

Go to page (83 pages in total): [First page] << < 26 27 28 29 30 31 32 33 34 35 36 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 3001 |
PF17340 (DUF5370) |
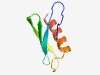 Estimated TM-score=0.58; hard target. |
>Family of unknown function (DUF5370) (Length=62) MGAIERSGYRFEPEFSVINQNGAVHVYQNGEFIEEIKFTFSGKFPEHDQIEELVDKYCEQHQ |
| 3002 |
PF17228 (SGP) |
 Estimated TM-score=0.58; hard target. |
>Sulphur globule protein (Length=94) WWGSPWDNNGYGNGNGYGAGNGAGNGSGNFGMNFNMSGNAQTAANGYNGYNGYNGYNGYNGYNGYTAPMPPPWAYAAPVA PVAPAAPQAPEAAK |
| 3003 |
PF17151 (CHASE7) |
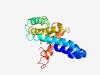 Estimated TM-score=0.58; easy target. |
>Periplasmic sensor domain (Length=186) TWREVVVLESAYRASQRNHLENVANVLDRQLQFSVDKLLFLRNGMYEALVTPLDFADLHDAVAQFAQLRGKSFWQLELDR RRTLPLNGVSDAFVNRTTLLSRDNDALANELSAALELGYLLRLAHSSTMLAQRTMYVSRSGFYVSTEPTTNGGDIMSRYY QYVIQPWFSGQSQRQNSARGVRWFSS |
| 3004 |
PF17150 (CHASE6_C) |
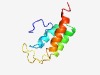 Estimated TM-score=0.58; hard target. |
>C-terminal domain of two-partite extracellular sensor domain (Length=86) MDTSRRFEGIWTFDRTVSAKAAELLLDRIATYRPELADKIEQAKAEYISSGSTQPSDRNRQDSPNPDPFVQRLVTYLQAG QYKLLK |
| 3005 |
PF17113 (AmpE) |
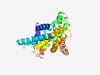 Estimated TM-score=0.58; hard target. |
>Regulatory signalling modulator protein AmpE (Length=284) MTLFTLLLVLAWERLFKLGEHWQLDHRFEVIFRRGRQASLAKTLVITIVWMVIIWAVLRAVQGIFFGVPTLILWVILCLL CIGAGIKRKHYRAYLKSARQGDADACKEMAEELALIHGLPTDCSEEARLRELQNALLWINYRYYLGPLFWFVVCGPYGPI ALGGYAVLRGYQTWLARHMPPLKRSQSGVDALLHILDWIPVRLAGVAYALLGHGEKALPAWFASLGDIHSSQYHVLTRLA QFSLARDPHTDPVQTPRAAVSLARKVTMIILVVVALLTIYGTLI |
| 3006 |
PF17100 (NACHT_N) |
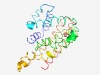 Estimated TM-score=0.58; hard target. |
>N-terminal domain of NWD NACHT-NTPase (Length=222) ERLWNQAYDELKASEPKLVEAYEKILSVGLHRNDPSSVTCESTENEIEGAHETRCRQMQQLVRDGLDRTQKAASIKRGID EGLQAVQAVRGIVDKAIQAAPEAAIAWVGVCLGLEILSNPVTEARYNRNGIAYVLSRMEWYWNLVSLLLDENKAEQSSAG LRVQLEKHVVQLYEKLLSYQVKSVCLYHRNWAAVVLRDVFQLDDWAGQLDDIQKAEAAVRTD |
| 3007 |
PF16971 (RcpB) |
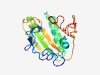 Estimated TM-score=0.58; hard target. |
>Rough colony protein B, tight adherence - tad - subunit (Length=167) MRKLVTSIVFVVLMYSAQGVAFSKNELVTNTNVAQPVQLDDFSRTQLVNRYVMSEYSDVVYHNVLYSVLRDVGSDRSKRV HIIWSNKESQKKATGMRNFLINKGVEAASIKLVRSEYKKALYPIYVEVSRIGTKRANCRIDTAEDMMSWDGLNPCATKSN QRIQLKY |
| 3008 |
PF16910 (VPS13_mid_rpt) |
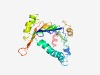 Estimated TM-score=0.58; hard target. |
>Repeating coiled region of VPS13 (Length=233) FEYKPLDNRADNAAALYMRNIDIIYNPEIIRELIVFFTPPETSADSINALIEVAGDTFEGIKKQTKASLEYALEQHTTFD LKVDMDAPVIIIPENCTSTSCRAIVIDAGHINVESNLSSPEMRTQMKSKSGAEMSAEDNLQFRSLMYDRFTIQLTETKVL VGDSLDTCLVQIRNPRPELSYLHLIDHIDMMFLLELCIIRKSVDMPRFKVSGHLPLLKVNFSDTKYKALMQLP |
| 3009 |
PF16907 (Caskin-Pro-rich) |
 Estimated TM-score=0.58; very hard target. |
>Proline rich region of Caskin proteins (Length=85) KKRSHSLHRYALSDGEHEEEDGTTSTLGSYATLTRRPGRGQMPRACLQADAKVTRSQSFAIRAKRKGPPPPPPKRLSSVS STPTA |
| 3010 |
PF16888 (DUF5082) |
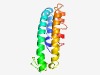 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF5082) (Length=121) KYEINQANYRISECQNEIQGLEKKIERLEGAKQKLQTYKLNIESEKFDITQKLSCSSWKGSNKEEYEGIAEEQLKPCYQT YYDETDQAVDAIMDEITRLENQIYDQEGVIGWLKSQINSLG |
| 3011 |
PF16634 (APC_u13) |
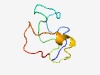 Estimated TM-score=0.58; very hard target. |
