

Go to page (83 pages in total): [First page] << < 21 22 23 24 25 26 27 28 29 30 31 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 2501 |
PF10903 (DUF2691) |
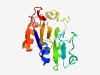 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF2691) (Length=153) KRGVLFEIPNGYGRFLLDILKPVEPADFNWLLADGECYLVENGELSDENLFPQDPQVMTGEAFKKRLEDARYYLIFADLK AFPKDGHPEDVSTYESYMKSSCELIVLTADSSFVTIYCKDQKNLELLYQNAKDCGFKRIEYITDHNDARTGMH |
| 2502 |
PF10876 (Phage_TAC_9) |
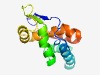 Estimated TM-score=0.61; easy target. |
>Phage tail assemb.y chaperone protein, TAC (Length=133) FEIEGLTYVMTPANAMAAWQSLKRAGVLLRGMDADALANAQGAASVALGTLLSHLGDPAVTEIEALVFEQTAIKTPEGTT YRLSPDRLNEHFNTRRTHLLRVLMEGVKYQYSDFFVGGMAAFQDLIPMPSAEQ |
| 2503 |
PF10844 (DUF2577) |
 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF2577) (Length=106) IKSAAMGAVNDSKPMSVLFGKVVSTSPVKIFVEQKLTLNEAQLIFTNNVRDYDLEMTVEHFTEESNSHVHPYKGKKKFRV HNGLAAGDEVIMLQMQGGQKFVVLDK |
| 2504 |
PF10748 (HofP) |
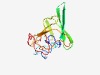 Estimated TM-score=0.61; easy target. |
>DNA utilization proteins HofP (Length=132) TKRWLLVCSCLLMLTGMRDPFKPPEDRCRIAELPMWRYQGVVSQGERLTGILKDSQQKWRRVEQYQVLENGWTISRLTAD SITLETGKGCEPSQWRWQRQGEVNEAMDSRSVDRRDARRTGGESAKRDAGGG |
| 2505 |
PF10735 (DUF2526) |
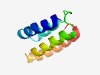 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF2526) (Length=80) MSHFDDVKVQVDAALQENVIAHMNELLVALSDDDQLSREQRYQQQQRLRVAVAHHGKSEREDARAPEDERREHLTRGGKI |
| 2506 |
PF10710 (DUF2512) |
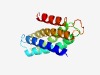 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF2512) (Length=139) MDHIKALAIKFVAVAVVLYSILGIFYTATLGEIFMISLLVTGVAYVVGDLFILPRFGNLTATIADFGLSFVAVWFLSAML IEGDFAFLSAALFSALFITAIESLFHMYMQRHVLDEQQDNKEEKNRMTTQQLQTEFAEE |
| 2507 |
PF10704 (DUF2508) |
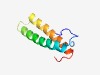 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF2508) (Length=70) MFFRRKGWLRSEFDEKLLEQLNRLKTDWNNQKSLVEKSFDPSPEAISQAKLAEAKYFFLFREAKKRNVRV |
| 2508 |
PF10689 (DUF2496) |
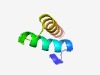 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF2496) (Length=43) SLENAPPEVKLAVDLIMLLEENQIEPRIALAALEIVRTDFEKK |
| 2509 |
PF10675 (DUF2489) |
 Estimated TM-score=0.61; very hard target. |
>Protein of unknown function (DUF2489) (Length=133) ILGLACYAGYLLLQLKKQKELQQQHQKLAIEKRNANIFDNVNILCMAGIQGQCDLSEISIRIYCIMDYVQGEQRVDFDAE YPAIAELYHIVKDMPRGEARQALEKKERMKQNLERHKAEGRLSEAIVEELKAL |
| 2510 |
PF10668 (Phage_terminase) |
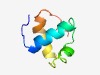 Estimated TM-score=0.61; easy target. |
>Phage terminase small subunit (Length=59) MPRQRSPDRDKAYEIFKEHGGNIQNRKIANILDIPEKTISGWKVKDKWNEKLNGVLQTK |
| 2511 |
PF10620 (MdcG) |
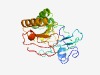 Estimated TM-score=0.61; hard target. |
>Phosphoribosyl-dephospho-CoA transferase MdcG (Length=200) RHALVWLRADAPSQALTPGAQPRLQAWFAAGFPAVVARRDGAEQPGQVRLGVPLPPAQGKQRIALCAQVSDIARSVPALA LPEVISHAPPQWQAALHALQAQATAIGMQPRVFGSFAFQAVTGLPYVHAASDVDLLWTLDTPTQAHALVTLLQQWEHVTA RRADGELLLPDGNAVNWREYAGDAQQVLVKRNDGCRLLPR |
| 2512 |
PF10601 (zf-LITAF-like) |
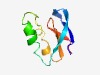 Estimated TM-score=0.61; hard target. |
>LITAF-like zinc ribbon domain (Length=69) EPVTTTCPHCGSNITTTVNQINGFAPWIVCGGMALIGCMLGCCLIPLCINTLKDTEHCCPSCQQVIGRH |
| 2513 |
PF10534 (CRIC_ras_sig) |
 Estimated TM-score=0.61; hard target. |
>Connector enhancer of kinase suppressor of ras (Length=93) NLRTLSHKLNASAKNLQNFILGRRRGGHYDGRASRRLPNDFLTSVVDLIGAAKNLLAWLDRSPFASVTEYSLLKNNIVQL CLELTTIVQQDCT |
| 2514 |
PF10336 (DUF2420) |
