL2047162 | C31H31NO5S | 529.651 | 7 / 1 | 6.4 | No |
| 133046 | 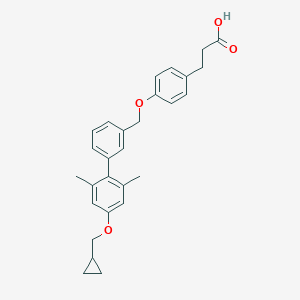 CHEMBL2048615 CHEMBL2048615 | C28H30O4 | 430.544 | 4 / 1 | 6.2 | No |
| 142374 | 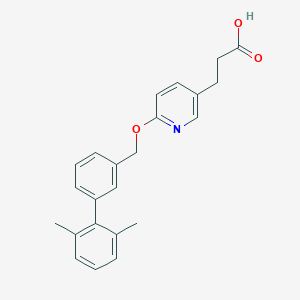 CHEMBL3288344 CHEMBL3288344 | C23H23NO3 | 361.441 | 4 / 1 | 4.8 | Yes |
| 561874 | 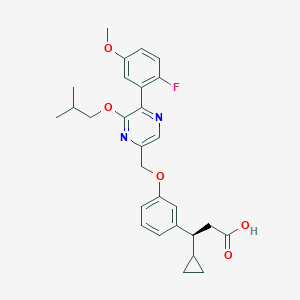 SCHEMBL17418589 SCHEMBL17418589 | C28H31FN2O5 | 494.563 | 8 / 1 | 5.3 | No |
| 153595 | 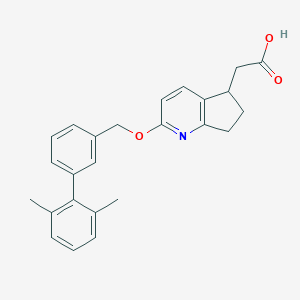 CHEMBL2022250 CHEMBL2022250 | C25H25NO3 | 387.479 | 4 / 1 | 5.0 | Yes |
| 156976 | 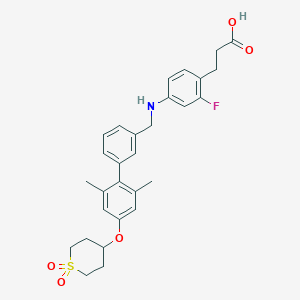 CHEMBL2048626 CHEMBL2048626 | C29H32FNO5S | 525.635 | 7 / 2 | 5.5 | No |
| 157379 | 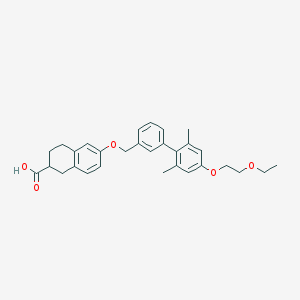 CHEMBL2022580 CHEMBL2022580 | C30H34O5 | 474.597 | 5 / 1 | 6.1 | No |
| 158501 | 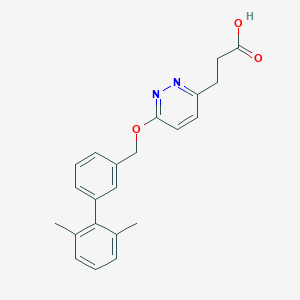 CHEMBL3288349 CHEMBL3288349 | C22H22N2O3 | 362.429 | 5 / 1 | 3.8 | Yes |
| 562355 | 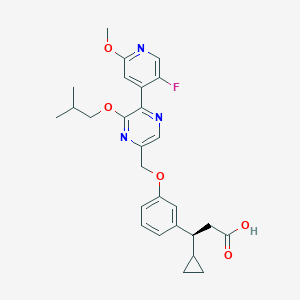 SCHEMBL17419073 SCHEMBL17419073 | C27H30FN3O5 | 495.551 | 9 / 1 | 4.6 | Yes |
| 161463 | 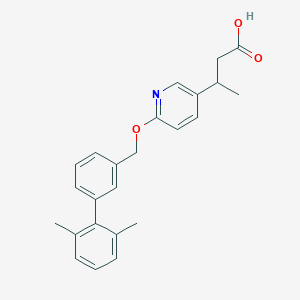 CHEMBL3288358 CHEMBL3288358 | C24H25NO3 | 375.468 | 4 / 1 | 5.1 | No |
| 562422 | 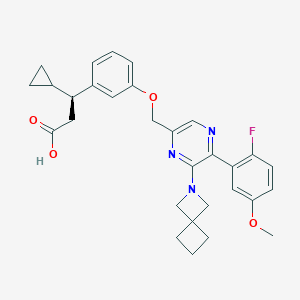 SCHEMBL17418743 SCHEMBL17418743 | C30H32FN3O4 | 517.601 | 8 / 1 | 5.1 | No |