>Unstructured region on APC between APC_crr and SAMP (Length=54) IESPPNELAAGEGVRAGAQSGEFEKRDTIPTEGRSTDEAQRGKVSSITIPELDD |
| 3012 |
PF16142 (DUF4850) |
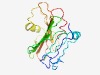 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF4850) (Length=187) HVQVPLYGITALNPVDDGLLKNFKYCNPKNCQFNFKLPAEQAKNLKLIAIPEIGVVLVPRTWQDIQADAGANGTGYALII SPDQKQAIQLYDSSLCVGCGLPYASLYFPELLKESIENEFGGYQDSQKLMNVVHPSKHTAFFSYQIPKLNNKTHGVAKYH DDGDFNFREIKVTLDKSQQHLVGPILN |
| 3013 |
PF16086 (DUF4816) |
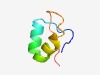 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4816) (Length=42) VWKKKLVWKPEWKQIWKTESKLIWKQEWKKVQVPVWKEIQVP |
| 3014 |
PF16084 (LydB) |
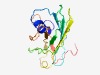 Estimated TM-score=0.58; hard target. |
>LydA-holin antagonist (Length=147) MIGWGVCVLALALADRYLLKRKDITHLELGDVEIKPGFIRVPFKYRSKFPFLRGATVRYWIRDVQKPTTVIEGEQRCLAS AEQGENSEWLYIPTEYMGKGERLWHFNVMVTHGDSFINPLYRIFPVTQQIRSSYVINLAQDVSDDEK |
| 3015 |
PF16083 (Phage_holin_3_3) |
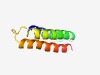 Estimated TM-score=0.58; easy target. |
>LydA holin phage, holin superfamily III (Length=77) VIGFFAAWGGVVKYLMDRQKADRKWNWVSVINQIVISSFTGLLGGLLSLDSGASHYMTLIIAGLFGAMGSRVLENLW |
| 3016 |
PF16080 (Phage_holin_2_3) |
 Estimated TM-score=0.58; easy target. |
>Bacteriophage holin family HP1 (Length=55) DKYTSPTAYTWGAFTAMLGALSLNDWAIVIGIICTIGTFAVNWYYKHREFNRHEK |
| 3017 |
PF15953 (PDU_like) |
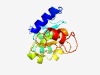 Estimated TM-score=0.58; easy target. |
>Putative propanediol utilisation (Length=153) DNLVQQVMQRLEERKHTSVEVTFNHQVAPPSEQIFLRNGKVILKDISIELITDLYSMEKTNAWVKWVLEGISYDVKFYFL INEQMVNFIPRMMILDWPILFVVNNESPVIASYNRIITREEIAAKPDKSILVRYQKQHITDEALDICNYKKIK |
| 3018 |
PF15933 (RnlB_antitoxin) |
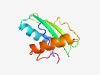 Estimated TM-score=0.58; hard target. |
>Antitoxin to bacterial toxin RNase LS or RnlA (Length=94) SYIVLSTSYMSPIEWVEELEACLMKEEYVGNILLDLLLSNGYDRDRFYEIFFDGKKLNFQTLLNVTKVSEELKQVSGEYY SRNSEILDNSVLTK |
| 3019 |
PF15866 (DUF4729) |
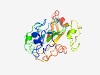 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF4729) (Length=215) CCPLAHNGVTPCPLLLNESLLEHFIKSHLDVPGLELCEIFERDRLLIIFNPFTFELGKNFCMSAIVYGGFRDSPYSLPLM RFMPIYNVNLSDTHGKYACHLPLFLLICRTRLSAIDDDPPITTTMRTRHMGRQPVDEDVLALWVVSMELPQPIHVTISLF NRRLDVTRSAIMKVRALKESQNCADFMRTSENYMRLTEQDMQILTNNNKESIYME |
| 3020 |
PF15635 (Tox-GHH2) |
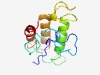 Estimated TM-score=0.58; very hard target. |
>GHH signature containing HNH/Endo VII superfamily nuclease toxin 2 (Length=118) QTAHHLIEASALHDKGRGGKGSVPLKGISNYSENKAPCVCAEGVNQNVGTHGLMHTFQSAAAAKSRSGILQLSNGSSISA KKTTYGTAKRQSMAAMGKVFPQSKCSKECLSAQLDNYH |
| 3021 |
PF15592 (Imm41) |
 Estimated TM-score=0.58; hard target. |
>Immunity protein 41 (Length=115) QVLTSNYQLKEGSFLYSLLEEASFNEIAFWNYYNAVIEIIDQTKDKQLDRELTKIIHWTYKRVMRSFLCHFDPSDVFKIS DFPDDRFIYFYERLDFMIDGYYEGFIMSEKQFGDN |
| 3022 |
PF15541 (Ntox4) |
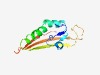 Estimated TM-score=0.58; hard target. |
>Bacterial toxin 4 (Length=106) LKGYERAHLWGHGFGDEAKLGLMYAPKALNQEVQNFGFEKFIRELQVLSQQEGRKVFCRCKAFSHPPKGKSLNAAKGELL ISEVFYEVVYEAKDGRKIVGKFAASI |
| 3023 |
PF15524 (Ntox17) |
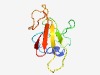 Estimated TM-score=0.58; very hard target. |
>Novel toxin 17 (Length=94) WSITARIQYAKLPRQGRIRYIPPKNYSPSAPLPKGPNNGYLDKFGNEWTKGPSRTKGQEFEWDVQLSKTGREQLGWASRD GKHLNISIDGKITH |
| 3024 |
PF15460 (SAS4) |
 Estimated TM-score=0.58; hard target. |