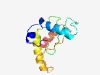 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF2420) (Length=109) VCEDESMLHQNCGTLMKVLREFMENYYGKLHFVTKELMMEIPCLELTLCEDNIYNEKITFNDILSIFEILRERSIERGEN GIPDHLNVQIAVRPRFVSRYNSLVELTES |
| 2515 |
PF10233 (Cg6151-P) |
 Estimated TM-score=0.61; hard target. |
>Uncharacterized conserved protein CG6151-P (Length=113) TGVVCIVLCFALGIANIFSFNAVRIIFSVLCLVSSFILIFIEVPFLLRICPTSSKFDEFIRRFTTNWMRAAMYTIMSVVQ WLSLLSGASSLIAAAVFLILAALFYALAGLKSQ |
| 2516 |
PF10180 (DUF2373) |
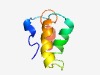 Estimated TM-score=0.61; hard target. |
>Uncharacterised conserved protein (DUF2373) (Length=62) EYLTCWSERREEWKFHKSRQIWLLQHMYDTTKVTDDHFKGLLTYLEGLRGASRGITLQKAEA |
| 2517 |
PF10081 (Abhydrolase_9) |
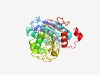 Estimated TM-score=0.61; hard target. |
>Alpha/beta-hydrolase family (Length=288) IRVYVGLDTAPTPEERIDLVLAELERTGAFDRSTLVLIPTTGTGWVNPTAARAMELVHNGDLAMVAWQYSYLPSWISFLA DREKAAAAGKMLVERVHERWSQLPEDDRPDLFLYGESLGTQSGEGAFDSLAEIRAQMDGVLWVGPPNSNRLWRQLVERRD PGTTEVQPVYADGLVVRFADSREAIHEPETPWLSPRVLYIQHASDPVVWWSPDLLFSRPDWLSEPPGRDRLPQMRWFPFV TFWQVSADLTNAAGVPDGHGHNYGSAVLEGWIAVTQPEGWTENDTERV |
| 2518 |
PF10058 (zinc_ribbon_10) |
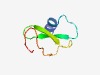 Estimated TM-score=0.61; hard target. |
>Predicted integral membrane zinc-ribbon metal-binding protein (Length=50) LDRIVDFIVGDSPKDRFGMICKACHAHNGMLPKEEYEYTTFRCAFCNVLN |
| 2519 |
PF10050 (DUF2284) |
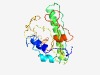 Estimated TM-score=0.61; very hard target. |
>Predicted metal-binding protein (DUF2284) (Length=178) PISTDLIVVADWVRLKCRYGCRAYGKHLCCPPFAPSPEEMRRVVSEYKTAILARFLAEPGPGNDPRHIHHYLWDSINNLH DTVYQLETRAFLLGYYKALGFYALPCTLCETCVGEEMQESGEKMNPLDVLKCRHKDRMRPSMEGCGVDVFKTLENAGYSP QVIRSYSERVVLFGMVLL |
| 2520 |
PF10043 (DUF2279) |
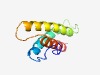 Estimated TM-score=0.61; hard target. |
>Predicted periplasmic lipoprotein (DUF2279) (Length=90) DEGWFGKDTINLGMDKLTHAWNTYWITDMIEARIRRKTGATNAGLTAASLSMGVMLYAELWDAHKKSSGFSFQDIAMNAS GAGFSLLRNA |
| 2521 |
PF09931 (DUF2163) |
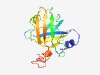 Estimated TM-score=0.61; hard target. |
>Uncharacterized conserved protein (DUF2163) (Length=163) MKTLPAGMQAHLDTGTTTLAWCWKLTRTDAQVMGFTDHDRPIQFDGVIYEAASGFSASEIASSLGLSVDNLDVDGALSSA AITEADIRAGLYDGAEVEIWRVNWADPDQRVLMRRGTIGEVIRGELAFTAELRGLAQALDQTQGRTYQRICDADLGDARC KVD |
| 2522 |
PF09888 (DUF2115) |
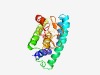 Estimated TM-score=0.61; hard target. |
>Uncharacterized protein conserved in archaea (DUF2115) (Length=172) TKGDLGLILGEEVSRYSLADLQIIGGRLHAEMVRLPEPYRKQVHPYITEQLFGAHHQLLSMYRSGRFLSMTEPITDCATF VEFCNMIPDGCLRWDESTQRLPFRYTPRHRAFYYLISAFTIFVLDRPGHPVGTPFPGGFRVEERNGSYYCIIRDREKDVP FSICNFCPALQT |
| 2523 |
PF09862 (DUF2089) |
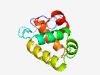 Estimated TM-score=0.61; easy target. |
>Protein of unknown function (DUF2089) (Length=115) CPVCSHRLTITKLACDHCQTKIEGEFSPCKFCRLPNDQLQFLEAFLKCRGNIKDVEKELGISYPTVRNRLDGLLQALGYQ VDKEENRQENLSRQEIVRALEKGEITPEQATQQLR |
| 2524 |
PF09858 (DUF2085) |
 Estimated TM-score=0.61; hard target. |
>Predicted membrane protein (DUF2085) (Length=90) CHRMPERSFFIKGHQFPVCARCTGFYTGLMIYLIVNLFYKHPYDWNMLFISMVLMVPVAIDGFTQYFGPRESTNTLRFIT GFIGGIGLII |
| 2525 |
PF09855 (zinc_ribbon_13) |
 Estimated TM-score=0.61; easy target. |
>Nucleic-acid-binding protein containing Zn-ribbon domain (DUF2082) (Length=62) CPKCGNTEYESDQFQATGGNFSKIFDVQNKKFITISCKKCGYTELYKAQSSAGWNILDFLIG |
| 2526 |
PF09838 (DUF2065) |
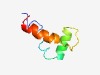 Estimated TM-score=0.61; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2065) (Length=52) IAAVGVMMLLEGLPLLLAPEQWRRYVNYILTLAPGQLRFIGLAMLLLAVVLV |
| 2527 |
PF09835 (DUF2062) |