| 562517 | 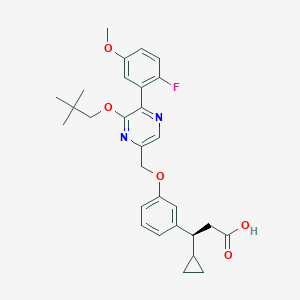 SCHEMBL17418558 SCHEMBL17418558 | C29H33FN2O5 | 508.59 | 8 / 1 | 5.7 | No |
| 550002 | 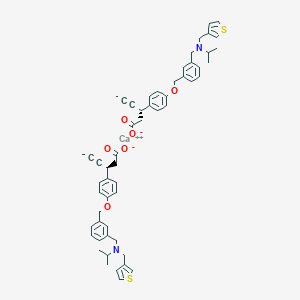 CHEMBL3930086 CHEMBL3930086 | C56H60CaN2O6S2 | 961.302 | 10 / 0 | N/A | No |
| 170298 | 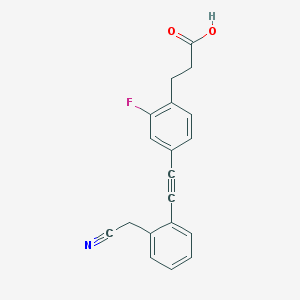 TUG-770 TUG-770 | C19H14FNO2 | 307.324 | 4 / 1 | 3.5 | Yes |
| 173664 | 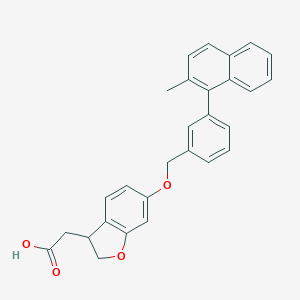 CHEMBL2022252 CHEMBL2022252 | C28H24O4 | 424.496 | 4 / 1 | 5.9 | No |
| 562815 | 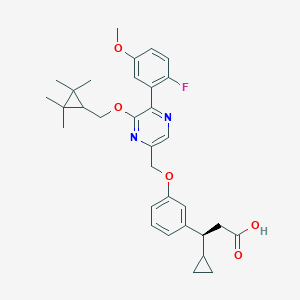 SCHEMBL17418799 SCHEMBL17418799 | C32H37FN2O5 | 548.655 | 8 / 1 | 6.4 | No |
| 550186 | 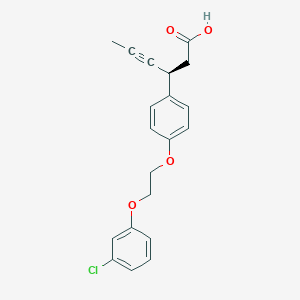 CHEMBL3927863 CHEMBL3927863 | C20H19ClO4 | 358.818 | 4 / 1 | 4.5 | Yes |
| 185499 | 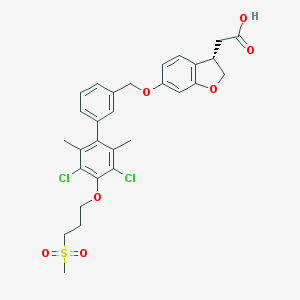 CHEMBL2047170 CHEMBL2047170 | C29H30Cl2O7S | 593.512 | 7 / 1 | 6.0 | No |
| 185523 | 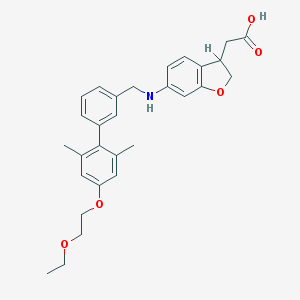 CHEMBL2022584 CHEMBL2022584 | C29H33NO5 | 475.585 | 6 / 2 | 5.2 | No |
| 189505 | 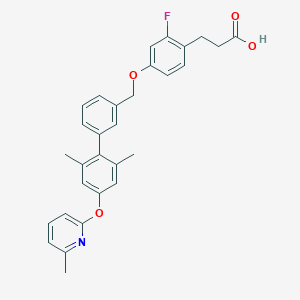 CHEMBL2048620 CHEMBL2048620 | C30H28FNO4 | 485.555 | 6 / 1 | 6.8 | No |
| 193395 | 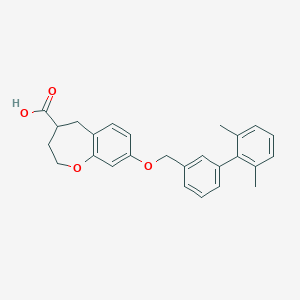 CHEMBL2022248 CHEMBL2022248 | C26H26O4 | 402.49 | 4 / 1 | 5.6 | No |