>Something about silencing, SAS, complex subunit 4 (Length=95) ALPDALYLPVHKRAERLERSIRNTEKGRAQHEKDQIVRLLEGLQGHDWLRIMGVSGVTESKKKSFEPARRHFIKGCQAII DKFRQWAQEEKRRKA |
| 3025 |
PF15404 (PH_4) |
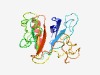 Estimated TM-score=0.58; easy target. |
>Pleckstrin homology domain (Length=188) QKGILYQKPKKHAVFSKYYIVLIPGFLILFNCFHRSTTGFAKNVIDYNHYLTIPISECYLYSGTTTELDLLRRDHTFDII NPGNHAIPRAYDDGWKSTENESSRCFTLWFGTKRAIAKYSKMQEHKAEHAEESVTDSDGEYVKNKNYESNPGTLRLVNRL GVSGRSMVFMARSRQERDLWVVSIYNEL |
| 3026 |
PF15241 (Cylicin_N) |
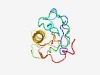 Estimated TM-score=0.58; very hard target. |
>Cylicin N-terminus (Length=107) EVNMRTYENSIPISESNRKSWNQEYFALTFPKPPQPGRKKRSRPSESQVTVPRHDKRKLQEGQKPAHIWIRHSLRKFFQR PSIYLAVRRQAPYRHPYTPKTNLKKAE |
| 3027 |
PF15120 (SPACA9) |
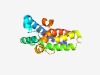 Estimated TM-score=0.58; hard target. |
>Sperm acrosome-associated protein 9 (Length=158) EAAEALRNIEQKYKLFLQQQFTFIGALQHTREAARDTIRPVASISQVQNYMDHHCHNSTDRRILTLFLSICNDLSNLCTM LETRYPDNTTTKDILEKCKLLLSHNNDLSTIRAKYPHDVVNHLSCHEARNHFGGVVSLIPIVLDYLKEWVAHVEKLPR |
| 3028 |
PF15043 (CNRIP1) |
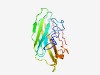 Estimated TM-score=0.58; hard target. |
>CB1 cannabinoid receptor-interacting protein 1 (Length=154) VHATVSLKIQPNDGPVYFKVDGQRFDQNRTIKFLTGAKYTVEVVLKPGVVHATTMGIGGVNIPLEEKSRDPQVVCYTGIY DTEGVPHTKSGQRQPLQVNIQFSDIGTFETVWQVKFYDYHKRNHCQWGNAFGSIEYECKPNETRSLMWINKEMF |
| 3029 |
PF15001 (AP-5_subunit_s1) |
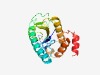 Estimated TM-score=0.58; easy target. |
>AP-5 complex subunit sigma-1 (Length=198) MVHSFLIHTLCPVGALSCTESRVLYSRVFGADLTQTGPAGDREQESEARRLLQKEQLAVVARQVRSVCALQREAAGRPGL HETGAVLGEEEAALQEADSGVLRLAEGDPFPREHTVVWLGVQALGFVLVCEAHENLLLAEGTLRSLARHCLEHLRLLGTG SEVLLKADRIEVLLHRLLPHGQLLFLNHRFKHSLDKEL |
| 3030 |
PF14997 (CECR6_TMEM121) |
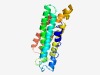 Estimated TM-score=0.58; hard target. |
>CECR6/TMEM121 family (Length=193) PFAFISWFVYACLYVPRVAIIFKWEIADELTDHDWFGPNALKTTLALSAPLFLLLLYGHHDAKPHTNRKNYVDRLALGVT FDILDSLELLDILFVQESRVILTFELENAILSFACIVFFLPTVALFELRKSRSDGEVASVNFGVLYSLLFMLLVNVPFLV IRMLLWNIYNQDVSVFLIKNVMLLIIGFKDIWE |
| 3031 |
PF14846 (DUF4485) |
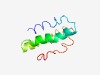 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4485) (Length=81) NIEFVYYTAMLKKFAPNLSKEKDKEKVIPWIRKLYAAEYATEFFKVKRNRYLFNMILSILNDEMYGVFKSIPPIGPLKSP E |
| 3032 |
PF14828 (Amnionless) |
 Estimated TM-score=0.58; easy target. |
>Amnionless (Length=446) YKQWIPDTNYENKTNWDKGKVPCGNDVVVFPAQGKVSVFVETTHAVQEMRLPVDGELILNSGAGFYVLSGKDPACGAGAT VQFQDSESLQWFNPAMWQAAATQNDFQLGKFLFSVHEESVPCQQDDVVFKALSSFRVDTTSSQSTIPVTSVSVLGKSFNS KSEFSEYLSTRTGQMQFHGSSAVTVGNPGCGDPSGCDCGNSVNHQLICSTVQCATVSCKQPLSPMGHCCEVCGAIVTIQY TVGFNMQTYRERIHHLFLILPEYRSIQLGVSKVFKSQKLLGVIPFSTTPEIQVVIVDGEEGKLSEALAQNIMTDIRSHGS ELGITESEFQASSGNSSDPSGGNAGMVAGIVIAVLVLIALVAVGVVLIQKGVVRLPPMPTIPMPSLSSFKGNTDVGDLGG TIDRGFDNPIFDHPDMLSNNPGLYGTESQSISMRQTGVHFINPVYD |
| 3033 |
PF14741 (GH114_assoc) |
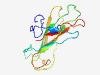 Estimated TM-score=0.58; hard target. |
>N-terminal glycosyl-hydrolase-114-associated domain (Length=134) QAGDDVPVSFENMASYVRMLTLFGNAQTDFYFSPPSSVADGTAKSIVVGASALVPTEGSGGVWSVGVSSIDADGNSTLHT VGDFAAAGSEDWTPVTLTMPVTADVADYVDDNNEMLLTVSCSEPGTTQALFIDF |
| 3034 |
PF14625 (Lustrin_cystein) |
 Estimated TM-score=0.58; hard target. |
>Lustrin, cysteine-rich repeated domain (Length=41) CAQGQAYRDTSGNFVQCGHSPKSCPQNYECYYDGNLWGCCP |
| 3035 |