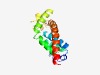 Estimated TM-score=0.61; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2062) (Length=143) LRFLGRLLHDANLWHLNRHSVARAMAVGLFAAFMPIPLQMLLAAALAILVRGNMPIAVSLVWLTNPITMPPVFYCTYKVG SWLLGQPPRQLPDSYSWDWVMSQMSTLWQPFLLGSVVTGVVLGALAYFLTMLYWRWWVSRQWR |
| 2528 |
PF09821 (AAA_assoc_C) |
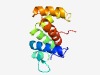 Estimated TM-score=0.61; hard target. |
>C-terminal AAA-associated domain (Length=118) VIATSLHMEIDDLFPVAETLQMLRFAEVEGGDIKLTPEGRRFAETDTNERKHLFAHHLLTHVALAAHIRRVLDERPSHRA PWSRFSDELEDHMAADDAEQTLRAVISWGRYAEVFAYD |
| 2529 |
PF09801 (SYS1) |
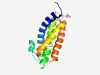 Estimated TM-score=0.61; hard target. |
>Integral membrane protein S linking to the trans Golgi network (Length=143) MAELPPLRIATQIATLQALYYVGAIILMLFTALVAGISFTLDVVFGWDAVRGDTTQGWLCGFVLVLDGGFCMAIAIILLV SRSKLVPDFAVTIHALHLVITTLYTGRVPRNAMWWACMMASAGLCAALGVWGSRYRELRPIYF |
| 2530 |
PF09493 (DUF2389) |
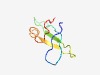 Estimated TM-score=0.61; hard target. |
>Tryptophan-rich protein (DUF2389) (Length=59) SKWTATQKTWGWRHFQVVNRKNQGQLVFAELVASCDPTVRFWLNAKQLKDRNLWEPGWQ |
| 2531 |
PF09317 (DUF1974) |
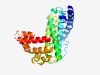 Estimated TM-score=0.61; hard target. |
>Domain of unknown function (DUF1974) (Length=279) IRCHPYVLEEMAAAQNNDVDAFDKLLFKHIGHVGSNKVRSFWLGLTNGLTSRSPTRDATKRYYQHLNRLSANLALLSDVS MAVLGGSLKRRERISARLGDVLSQLYLASAVLKRYDDEGRNEADLPLLHWGVQDALHQAEQAIDDLLRNFPNAAVAGLLR VVIFPLGRRYDAPSDRLDHKVAQILQTPSATRSRIGRGQYLTPSEHNPVGLLEEALQDVIAAEPIHERICKALGKNLSFT RLDLLAKQALEKGQITQEEADILVKAEESRLRSINVDEF |
| 2532 |
PF09207 (Yeast-kill-tox) |
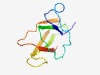 Estimated TM-score=0.61; trivial target. |
>Yeast killer toxin (Length=87) DGYLIMCKNCDPNTGSCDWKQNWNTCVGIGANVHWMVTGGSTDGKQGCATIWEGSGCVGRSTTMCCPANTCCNINTGFYI RSYRRVE |
| 2533 |
PF08878 (DUF1837) |
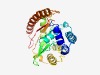 Estimated TM-score=0.61; hard target. |
>Domain of unknown function (DUF1837) (Length=244) KDLARAVAARLIDYAIPRSEIMEAQEKDQKSNTTVNTVELKMKAKQLFTSLKKSGEGGEMLLYMLVQAFLGLPQLLCKMP LKTSSEMHYHGADGIHIGYDRNTKKLALYWGESKLYKSFDSALKNCLDSLKPFLFDDGGTQASQNRDLQLITDNLDLCNE ELEDAILDFLNPNSPSFKKMQYRGVCLIGFDNGAYPLVPNSKDEDSVLNEIKDELGRWEKKLEKQIKNRTPLDSFILEVF LVPF |
| 2534 |
PF08747 (DUF1788) |
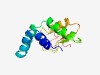 Estimated TM-score=0.61; hard target. |
>Domain of unknown function (DUF1788) (Length=122) EFDLYKMLIDITKQKRIFDRIFEMEERQGKDQLFRAMTTFAKPEVFLEKIKEKITMDDYNVVFLTGIGKVYPFVRSHNIL NNLQEVLDNVPVIMFFPGKYDGQSLQLFGKFKDDNYYRAFRL |
| 2535 |
PF08200 (Phage_1_1) |
 Estimated TM-score=0.61; hard target. |
>Bacteriophage 1.1 Protein (Length=46) MRTNFEPITKRNNVINEHGTEWQERKDHMKKRHKTQRGNSEKRNWK |
| 2536 |
PF08186 (Wound_ind) |
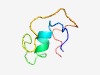 Estimated TM-score=0.61; hard target. |
>Wound-inducible basic protein family (Length=45) MIYDVNSTLFRSFLRVSAAHGTGKEGKARAGPGEKKSDNKPVMND |
| 2537 |
PF08170 (POPLD) |
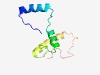 Estimated TM-score=0.61; trivial target. |
>POPLD (NUC188) domain (Length=92) GWTLIVPSGWAMPFWMSLTHTGTRVGGQRERQTQVFEAGVACFPFDYPFTSSYEQQVNATAAEEQENWEKKPPAKRVNYD ALGTRSPWKPDW |
| 2538 |
PF08056 (Trp_leader2) |
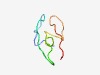 Estimated TM-score=0.61; very hard target. |
>Tryptophan operon leader peptide (Length=41) MLQEFNQNQKAKVAVCLNKTNSTDLAWWRTWTSSWWANVYF |
| 2539 |
PF07841 (DM4_12) |
 Estimated TM-score=0.61; hard target. |
>DM4/DM12 family (Length=83) RTYFYDQLEERMELYGFNASGCMERLICEVSELPLHEHNGVFGDVLSVIFRPSSSVREALPVSYYEAESRGSIEGCQQYR AFC |
| 2540 |
PF07613 (DUF1576) |