| 194006 | 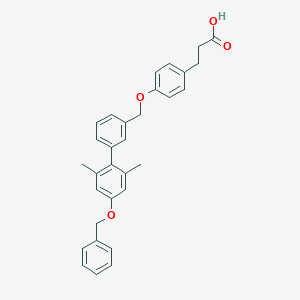 CHEMBL2048616 CHEMBL2048616 | C31H30O4 | 466.577 | 4 / 1 | 7.0 | No |
| 194243 | 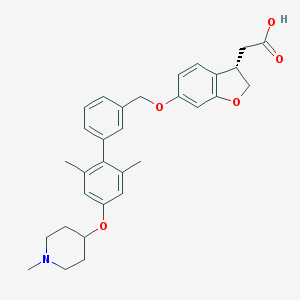 CHEMBL2047161 CHEMBL2047161 | C31H35NO5 | 501.623 | 6 / 1 | 3.3 | No |
| 197811 | 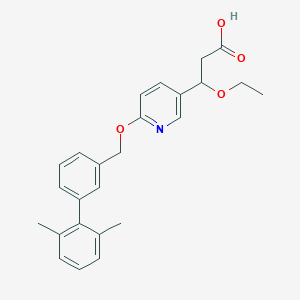 CHEMBL3288359 CHEMBL3288359 | C25H27NO4 | 405.494 | 5 / 1 | 4.6 | Yes |
| 563543 | 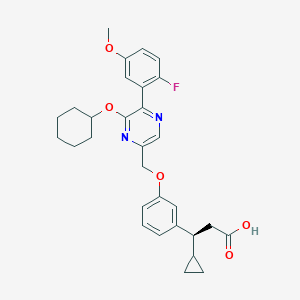 SCHEMBL17418585 SCHEMBL17418585 | C30H33FN2O5 | 520.601 | 8 / 1 | 5.8 | No |
| 563612 | 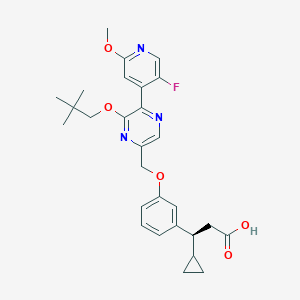 SCHEMBL17418796 SCHEMBL17418796 | C28H32FN3O5 | 509.578 | 9 / 1 | 5.0 | No |
| 563621 | 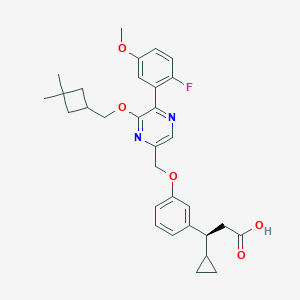 SCHEMBL17418785 SCHEMBL17418785 | C31H35FN2O5 | 534.628 | 8 / 1 | 5.9 | No |
| 201662 | 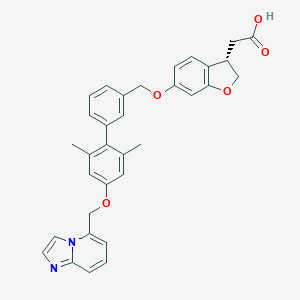 CHEMBL2047163 CHEMBL2047163 | C33H30N2O5 | 534.612 | 6 / 1 | 6.4 | No |
| 211731 | 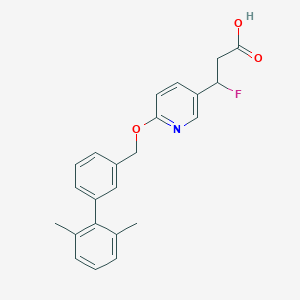 CHEMBL3288357 CHEMBL3288357 | C23H22FNO3 | 379.431 | 5 / 1 | 4.7 | Yes |
| 564008 | 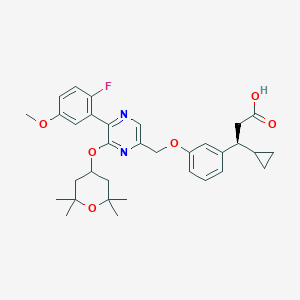 SCHEMBL17418721 SCHEMBL17418721 | C33H39FN2O6 | 578.681 | 9 / 1 | 5.6 | No |
| 213061 | 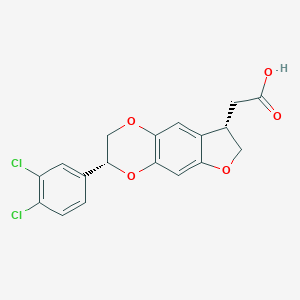 CHEMBL2381287 CHEMBL2381287 | C18H14Cl2O5 | 381.205 | 5 / 1 | 3.7 | Yes |