PF14477 (Mso1_C) |
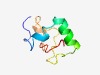 Estimated TM-score=0.58; hard target. |
>Membrane-polarising domain of Mso1 (Length=63) RSQHQETQSAPARTYTRSSSRLQDMYNRSREQSVPGAGYSSQSYQPARTNSASGNGSSSRLRD |
| 3036 |
PF14469 (AKAP28) |
 Estimated TM-score=0.58; hard target. |
>28 kDa A-kinase anchor (Length=122) WMKCEQFTVEKGIQKIEEFIKQTWEYQGAWLYCIDFLREEDDKYSKIYRYQVRWSIPTRRKPIPRATACVYFTIEVSKIK PGHFPVDVYYVFEGNRLVHKPGNSRFREKWLKDIIDSKIIMM |
| 3037 |
PF14468 (DUF4427) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF4427) (Length=132) LSASKVKNYADSINDYVSELYSKKDFLNDSYAMEFGNAWVWIHDNQSQVVRALLQAGMIKVNKEGRYLLDVNLASVDWPL RRKEAFASHVAGWLKHRFDIEAGRYSVRGKDDYDAIPSYETPLKDQHPFYNH |
| 3038 |
PF14461 (Prok-E2_B) |
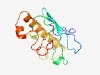 Estimated TM-score=0.58; easy target. |
>Prokaryotic E2 family B (Length=141) CGFRYTPAKQMPKGILLDTKSRRKGYYVKEYSTKGGVFVIALVLWNDPHIQLPFAYILQQPEQYKGRLLPHINFGFCLCY VTQMEADWNSNDLKSTYQDVDEQIQLTLDNSVASVESGTSNDVELEGEFSAYWQSEEELYL |
| 3039 |
PF14298 (DUF4374) |
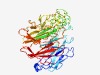 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF4374) (Length=436) TSCSEDDQSGSNNNSNTSTGNYVIAATVTSSSTDTYVLLTAPSLDEGEVTTQGNGLVNEGATYWVFYTNRYLYALNYNQG EAGTTQSFILNNTGALEKRSQEYYVNRFTTYGYYDRYIMTTSTGDGPTELKDENGYVPQSFLASYLDVENQTYTSNTVNA STPFLSENFLGNGEYVTLAGLAQVGTRLYAAAVPMGLSQYGCMQKNDDGSYKWVLPGNEDLIKTESGGSGSGAYDKDELQ WTQYPNECWMAIFTDQTLTDWKLIKTDKISYACGRNRSQYYQMNWLADDGYMYVFSPSYAKTMKDVRQQTTLPAGVVRID TKAEDFDPDYYYNLEAKADGASFLRSWYIGGDYFLLLMYDKAITASDKVANRLAIFNVSNGDLSYVSGLPTDVSGFGNTP YMENGSAYIAVTTTSGYPAIYRIEPSTATASKGLVV |
| 3040 |
PF14207 (DpnD-PcfM) |
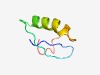 Estimated TM-score=0.58; easy target. |
>DpnD/PcfM-like protein (Length=46) YDVTITETLKMTVTVEAESQLEAEQMVSDSWRNQEYILDADNFTGV |
| 3041 |
PF14128 (DUF4295) |
 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4295) (Length=48) AKKTVATLQTGKEGRSYTKVIKMVKSPKTGAYIFDEQMVPNDKVQDFF |
| 3042 |
PF14101 (DUF4275) |
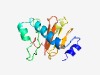 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4275) (Length=139) MSINEILKNKSIKVIEVPKWGTFLRKQWENVFANHLSHNEKKEIFMYDDDGFCGYLWHVFSFEKRVCLKEQEAETAFNNE RKNSCYIFYQHTDDVFIVEEAANLNANDLLNEFDVYVVDKEFNWTYVKTHETGWCGPYF |
| 3043 |
PF14093 (DUF4271) |
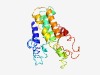 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4271) (Length=204) ITSILLGCFILATLVFSKSRRFIEKQAKNFFYIPRSENSTMTETSGEVRFQIFLVLQTCLLASIVFFCYTRTFVGDTFTI DQYQIIGIYTGIFAAYFLVKTIAYAFVDWVFFDKKKNRQWIQSFLLLISTEGVALFPLVMLLAYFNLSMESAVIYSIIVV ILSKLLSFYKSYIIFFRRSGDFLQIILYFCALEIIPLFSLWGLL |
| 3044 |
PF14069 (SpoVIF) |
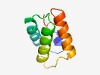 Estimated TM-score=0.58; hard target. |
>Stage VI sporulation protein F (Length=71) FDNIEKKTNVKQEDIFKLAQSVSNANLKDEKTVRQLIAQVARLANKPVSKEKEDQIVNAIINNNMPMDLGS |
| 3045 |
PF14041 (Lipoprotein_21) |
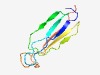 Estimated TM-score=0.58; hard target. |
>LppP/LprE lipoprotein (Length=84) DPCATLSWALAALDGGTGSSPWTVLFFHDDTYLGTATALPYSFTDVASQTDDTVTVSYGWLREGESTSQQSGGPGLVRYT WNGS |
| 3046 |
PF14038 (YqzE) |
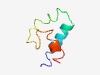 Estimated TM-score=0.58; hard target. |
>YqzE-like protein (Length=54) KTNDYVKYMTQQFVQYMDQPKEERKTKKIQKKEERAPFANRWFGVIPLSFMLWY |
| 3047 |
PF14034 (Spore_YtrH) |
 Estimated TM-score=0.58; hard target. |
>Sporulation protein YtrH (Length=99) NFISCYFIALGVLLGGALIGGIGAYLSGQPPLTIITSLANRLKIWALVAAIGGTFDAVYSFERGIFEGNTRDIVKQLLLI LSAMGGAQTGYLIINWLTQ |