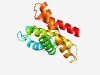 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1576) (Length=175) AKVFSGLYNILTQTDILITDYIGVGGIGAAFVNAGTLTLMFILLLHVLKVTFNGASIASLFLIAGFGLFGKNLFNVWFIV LGVYFYSRVQGDKFSKHIYVSLFGTALAPIVSEIMFNYSLPVGYRAPLAVLVGVSVGFVLPPLATYLLRVHQGFNLYNIG FTAGIVGTIFVSVFK |
| 2541 |
PF07552 (Coat_X) |
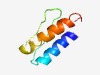 Estimated TM-score=0.61; hard target. |
>Spore Coat Protein X and V domain (Length=58) DDVVQEGTQVSSNFQLNEELIFIKDSCDVTVTTTDTKAALNLQAALQAAIALVVSISI |
| 2542 |
PF07291 (MauE) |
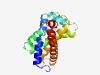 Estimated TM-score=0.61; hard target. |
>Methylamine utilisation protein MauE (Length=184) MALLAEPVVTTFVRAFLILLLASAAIPKLRHGEEFFGVVRNFRLMPEWLARPFALVLPWLELGIAVGLVLPVTAPLAAGL AGGLMVLFGIAIAINVARGRTAIDCGCFRNGMKQKLSWLLVGRNAGLALAAFGLAWLLPVAPAAGPFDLAIGFAAAGLTM LLIYGASLLSGLQSGARSSQLSKG |
| 2543 |
PF07179 (SseB) |
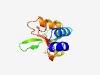 Estimated TM-score=0.61; hard target. |
>SseB protein N-terminal domain (Length=125) LVDAMVAFKANMNKSTEKAMIEAVKNAKLIAPVIMDDLPDDLEPGQEYQANAKFMMVQQKNGKRFFPAFTEWLELLKWKN DPDCKTVTLSFDQYCALILRQNGEASGFVINPTEHNMVFTKEKLA |
| 2544 |
PF07176 (DUF1400) |
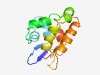 Estimated TM-score=0.61; hard target. |
>Alpha/beta hydrolase of unknown function (DUF1400) (Length=119) AAETVLLQYRDLRLSVSLAEFQTFAADGNASPALQDFLQQTEEDPEAVRQWLTTEISPGFSSVLPQEFILLQISKTLGEP LEREDLGALGTALRAAFENDQAFSVMEVLEQYPGPNVRL |
| 2545 |
PF07099 (DUF1361) |
 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1361) (Length=165) WNLFLAFIPLALSVWLFRRSSGNRNLLWWVGFIVFMAFLPNAPYLLTDIIHLIDAVRRDYSVWVITLVLIPQHLSVILLG FEAYVISLINLGYYLQKQGMGKFTIPAELITHALCAVGIYLGRFLRFNSWDLVTQPEDLAQTIVDDLTAKRPLLVMFVTF IVLTV |
| 2546 |
PF07040 (DUF1326) |
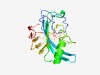 Estimated TM-score=0.61; very hard target. |
>Protein of unknown function (DUF1326) (Length=184) ENCNCDVVCPCLLSTNAQLTSKPTQGVCDVALVFHIDQGNYGDVRLDDLNVAMIAHTPGPMAEGNWTAAAYIDERADDRQ MEALGAIFTGAAGGPMAAFAPMIGNNLGAKKTPIRYQVDGKKRSAEIPGVMHMAVEPLPTMREDGEMWAATGHPVNPDRL TMAMGVQGSTFSDHGMNWDNSGRN |
| 2547 |
PF07027 (DUF1318) |
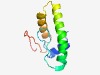 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1318) (Length=89) AFALELDQAKTQGLVGEQPNGYLGVVKATPEAVQLVSDVNEKRRQAYERIARQNGITLEQVANLAGQKAIEKTSGGEYVK TPDGQWIKK |
| 2548 |
PF06980 (DUF1302) |
 Estimated TM-score=0.61; easy target. |
>Protein of unknown function (DUF1302) (Length=597) SFNIGEIEGQFDSSLSIGASWSTANPNKNLIGVNNGGKGLSQTSDDGHLNFKKGETFSKIFKGIHDLELKYGDTGVFVRG KYWYDFELKDENREFKDISDSNRKEGAKSSGAELLDAFVYHNYSIGDQPGSVRFGKQVVSWGESTFIGGGINSINPIDVS AFRRPGAEIKEGLIPVNMFYVSQSLTDNLSAEAFYQIEWDQTVVDNCGTFFSQPDIIADGCSDNLRVLNSSRALSPQALA GLSAFGVDVNSEGVKVPRGADRDARDSGQFGVAFRYMFEPLDTEFGAYFMNYHSRAPIFSATRAPQALFNLVNNPLASPA VRALAPLIIAGNSQYFVEYPEDIRLYGLSFSTTLPTGTAWSGELSYRPNAPVQLNSTDILFAGVRPIDRAPYNNASVLTG TPGQDLHGYRRKEVTQFQTTFTHFFDQVMGASRLTVVGEVGVTHVGGLESNSKARYGRDPVYGPGPLPNGTCATLNGATI VGSGLGSSTANLDRNCENDGFTTSTSWGYRARAIWDYNDVFAGVNLKPSVAWSHDVSGYSPGPGGNFEEGRKAVSLGLDA EYQNTYTASLSYTNFFDGKYTTVDDRDFVALSFGVNF |
| 2549 |
PF06900 (DUF1270) |
 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1270) (Length=53) MSDTYKSYLVAVLCFTVLAIVLMPLLYFTTAWSIAGFASIATFIFYKEYFYEE |
| 2550 |
PF06891 (P2_Phage_GpR) |
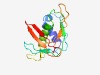 Estimated TM-score=0.61; very hard target. |
>P2 phage tail completion protein R (GpR) (Length=130) MMKPASLRDALTASVPHLAANPDALHVFVDEGNVVGTGARSLSFEYRYTLTLIVTDYPASPDTIVVPVLAWLRTNQPELF TNDEKRRDGFKFEVEILNHCTVDLAIKLKLTERVTVKPAGGGYQVEHHPE |
| 2551 |
PF06889 (DUF1266) |
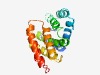 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1266) (Length=174) GHYAVQGPEGLAEFIDYLLEVGDRQEYQINYAPYTLNPARLASEIATLESDECGEEERNHLLRLKRVRANEDGCNDLDLT AWDLAQAVDLAIAGRQLDWLSEEAFFARLRRAHALAASHYGGWEEYARGLYAGFSFFMGETPEREAFLGGFRQALASWLA AAPPLAGPWATLDF |