| 217876 | 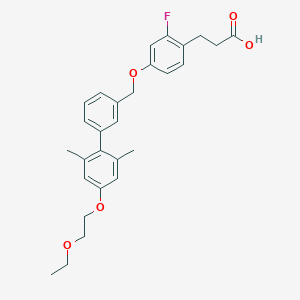 CHEMBL2048618 CHEMBL2048618 | C28H31FO5 | 466.549 | 6 / 1 | 5.8 | No |
| 564244 | 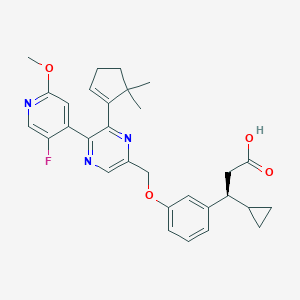 SCHEMBL17418947 SCHEMBL17418947 | C30H32FN3O4 | 517.601 | 8 / 1 | 5.0 | No |
| 564374 | 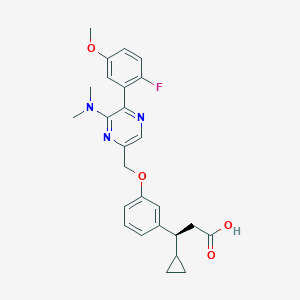 SCHEMBL17418582 SCHEMBL17418582 | C26H28FN3O4 | 465.525 | 8 / 1 | 4.1 | Yes |
| 564396 | 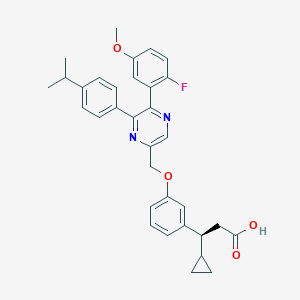 SCHEMBL17418793 SCHEMBL17418793 | C33H33FN2O4 | 540.635 | 7 / 1 | 6.5 | No |
| 226653 | 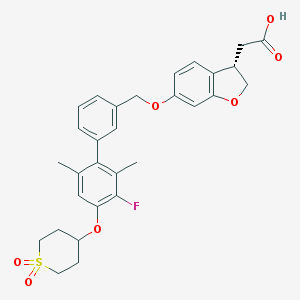 CHEMBL2047167 CHEMBL2047167 | C30H31FO7S | 554.629 | 8 / 1 | 5.0 | No |
| 564538 | 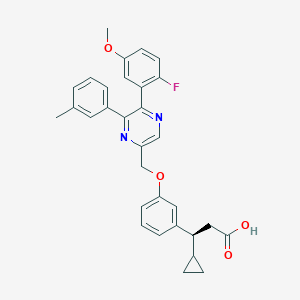 SCHEMBL17419029 SCHEMBL17419029 | C31H29FN2O4 | 512.581 | 7 / 1 | 5.7 | No |
| 231693 | 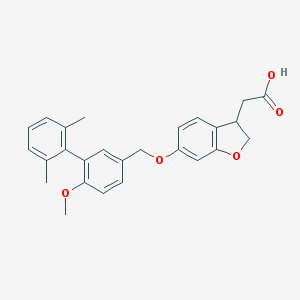 CHEMBL2022255 CHEMBL2022255 | C26H26O5 | 418.489 | 5 / 1 | 5.0 | Yes |
| 564582 | 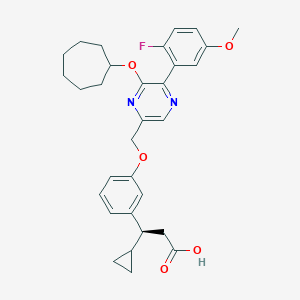 SCHEMBL17418762 SCHEMBL17418762 | C31H35FN2O5 | 534.628 | 8 / 1 | 6.3 | No |
| 236192 | 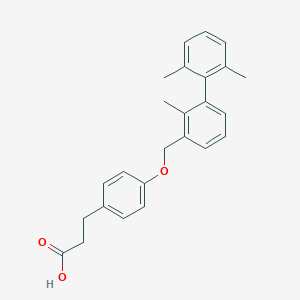 CHEMBL2048610 CHEMBL2048610 | C25H26O3 | 374.48 | 3 / 1 | 5.9 | No |
| 238427 | 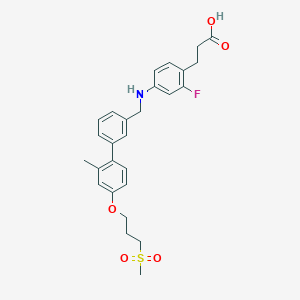 CHEMBL2164242 CHEMBL2164242 | C27H30FNO5S | 499.597 | 7 / 2 | 5.0 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417