| 3048 |
PF14019 (DUF4235) |
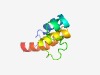 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF4235) (Length=77) YKPVGVLLGLAAGMLAGNIFRQVWKLTAGDGEAPNPTDEDRRWGEILAAAALQGAIFSVVRAAVDRGGAVGVRRLTG |
| 3049 |
PF13889 (Chromosome_seg) |
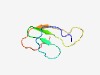 Estimated TM-score=0.58; hard target. |
>Chromosome segregation during meiosis (Length=53) VLRYLIHLNICCPAKGRFYLYSTIRVVFANRVPDGKEKLRNEIQYPEPRYSPY |
| 3050 |
PF13885 (Keratin_B2_2) |
 Estimated TM-score=0.58; hard target. |
>Keratin, high sulfur B2 protein (Length=45) CCGSVCSEQSCGLETCCRPTCCQTTCCRTTCCRPSCCISSCCRPS |
| 3051 |
PF13800 (Sigma_reg_N) |
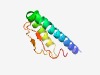 Estimated TM-score=0.58; hard target. |
>Sigma factor regulator N-terminal (Length=86) ALKKAKRKQLLKIVITSIIVVLILLPILYKTGNYFAAKSSTKLHEQLFLHNEIAEPNIQIDSQVTSNTSMFGGNIVTNRS KNINGY |
| 3052 |
PF13782 (SpoVAB) |
 Estimated TM-score=0.58; hard target. |
>Stage V sporulation protein AB (Length=109) LLAVGLIPRFAGKTHTGDHIFLYEEMVVAGSIIGNLWSVFQLSINLGQWFLILFGFFAGIFVGCLALAIAEMLDTIPIFA RRIGFRHGLGIVVLSVAFGKMLGSLIYFF |
| 3053 |
PF13708 (DUF4942) |
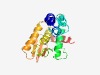 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4942) (Length=193) NTVDKGAWEYLMKESGLRSFMDSKARQDWDNQIKEGYLKVPEFNFDNIKATFTTLHEARGEMFERGVIECFKNLSWDYKT NQPCKFGKRIIMYYLFNEKSRNGFCSQYPKYENANKVDDILRAFHIFDGKPELDHRNSFQQMVTKAREFDTDVTEIENEY VIIKWFNNGNGHINFKRLDLVDKMNAIIAKYYP |
| 3054 |
PF13274 (DUF4065) |
 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF4065) (Length=111) LKLLWLADRLHARMYGRTITGDCYKAMHNGPVASATRDILEYGITKSIKTEATTYLKSIEEDRYLFGSVGEVNYKVFSKT DIDCLDMVMETFGDQEEFQLSKLSHSFPEWK |
| 3055 |
PF13013 (F-box-like_2) |
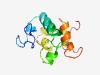 Estimated TM-score=0.58; hard target. |
>F-box-like domain (Length=102) ASPKSNLYYRYSSESPKYAKILDCPDEILQLIFSYCYDASYIEKLPFAFSYRKQRHTLIHDLPNTCLRFKKILSPRNVSF WKRLLKVHKKNPNSAVTCSESI |
| 3056 |
PF12789 (PTR) |
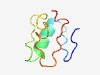 Estimated TM-score=0.58; hard target. |
>Phage tail repeat like (Length=60) EYSKKTHIHYINQIPSLKETLETKADKTHTHSISDITNLQETLNRKSAVGHTHTIANITN |
| 3057 |
PF12725 (DUF3810) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3810) (Length=323) YANTIYPVLSRGVNRVTSLVPFSVMEALIVLLCAGAVFYMIFFIAKIIHNRGKRRKIFSLFAINLICIASVLYFVFVVSC GINYYRYPFAQTCGLEIRPSSKAELTALCTDLADRANELRKGVQTDRNSVMRLNTEDIETTAKKARTAFDAVSGDYPLLR AGYGPPKPVFFSRLMSYCDTTGIFFPFTFEANVNVDIPDYSIPLTMCHELSHLRGYMREDEANFIGYLACEKSGDADFRY SGTMLAYIYASNALYGTDPSAANQIYGTLSDGVRRDLENNSAYWKQFEGPVATAAEHVNDSYLKANKQTDGIKSYGRMVD LLL |
| 3058 |
PF12716 (Apq12) |
 Estimated TM-score=0.58; hard target. |
>Nuclear pore assembly and biogenesis (Length=54) QPDVASILALLAILFISLKILDMMYRAVMFWVTMALRLVFWGAILALGFWIWNR |
| 3059 |
PF12709 (Fungal_TACC) |
 Estimated TM-score=0.58; easy target. |
>Fungal Transforming acidic coiled-coil (TACC) proteins (Length=77) DTNKRLKLLAEDLYVQYSSKHEQKVKLLKKGYDTKYQSKIDQISLQNQGLVQEIEQIKNQLSTERKEKQKLLELLDQ |
| 3060 |
PF12676 (DUF3796) |
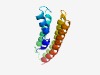 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3796) (Length=126) PKLGLLGLFGLFGFLGFVPQFFGESGVLDVPFPLIFFCFFGFFGFYYEGKMSGTMIDERYEANVNRAAAIANRVSLTLII MAAILALSLFRIHDSYGMLKLLLAVIGFAFGLSLFLQEYLLYRFEN |
| 3061 |
PF12657 (TFIIIC_delta) |
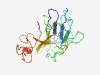 Estimated TM-score=0.58; easy target. |