| 2552 |
PF06853 (DUF1249) |
 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1249) (Length=119) RLMRLLPDMRSGCETRRIAISQGDQLLGVLVLEVLESCPYTTTVRVSQEHCLSWLPVPKMEVRVYHDARMAEVVRAENAR RFRGIYHYPNAQMHQPDEKNQLNLFLGEWLGHCLACGHE |
| 2553 |
PF06645 (SPC12) |
 Estimated TM-score=0.61; hard target. |
>Microsomal signal peptidase 12 kDa subunit (SPC12) (Length=70) IDFEGQRLAENLSTGLLSIVGVVAFLVGYITQNITYTLWIGLAGTALTFLVVVPPWPFFNRHPVKWLPST |
| 2554 |
PF06477 (DUF1091) |
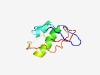 Estimated TM-score=0.61; easy target. |
>Protein of unknown function (DUF1091) (Length=81) LTIRSKDGEFQRTLYNNTLDLCEFLAKPFRHRILQFVQVEIMQNSNMPTKCPILAGSYYIRNATFARARVPSFMPPSYFR V |
| 2555 |
PF06476 (DUF1090) |
 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1090) (Length=108) MSTPLLAAEDAQPLTGCAAKRQAISSQIEQAKAHGNSYQQAGLEKALAEVTANCTDAGLKKERENKVLDAKHDVSRRQAD LDKAMKKGDPDKINKRKEKLAEARKELQ |
| 2556 |
PF06419 (COG6) |
 Estimated TM-score=0.61; easy target. |
>Conserved oligomeric complex COG6 (Length=659) LNTLSTFYTDNTPQARRNLRSTIEKRSLQINLDFLRASQAAQLALDRVEDEVNSLADCCDRIAKALSSCSAATGDIINTT ERLKQELEVTTQKQEIVSCFLRDYQLSPQEINALRDEELNENFFKALSHVQEIHANCKILLRTHHQRAGLELMDMMAMYQ EGAYERLCRWVQAECRKLGDTDNPEVSELLKTAVQCLKERPVLFKYCAEEVANMRHNALFRRFISALTRGGPGGMPRPIE VHAHDPLRYVGDMLGWLHQALASERELVLALLDPDALIDAGSTANRFSKNVENDSGKIEADLTFVLDRIFEGVCRPFKVR VEQVLQSQPSLIISYKLSNTLEFYSYTISDLLGRQTSLCNTLWALKDAAQKTFFEILKSRGEKLLRYPPLVAVDLSPPPA VREGVSVLLEIIETYNSMMVPASGKKPAFDPVISALLDPIIQMCEQAAEAHKSKGAGHSSRRSRMSSDSGQLSKSAVDAI LSNNNSATFSQNSEAPSKIFLINCLCAIQQPLLGHEVAAEYAKKLGTMIDNHVHDLVEKEVDRVLRRCGLSMKMHHFHNS LNKDTTATAPLAELEDTSPASLSECLKAFFGFILGSESSLPEFEQMQVPKLRSDACIQVARSLADAYELIYNAIMDPENS YPDPKSLARHPPDQIRTIL |
| 2557 |
PF06271 (RDD) |
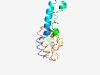 Estimated TM-score=0.61; hard target. |
>RDD family (Length=136) LASRWSRLWASIIDAFVYAIFIVPVFLYGGLYDKLLSNETIELTEQVALTVYSWVVYFLCQGYLLHKKGQTIGKNLMDIA IVDMEGKQIGLLNIVGKRIIPMVAVAYIPFIGPFISTFDYLFVFRKDKRCLHDLIA |
| 2558 |
PF06237 (DUF1011) |
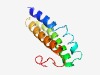 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF1011) (Length=97) RNVYLLALLAASNALTNGVLPSVQSFSCLPYNNMTFHLSVVLGNIANPLACFLAMFVVLRSPVGLGLVSLAAAVFAAYLL ALAALSPCPPLQASLTG |
| 2559 |
PF06233 (Usg) |
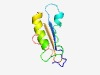 Estimated TM-score=0.61; very hard target. |
>Usg-like family (Length=78) QMDGYGLTTAEIHYHLPDHPSLLQLYVWQDYDLAPEFPELKGFLDYWERELEGPLHSVRVAHHRLIKPSEWRAVDGII |
| 2560 |
PF06062 (UPF0231) |
 Estimated TM-score=0.61; very hard target. |
>Uncharacterised protein family (UPF0231) (Length=117) MDYEFQRDLTGGILARFSMDHEAIGYWLNDEIQGDISLIDQILTEMDEIKGSEKQWQLIGKEYSLSIDDEEIMVRANTLH FETDDLEEGMSYYDNESIAFCGLDDFATMLQKYRLFI |
| 2561 |
PF05926 (Phage_GPL) |
 Estimated TM-score=0.61; hard target. |
>Phage head completion protein (GPL) (Length=139) ITNDAFYPDVSLEHARDTMRLDGTVTDARLRHELLAAIASVNDELRAARSAWREAGITRLADVPADQLDGESVLLQHYRR AVYCLAKATLIERYRDYDTTGDGARRADELEPQSDELRRDARWAISDIVGRPRVTVELI |
| 2562 |
PF05875 (Ceramidase) |
 Estimated TM-score=0.61; easy target. |
>Ceramidase (Length=286) GYWSPVTSTLNWCEEDYYATIYSAEIVNTLTNLLFVWLAFRGINNCIQNGHDRIFLVTFFGYLVVGTGSFLFHSTLKYPM QLVDELSMIYTTCLMFYATFSHKQTGPIRVLLAVFLVALSIFITAYYHYLGDPVFHQNMYALLTAIVLFRSMYVMERDIR PKPREREAAPGQSLISDKEQQRRDDRDRKILKTMWLMIACGLSIFLGGFGIWNLDNMYCSRLRKWRHEIGLPWGIFLEGH GWWHLMTGTGAYFYIVWGVWLRHCLNGRQEEYKLVWPSVFTSLPSV |
| 2563 |
PF05862 (IceA2) |
 Estimated TM-score=0.61; easy target. |
>Helicobacter pylori IceA2 protein (Length=55) MAVVIKVVNGKIQEYENGIHKRTYGSNIVAADTDGHIVAAVTAKGRIEEYENGIH |
| 2564 |