>Transcription factor IIIC subunit delta N-term (Length=174) WPSALDCLAWSADCELAVAAGEVVELLIPNSKAQLQSADGALDGTTNPSWENIRIRTNIFSEAELPLHEPAPSRTYSTGE EISSGLVAGLAWSPPGLAKHRRCALAVQTQNLVLSIWESSVNPRQPGSWKRSLIVNRELERYFAKHLTSEEELHGRDAAE VLRLRRRIRTFSWC |
| 3062 |
PF12652 (CotJB) |
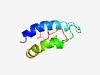 Estimated TM-score=0.58; hard target. |
>CotJB protein (Length=77) ELLNHINQVSFAVDEVKLYLDTHPCDQEALVYFHEYSRKRNEALNEYARAYGPLTIDTASESCTERWNWINEPWPWQ |
| 3063 |
PF12635 (DUF3780) |
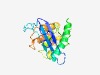 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF3780) (Length=182) HQTLGFGVPATSNPHHFIVEIPRTNTGSVGIVENLGMQTHDQSQSLIVRVLLERPRWTAIRGEVQRAFNARLKERNLKTS TWKVGANPVDRLLGKELCVLAWAVEGMEFDHIPIAVRNWLALRPEERWWLFGMTALSTGGINDGNKGWRLALRHALGDVA QHELFQHRPPQSKVKSTEKDTL |
| 3064 |
PF12618 (DUF3776) |
 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF3776) (Length=84) AKKEEDEKTLKTEKRVKQPDKEGKFIVISEKGKISEKNISKNGKEDKENISENDMEYSVDTQIEKKPENDKVKSPQENVK ETKR |
| 3065 |
PF12550 (GCR1_C) |
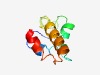 Estimated TM-score=0.58; hard target. |
>Transcriptional activator of glycolytic enzymes (Length=81) YKLSRNLSNVTSLWREWSVGLGDGNPSVATLNEEYGSKWRQEAKERKYYSKRLVIINEIQKVANNNGLSLEDAAQSVENE R |
| 3066 |
PF12402 (nlz1) |
 Estimated TM-score=0.58; hard target. |
>NocA-like zinc-finger protein 1 (Length=57) ASMTYPGTLAGAYAGYPPQFLPHGVALDPTKSSSLVGAQLASSLGCSKPAGSSPLAG |
| 3067 |
PF12400 (STIMATE) |
 Estimated TM-score=0.58; hard target. |
>STIMATE family (Length=126) CAFYFVNFTLDTTFGVLLNYAFLQLLVIIADKCNIQSLQVPGDYGNPIQIKIWLLQLTTWIVIIFTTKLVIGTVIFCFER QLGDIAKWIFTPLASHPHIELIIVMVACPCLMNGLQFWIQDNFLKK |
| 3068 |
PF12394 (DUF3657) |
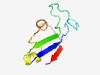 Estimated TM-score=0.58; very hard target. |
>Protein FAM135 (Length=66) LKFELIYIPTLGNAWTDVQDSSDDTDLIPVHEFRIPHRALLGLHSYCPVHFDALHSALVDLTIHIV |
| 3069 |
PF12322 (T4_baseplate) |
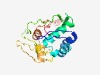 Estimated TM-score=0.58; hard target. |
>T4 bacteriophage base plate protein (Length=203) YKFDVRVGSKIINCRAFTLKEYLELITAKNNGSVEEIVKKLIKDCTNAKDLNRQESELLLIHLWAHSLGEVNHENSWKCT CGTEIPTHINLLHTQIDAPEDLWYTLGDIKIKFRYPKIFDDKNIAHMIVSCIETIHANGESIPVEDLNEKELEDLYSIIT ESDIVAIKDMLLKPTVYLAVPIKCPECGKTHAHVIRGLKEFFE |
| 3070 |
PF12315 (DA1-like) |
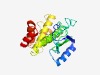 Estimated TM-score=0.58; hard target. |
>Protein DA1 (Length=212) GLNMQVKQQIPLLLVERQALNEAMEGEKNGHHHMPETRGLCLSEEQTVSTILRRPRIGAGNRIIEMITEPYRLTRRCEVT AILVLYGLPRLLTGSILAHEMMHAWLRLNGYRTLSQDVEEGICQVLAHMWLDSEIMSGIGSNVASTSSSSASSSSTSSKK GTRSQFERKLGEYFKHQIESDTSPAYGDGFRAGNEAVLKYGLRRTLDHIRMT |
| 3071 |
PF12309 (KBP_C) |
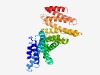 Estimated TM-score=0.58; hard target. |
>KIF-1 binding protein C terminal (Length=356) EWAINAATLSQFYINKLCFMEARHCLSAANVIFGQIGKIPATEETPEAEGDVPELYHQRKGEIARCWIKYCLTLMQNAEL SMQDNIGELDFDKQSELRALRKKELEEEERIRKKAVQFGAGELCDAISAVEEKVQYLRPLDFEEARELFLLGQHYVSEAK EFFQIDGYVTDHIEVVQDHSSLFKVLTFFETDMERRCKMHKRRIAMLEPLTVDLNPQYYLLVNRQIQFEIAHAYYDMMDL KVAIADKLRDPDSHIVKKINNLNKSALKYYQLFLDSLRDPNKVFPEHIGEDVLRPAMLAKFRVARLYGKIITADPKKELK NLAASLEHYKFIVDYCEKHPEAAQEIEVELELSKEM |
| 3072 |
PF12290 (DUF3802) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3802) (Length=111) MVTDKDGYMHLIQYLTEHLGLFESSQNNAAATETVMELFEEQLSAQIIMVCGQNPQLSFAQRNMVIREIDAIVYDLEEIL ASVSNQQATPEQSIFITEFSGLIKNLFDQEV |
| 3073 |
PF12247 (MKT1_N) |
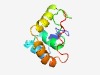 Estimated TM-score=0.58; hard target. |
>Temperature dependent protein affecting M2 dsRNA replication (Length=89) PNDIHEFIGTRLPEEIYFYLSRGIVGPHVINILTSGMLIENTPLDNRDSQEYRDFLKDIFPIQTQSLNLLTQPLHRFYQA KNVSAYYWF |
| 3074 |
PF12159 (DUF3593) |
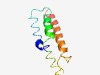 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3593) (Length=86) SQLFAFSLFPYIGFLYFITKSKSSPKLTLFGFYFLLAFVGATIPAGIYAKVKYGTSLSNVDWLHGGAESLLTLTNLFIVV GLRQAL |
| 3075 |
PF11966 (SSURE) |
 Estimated TM-score=0.58; hard target. |
>Fibronectin-binding repeat (Length=81) DNGTAKNPALPPLEGLTKGKYFYEVDLNGNTVGKQGQALIDQLRANGTQTYKATVKVYGNKDGKADLTNLVATKNVDINI N |
| 3076 |
PF11961 (DUF3475) |
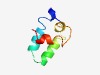 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF3475) (Length=57) ILAFEVANTIVKGANLMQSLSKESVSYLKEMVLPSEGVRRLISDDISELLRIAAADK |
| 3077 |
PF11956 (KCNQC3-Ank-G_bd) |
 Estimated TM-score=0.58; very hard target. |
>Ankyrin-G binding motif of KCNQ2-3 (Length=93) RHETALRDSDTSISIPSVDHEELERSFSGFSISQSKENLDGLHSCYAAAAPCATVRPYIAEGESDTDSDLCTPCGPPPRS ATGEGPFGDVGWA |
| 3078 |
PF11444 (DUF2895) |
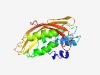 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF2895) (Length=201) NEITHLQAHVKTLRLAAGALFVVTLLLTFGWWSAPKNLTIHVPPDLRSGSTRRWWDVPPESVYTFAFYIFQQLNRWPTDG DADYPRNLHALSAYLTPACRAYLQQDYEYRRSNGELRHRVRGIYEIPGRGYGDDPATRVKVVSNNDWIVTLDVTADEYYG GDQVKRAFVRYPLKVVRMDVDPEHNPFGLALDCYARTPERI |
| 3079 |
PF11385 (DUF3189) |
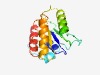 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3189) (Length=145) VIYHCFGGSHSSVTAAAIHLGLLSRHRLPTAAELLALPYFDGRSRGEEGDLKYMGTDAYGNKVYAVGKKNLGARFETFLY NLAAVIGIPRRNILLLNTSPLVNMSMRIGGFISRRMGLTFLGRPLVVWGTRRAFPRLGLFVAENR |
| 3080 |
PF11381 (DUF3185) |
 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF3185) (Length=58) KLVGIVLLVVGIILLYFGYQAYDSAASEVSQAVTGETTDEAMWYLIGGAVAVVIGLYG |
| 3081 |
PF11363 (DUF3164) |
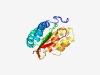 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF3164) (Length=193) YWKDADGNLKPEALVKEIDKERDELVRQFVSKAQALSKALGDFKQSVFDDVGAFVSLSAEKYGVKIGGAKGNITLFTYDG EYKLQLAVQENIRFDERIHAAKALIDECLHDWSEGAKPELKALIDNAFEVDKEGNLSTAKILSLRRVEIDDARWNQAMTA ISDSVQVIGSKGYIRFYKRDENGKYQPISLDMA |
| 3082 |
PF11345 (DUF3147) |
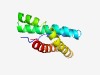 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3147) (Length=110) AILHFVIGGIAVALASIIADKVGGKLGGIIATMPAVFLAAIIALALDHRGTQLVEMSMNLSTGAIVGILSCILTVFLTSL YIKHKGYRKGTIFTVVCWFVISLAIFSIRH |
| 3083 |
PF11328 (DUF3130) |
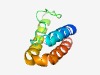 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF3130 (Length=82) MREIKVNEVTFQQHATKLASKSSGRYLPLKNGNMAYSRANSIDQLRSALIELVDVVEDFQHVTKQDAGRLKKMGIAYAKQ DX |
| 3084 |
PF11326 (DUF3128) |
 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF3128) (Length=73) RLSCSPCFDALWFCYSPVHQMQQYYRLGVLDNCSEKWSALVDCLTLKTKRSSQIQEILDTRERAKPPHIWTFR |
| 3085 |
PF11314 (DUF3117) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3117) (Length=50) LRPRTGDGPLEVTTEGQSIVMRIPLEGGGRLVVELTPDEAGALGDALTKV |
| 3086 |
PF11310 (DUF3113) |
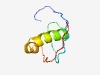 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3113) (Length=60) MQQQAYINATIDIRIPTEVEYHHFDDVDDEKDMLAKRLDDNPDELLKYDNITIRHAYIEV |
| 3087 |
PF11306 (DUF3108) |
 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF3108) (Length=234) TCFLSQAQKKDAAFETGEWFKFRIHYGFINASYATLEVKDDYLNGKSVYHVVGKGKTTGFASIFFKVDDNYESYFDKNSG KPYKFIRQIDEGGHTKDIEIDFNHEKEKALVNNKKHKTKEEINIEKNVQDMVSAFYYLRNNYDVSSIKEGDEVVLNMFFD KENFKFKLKFLGRETMRTKFGKVPCLKFRPYVQAGRVFKESESLTLWVSDDENKIPIRIKAKLVVGSLKADLDA |