PF05800 (GvpO) |
 Estimated TM-score=0.61; easy target. |
>Gas vesicle synthesis protein GvpO (Length=93) QSSQSSQEPRRPKPMEVLREARAQLAELTGMTAESVSSFERTEDGWVLEVEVLELERVPDTMSLMASYQVELDAEGQLTG YRRVRRYERGRAD |
| 2565 |
PF05753 (TRAP_beta) |
 Estimated TM-score=0.61; hard target. |
>Translocon-associated protein beta (TRAPB) (Length=172) FALSQLSVANEDNSARLLVSKQILNKYLVENRDIVVKYTLYNVGSGAAVNVQLVDNGFHPEAFTVVGGQLSATIDRIAPQ TNVTHIAVVRPKAYGYFNFTAAEVNYQPAEEVTELQFAVSSEPGEGAIVAEAQFNKQFSSHYLDWFAFAIMTLPSLAIPY GLWFSSKSKYEL |
| 2566 |
PF05647 (Epiglycanin_TR) |
 Estimated TM-score=0.61; very hard target. |
>Tandem-repeating region of mucin, epiglycanin-like (Length=62) HSSTVTNSASSETPCNSETQTSDGTFTMTVSSDSTTAGPSSTATNSASSETPCNSETQTSDG |
| 2567 |
PF05580 (Peptidase_S55) |
 Estimated TM-score=0.61; easy target. |
>SpoIVB peptidase S55 (Length=209) CYRLGIWVRDKTAGIGTLTFYDENTGIFGALGHGITDIDTGELLKIENGKIMSARISEIEQGSKGSPGEIKGAFYETQDA LGDITSNTEFGIYGKLYKKNLDNNKSKPLPVGFQEEVKEGKAYILTTINDNKVQKYEIEIVKAQNQPFPEQKSMIIRITD KKLLQKTGGIVQGMSGSPIIQDGKLIGAVTHVFVNDPTKGYGIYIDWML |
| 2568 |
PF05437 (AzlD) |
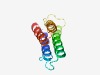 Estimated TM-score=0.61; hard target. |
>Branched-chain amino acid transport protein (AzlD) (Length=99) VFLVIVGTAIVTFIPRVLPLMVLSRFQLPEWATRWLSYVPISVMAALVAQEIFKHDEKISFSTNNVELLAAIPTFFIAIK TRSLLVTVLVGIIVVMVLR |
| 2569 |
PF05037 (DUF669) |
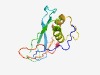 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF669) (Length=130) FKVDYNQVSEFEEFKKGEYEVTVVGYEMTQAKTGSNMVVLTYEVRSDVEQPCKGQKINYDNFVVSDKSMWRFHALSKAVG VPEGTPFESYTEWAKTMQNKPLRVVVGLREQNGRNYPQVNGFKPSEVGKP |
| 2570 |
PF05007 (Mannosyl_trans) |
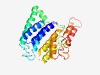 Estimated TM-score=0.61; easy target. |
>Mannosyltransferase (PIG-M) (Length=259) ISARGNADVLVCAAVLYTLYLLKKGQWVAAAFAHGALAVHLKIYPLIYLPSIFLYLCQFSFAGGLVASVKKLFSNWKGFT FVAISLGSFGVIVAFFYYIYGDLFLEEFLLYHIKRRDIKHNFSPYFYPLALVDQNVTLSKIIGFLAFLPQAYLVVYFAFK YYRDLPFCWFISTFAFVTFNKVCTSQYFVWYIVFLPLIVARIKMTTNEAARLILMWFASQAVWLFFAYLYEFQGWQTLEL VFVASVGFLLTNIYVMVNV |
| 2571 |
PF04971 (Phage_holin_2_1) |
 Estimated TM-score=0.61; hard target. |
>Bacteriophage P21 holin S (Length=63) DKLTTGVAYGTSAGSAGYWFLQLLDKVTPSQWAAIGVLGSLVFGLLTYLTNLYFKIKEDKRKA |
| 2572 |
PF04923 (Ninjurin) |
 Estimated TM-score=0.61; hard target. |
>Ninjurin (Length=102) INVYQQKKSLAQGMMDLALLSANANQLRYVLESCQNHPFFYVNLVFVSTSIFVQVVVGFGLIWKSQYNMNNPDDYRAATR INNLVSIGIFIITLVNVMLSAF |
| 2573 |
PF04887 (Pox_M2) |
 Estimated TM-score=0.61; easy target. |
>Poxvirus M2 protein (Length=196) CPPRQDYRYWYFAAELTIGVNYDINSTIIGECHMSESYIDRNANIVLTGYGLEINMTIMDTDQRFVAAAEGVGKDNKLSV LLFTTQRLDKVHHNISVTITCMEMNCGTTKYDSDLPESIHKSSSCDITINGSCVTCVNLETDPTKINPHYLHPKDKYLYH NSEYSMRGSYGVTFIDELNQCLLDIKKLSYDICYRE |
| 2574 |
PF04841 (Vps16_N) |
 Estimated TM-score=0.61; easy target. |
>Vps16, N-terminal region (Length=416) NTGEWFKVRPDYYRKVELATPDWPLDLDLEYMWVEVAPYGGPLAVTRDPTKLVPVKGNARPMIRIFDTTGREMGHILWNH GKLIAMGWSDMEELICIQENATVFVYDMFGREKESYSIGDEASVTKIVEGKVFQSSAGTGVAVMTTSGRVFLKQNSSKTE RKLPDIPNSSMNCSCWEIVTEGRNSYCLLGRDREVIKLFPGETVGTITANLFEKPHERIIKISVSYNHQHLALYTNTGLL WLGSVDMRQKYCEFDTGRKDMPLQMEWIMNSDNSEADAVVISYPSYLLIVNRNADRSDFPYDPVMFLVAEMDGVRIITQS SHEIIQRLPKCVENIFAVNSQEPASYLFEAQKKFEEKSYKSDEYLSMCREKIDLAVSECIEAASYEFCPETQKSLLRTAY FGKGFIRNHNPDEYMR |
| 2575 |
PF04802 (SMK-1) |
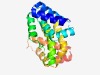 Estimated TM-score=0.61; hard target. |
>Component of IIS longevity pathway SMK-1 (Length=188) KFFDNLMNVFEMCEDLEKLDCLHMMFNIVKGIISLNSHQILEKILGDSLIMKVIGSLEYDPDAPQSLHYRNDVTAHVVFK EAIPIKNPMVLSKIHQTYRIGYLKDVILSRVVDDATATSLDSIINANKAAIVTLLKDDSNFFQELFARLRSPSTSVESKS DLVHFLHEFCSLSKSLEMELKLRLFKEL |
| 2576 |