| 3088 |
PF11256 (DUF3055) |
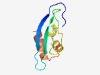 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3055) (Length=80) LYDDTENTKTRFVSFLGENHRFDFAIVQTNRHYGKHLVLDMQGNRFAIIGHDDLEEEGYLEHAFQLNEHDAEELRSFLSE |
| 3089 |
PF11237 (DUF3038) |
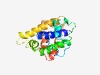 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3038) (Length=168) PRTRQQIDLILLAIEALELGGSEAMLFAAKELELQDIIKNRVVLWRLRCSNPLRRSYSRNPLALEEAKALVVIASHMARR MTVLIRQMLLAYQQLNEKQVPLENHFRLSEYLERFRAHFRSRMNPRRAKVAIYSSDDKLNELALSLLSQLLFCTGTSGVQ RFWISLFD |
| 3090 |
PF11210 (DUF2996) |
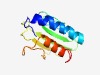 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2996) (Length=121) AVEDKPFAEFIQQDYLPALQTALTKQGVEDLDVSFAKQKIPVAGMSSAGDCWQVVGKFQSGQRQFNLYFPQENIQSQRAF SCAENNTKTSTLEPFLIDERKITLDLMVFGVVQRLNAQKWL |
| 3091 |
PF11201 (DUF2982) |
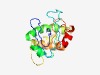 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF2982) (Length=150) FTLTASHFQQHLFKGGWVVRWRNIESIGICSYQQDGWHQPLPWIGIRLKDYSPYLDAICPRIATEILLSQRALLYLGARQ NHCEEKFEDMMLDPQPYTSKAGKQYDGLQAMLANRMKYQRKFYGYDVFISASDLDREADEFVGLTRRYLA |
| 3092 |
PF11153 (DUF2931) |
 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF2931) (Length=206) LLACTLALLSGCAASQAQPKLPFDAWYAGTFAPDHMEVWVESVDVIDRRGLAYERVNGGVPSYSSTIAGWPSHPAGGAGK GLHGIDLPEIIFVRWQSLVEPQTYNVRINIPEWIREEMLKPQAPYCRGEKRVVEGMYRRVITIGLAPGGIAKAWVGGPCL QSIEIGRFEAAVSKAGSSLGQNEGRYAYPLSEKAKAYIEQHGVPYG |
| 3093 |
PF11101 (DUF2884) |
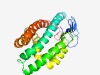 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2884) (Length=228) CEVSLNYDVTVEPKKLTVSDVGKEQYRIEMNKLFVNGEQISLNKQQQQLVSQYADEVSSQVPEVIELVNDAVAMASTAVS MALTPLLGDATGAKIDEMMDGLQDRVEDVAYQNGDKFYLGATESSLESAFGEEFEKEMEQLVQNSIGTMMMSLGSQMMSG DGDSFEAKMDAFSVKMDSIGEDIELQMEEQAQDLEARGERLCENFETLAALEKKLQAQIPELANYSLV |
| 3094 |
PF11087 (PRD1_DD) |
 Estimated TM-score=0.58; hard target. |
>PRD1 phage membrane DNA delivery (Length=54) MGEFGKTLITIVTAIIGVAIIAVIVSQRSNTAGVIQSATSGFSNILKSALAPII |
| 3095 |
PF10967 (DUF2769) |
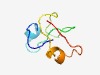 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2769) (Length=57) CICPKCPTYTNCAKEAKELLFCMKGKSFMCITNEKGCICPDCPVTKEYGLKNKFFCM |
| 3096 |
PF10925 (DUF2680) |
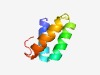 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF2680) (Length=56) LTPQQKQMISDYNEKIADLEKEFVNKLVDAGLITRQQADNIIKNIDERVSKANENN |
| 3097 |
PF10895 (DUF2715) |
 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF2715) (Length=168) FAAEVFVSPKVGQVGAHPWGKEPASRTEVLPYTPTLGLAVGVIAHNGFAFTTELDAGLSSLMFRAQFLVGWAIRSGKNFA FIPSLGVDVLSTRDDKLVGIPINLDFLIFFTKHVGLDFTFGSGVNVPLTKNLKKGANGNGQDLTWSSIWTGYCCTSLVMV KIGPVFRI |
| 3098 |
PF10835 (DUF2573) |
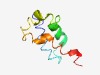 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2573) (Length=77) QKLQEQLDGLLEKYTELLLGETNDELKEEVKQWILYTHIAKSMPPLAKHWNATYPDAKQGIKEIIQHIKELNEAHRN |
| 3099 |
PF10834 (DUF2560) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2560) (Length=72) IPMTEEHKFQLEIYKLVMNQNAAAEEAFQFIGTDELKLELFKIHFQSGGANSDITTRTIEAVRKSKEALDLF |
| 3100 |
PF10656 (DUF2483) |
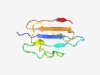 Estimated TM-score=0.58; very hard target. |
>Hypothetical protein of unknown function (DUF2483) (Length=72) MKQTVTYLIKHKDENLFITNRPTEVNDTVKYSTDMRDAREFDGLDKTVIDMSKHKAIKKTVTETIEYEKVEH |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218