PF04754 (Transposase_31) |
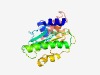 Estimated TM-score=0.61; hard target. |
>Putative transposase, YhgA-like (Length=202) TPHDAVFKQFLYHPDTARDFLDIYLPSTLRELCDLQTLKLESGSFIEDSLRASYSDVLWSLKTNEGDGYIYVVIEHQSSP DAHMAFRLMRYAMAAMQRHLDAGHKTLPLVIPMLFYHGALSPYPFSLCWLDEFDDPALARQLYSATFPLVDITVIPDDEI MQHRRIALLELMQKHIRKRDLMGLVEQLVSLLATGYANDSQL |
| 2577 |
PF04577 (DUF563) |
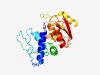 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF563) (Length=206) NVYFHWMVDVLPRLELLRQSGINFDQIDWFLVNSYQAPFQRETLTRLGIPEFKILESDQFPHIQAQQLIVPSFAGHTGWL QPWAIDTLRRWFLPKSSNSGKNYPERIYISRGDASYRRVLNEDEVIQFLRPWGFVTVQLETLSFTEQVALFAEAKVIMGA HGSGLTNILFCQPGTQVIEFMSPHYNRHYYWVISQYLGLEHYCLTG |
| 2578 |
PF04474 (DUF554) |
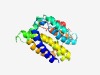 Estimated TM-score=0.61; hard target. |
>Protein of unknown function (DUF554) (Length=222) LGTIVNTIAILIGGFIGLLFGQALPDRMKKTVIQGIGLAVLLIGASMALQTKNTLVVIASLVIGGIVGELIDIEEQLKRF GQWLEQKFSRNSGQSGFTKAFVTASLIYCVGAMAIMGSLESGLKGNHTILYAKSMLDGISALVFASSMGIGVLASALPVF LYQGLITLSAGLLQGVLSPEIINEMGATGGLLIVGIGLNVLEIKEIKVGNLLPALFFAIPIA |
| 2579 |
PF04435 (SPK) |
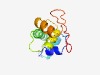 Estimated TM-score=0.61; hard target. |
>Domain of unknown function (DUF545) (Length=101) KFLAEETQNVRVPIPITTMCIQFKQTISSSASVNCLRKRLDKWRRYIHEWDGYDTDTKAKMMFGLSASVHPDFLIELKKE ADVELDSLHRIIAYKKKDGSL |
| 2580 |
PF04417 (DUF501) |
 Estimated TM-score=0.61; very hard target. |
>Protein of unknown function (DUF501) (Length=140) VSKRCSHNYPQVVVTAPILDKGSNIGIFPTTFWLTCPELNYQISKLEEEGWIKKIQDKINANQQLATALGAAHQDYARYR LNLISEQKLDDLEKNYTGQYLVLKESGVGGILEFDGIKCLHTHYAHYLATDNNPVGRLVD |
| 2581 |
PF04228 (Zn_peptidase) |
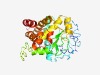 Estimated TM-score=0.61; easy target. |
>Putative neutral zinc metallopeptidase (Length=295) MRWKGRRVSTNVEDRRGGGGVKAGGISIIGLVVAFIAWKFFGVDPQQAYQATQAVTSETTSTAEAPQNLTLEQKEASEFV GTVLADTEDTWTPIFQQLGMTYKPPRLVLFSGVVKSGCGTAQSAMGPFYCPADQKVYIDTSFFKDMRQQMGISGEQNQTE LSRQDQAGDFAQAYVIAHEVGHHIQNLLGISGKVQQARAQSSQVEGNQLSVRLELQADCFAGIWANQNQQRTQFLEQGDI AEAMDAAEKIGDDYLQRRATGQVVPDSFTHGTSEQRMQWFELGYTTGDINQCNTF |
| 2582 |
PF04149 (DUF397) |
 Estimated TM-score=0.61; hard target. |
>Domain of unknown function (DUF397) (Length=50) AWFKSSYSGTEGGECVEVAPATTLVHVRDSKTATGPMLTVSRAAWTGFID |
| 2583 |
PF04071 (zf-like) |
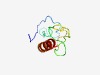 Estimated TM-score=0.61; hard target. |
>Cysteine-rich small domain (Length=83) NYKFFSHKECEYFPCHKTNDPDNFNCLFCYCPLYALKDKCGGNFKYTDSGIKDCSNCNLPHRRDNYDYIIGKFKDIINIT KKN |
| 2584 |
PF03862 (SpoVAC_SpoVAEB) |
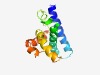 Estimated TM-score=0.61; hard target. |
>SpoVAC/SpoVAEB sporulation membrane protein (Length=116) MAKAFITGGIICTIGQFIYNYCELQNLSKDECSGWTSMLLVLLSVILTGINLYPKIAKWGGAGALVPITGFANSVASPAI EYKKEGQVFGIGCKIFTIAGPVILYGIFTSWILGFV |
| 2585 |
PF03858 (Crust_neuro_H) |
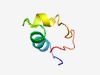 Estimated TM-score=0.61; hard target. |
>Crustacean neurohormone H (Length=41) RSAEGFGRMERLLTSLRGVADSSTPLGDLREAEEGAGHSLE |
| 2586 |
PF03692 (CxxCxxCC) |
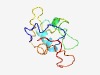 Estimated TM-score=0.61; very hard target. |
>Putative zinc- or iron-chelating domain (Length=93) KCTVCGLCCIDTKMELLPEDIERIETLGYRLEDFAVFDEDSGVWRLRNVNGHCVFFNEETKLCKIYENRPIGCRLYPLNY DDELGVVVDKYCP |
| 2587 |
PF03510 (Peptidase_C24) |
 Estimated TM-score=0.61; easy target. |
>2C endopeptidase (C24) cysteine protease family (Length=105) GYAIHIGHGVYISLKHVLTGNARILSEEPKGITISGELATFRLNNILPTAVPVGTNKPIKDPWGNPVSTDWQFKNYNTTS GNIYGACGSSCSLTRQGDCGLPYVD |
| 2588 |
PF03008 (DUF234) |
 Estimated TM-score=0.61; hard target. |
>Archaea bacterial proteins of unknown function (Length=93) WFRFVYPSRSDLESGQTDIVFSRIRAEFDSYVGRIFEQVCKEFLIDANNRGHLPFHFTNIGRWWHKDNEIDIIALDRETK EILFTECKWSNKR |
| 2589 |
PF02596 (DUF169) |
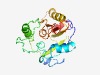 Estimated TM-score=0.61; hard target. |
>Uncharacterised ArCR, COG2043 (Length=207) LIDTLHLKTSPVAVRLVKADEELPADVQRITENVRHCQMVDNVRRKGSQFYALLEDHMCKGGAAVMGLTEMGEKLKTGEV YYHLNHFGSMDAAKSTMEKVPRVPAHSVKAVVYSPLEKTTFNPDVILIITNPKQVMELSQALLHKSGGRVNASFAGKQSV CADGVAYPYLSGEAGVTVGCSGSRKYTEIKDDEMIISIPVDMLPGLV |
| 2590 |
PF01963 (TraB) |
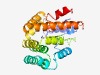 Estimated TM-score=0.61; hard target. |
>TraB family (Length=253) LYLLGSFHVLKPSDYPLSPDVMQAFAKADRLVFELPPSDAQSPELAGRMLQAARRSDGRRLQDALDPKTWQALTTYARKH GMPVDSLQTLEPWFVALTISLAEMSRQGMDPNAGLDQYLMAQAQAGGKPADGLERAEEQIALLDGMSPTEQHQLLEESLD DAGANDQLRQLHAAWRSGDVRTLSTQMAEEMRQHYPALYQDINVERNARWVPRLEQRLGKGSGTTLVVVGALHLLGSDGV VERLRARGYHVER |
| 2591 |
PF01927 (Mut7-C) |
 Estimated TM-score=0.61; hard target. |
>Mut7-C RNAse domain (Length=143) RFVLDTHLGKLASYLRMLGFDTLYQNDYTDDTLAHISAQEHRLLLTRDRGLLKRRQVTHGYYVRNTDPNAQVVEVLQRFD LFSSIKPFQRCLHCNKTLRPVEKQVIEDRLPPKVREHYDRFHTCDDCRRIYWEGTHYEKMNEF |
| 2592 |
PF01439 (Metallothio_2) |
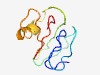 Estimated TM-score=0.61; hard target. |
>Metallothionein (Length=77) SCGGSCNCGSCGCGGGCGKMYPDLAEKITTTTTTATTVLGVAPEKGHSEGVGKAAESGEAAHGCSCGSSCRCNPCNC |
| 2593 |
PF18838 (LPD23) |
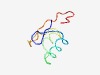 Estimated TM-score=0.60; hard target. |
>Large polyvalent protein associated domain 23 (Length=56) AFGGELGADLNLESLDVAQIMLAQGDDSETIRQATGWFTGIDGKWRFEIDDSQAEL |
| 2594 |
PF18823 (InPase) |
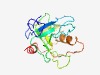 Estimated TM-score=0.60; easy target. |
>Inorganic Pyrophosphatase (Length=165) PTDAQKAAGNYKMGHATLHGLDITIENPRGSTRSGTDQDGKPWSVDMANHYGYIKRTEGADGDHVDAFIGPNPDSTHVFV VDQVDPRIGKFDEHKVMLGFDSLKAAREGYHANYEKGWKGAAAITPMSVDEFKAWLKNGDTTKPIDKKAGRSRTEKQAKA ARAAS |
| 2595 |
PF18810 (PBECR2) |
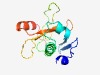 Estimated TM-score=0.60; hard target. |
>phage-Barnase-EndoU-ColicinE5/D-RelE like nuclease2 (Length=123) RFLDAFGVKPNSAGYYRDASGGVVTISRDLFSQAGATPSFSAGRYALMIAAAIQDPDEIWAAWALAGNASPMLQRTYIRR FDMGKDGNLLVQFVWTKKGWRAVTSFTATDAEIEALRVGAMLY |
| 2596 |
PF18733 (HEPN_LA2681) |
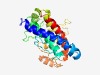 Estimated TM-score=0.60; very hard target. |
>LA2681-like HEPN (Length=223) TNIDEPPYLFGFYNQLKQEYVSARYLLWEGITESEKYNKHFSDNDVYLLNTLDSTAYGLAIEQIKIAFRSTYSLFDKIAY FVNDYWGLKIPEAKKINFQSVWVDTHKGVKQLRSEFMKYPNLSLRGLYWISKDFIEQDNSNELVLGKTMEPEADKLRTIR NHLEHKYLKVHDNMWGYTDEKWGSFFSDQLAYHISHEELSQKTIRLIKMARAALMYLSMAVHQ |
| 2597 |
PF18701 (DUF5641) |
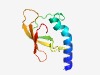 Estimated TM-score=0.60; hard target. |
>Family of unknown function (DUF5641) (Length=90) RWKLLQQSFQFFWRRWSREYLNTLQARGRWTKADTNLEVGTMVIVKVNDAPPLSWPLGRIIEVYPGTDKVVRVAKVITKQ GVFTRPVVKL |
| 2598 |
PF17446 (LtuA) |
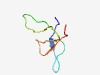 Estimated TM-score=0.60; hard target. |
>Late transcription unit A (Length=46) MFFIRARFIGFLDVHGYLAAKKGQQVMRSGSSIWVGSHGPIFYKVF |
| 2599 |
PF17364 (DUF5389) |
 Estimated TM-score=0.60; hard target. |
>Family of unknown function (DUF5389) (Length=104) MRNQNLPSGFSPFTWAIAGFCLPVLLWPLALLISPNLLNNPNLSDNQITAMSVFLWGYPFVLGIVARIFYKLHQRRPALA RKGLALSAVIFYGVLYYVATVGFN |
| 2600 |
PF17277 (DUF5342) |
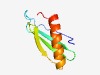 Estimated TM-score=0.60; hard target. |
>Family of unknown function (DUF5342) (Length=69) MLTHFQYKPLFENKELPGWRFSFYFRKERYAGVYHPDGKIEWVTAAPPEKAEDQVTGQIHELMLFHVYD |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
umich.edu
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417