

Go to page (83 pages in total): [First page] << < 19 20 21 22 23 24 25 26 27 28 29 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 2301 |
PF10505 (NARG2_C) |
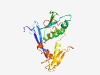 Estimated TM-score=0.62; easy target. |
>NMDA receptor-regulated gene protein 2 C-terminus (Length=210) EYVAPQEGNFVYKLFSLQDLLLLVRCSVQRIETRPRSKKRKKIRRQFPVYVLPKVEYQACYGVEALTESELCRLWTESLL HSNCSFYVGHIDAFTSKLFLLEEITSEELKEKLSALKISNLFNILQHILKKLSSLQEGSYLLSHAAEDSSLLIYKTSDGK VTRTAYNLHKTHCSLPGVPSSLSVPWVPLDPSLLLPYHIHHGRIPCTFPP |
| 2302 |
PF10343 (Q_salvage) |
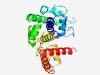 Estimated TM-score=0.62; hard target. |
>Potential Queuosine, Q, salvage protein family (Length=274) LHPKTADKAAADWIFLMATVNFSLWTNKEYSIEVNYKDKTYTGYYAAAACINKALDAGIPITDPKFMQNMTVKSVCDIFQ GHCKEGALVWALHKERTIAIIEAAKVLLAKFDGSFYNCIAQCGKSAVKLLAMIVENFPSYQDYAEYEGKKISFLRKAQIL VAHIYYCFKGKNDIADFYDIDQLVVCDHRVTQVLAFFLAFEYSPGLKQSLKIRPIFEIGEDIVLEMRAISIKSVYDIADR IAELRTENDTDLPKINAIDVGTLLWHYRRDNAEV |
| 2303 |
PF10303 (DUF2408) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF2408) (Length=140) EEIDPHLTTIMEKLFEIRRGLLSLAASSKRSNPSLEKSPLSQEHVHEQVEKLESQLTELESHRDENGKFKSLETDKVEDK GQNVLNGLLDDCHDLISDLSHQKNGGIELDAKLQPIYEQLIDIKTSLENLLVTRRWTLRE |
| 2304 |
PF10292 (7TM_GPCR_Srab) |
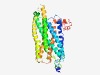 Estimated TM-score=0.62; easy target. |
>Serpentine type 7TM GPCR receptor class ab chemoreceptor (Length=331) ETNCQQMAQVATSSNLRITLFVSLICCLLCIPVNLYALYRIHISVKLHFNSKCVLFTHNFFVLIHCVARIGLHGKDLINY FDSWNSGCDIIPSRSRCDIRIFYKFSEYIIEISPFILTIERFVATFQAYHYENRYKWFGIVLNILHLSLGSLFLYIQNST NTGEVIIYYCWLANTGNRFLVNVPTFFIFFSQLATIPGLLYLLRKNEWNLQKFREASLQKHCTLTERYQISENLRTSSMF RIMSIVTWIFVVYNAGGAYIAHFYMGSMEFADQFALVEIIHAIPIYYIILSILIIRVDKKPQSEFTIRVNTYQPHYFIEL QKFFDEAFEKI |
| 2305 |
PF10203 (Pet191_N) |
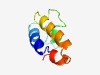 Estimated TM-score=0.62; easy target. |
>Cytochrome c oxidase assembly protein PET191 (Length=67) SSCQDIREALAQCLQESDCIMVQRHTPRECLTSPLAEQLPMKCQQLRKGFAECKRGMVDMRKRFRGN |
| 2306 |
PF10141 (ssDNA-exonuc_C) |
 Estimated TM-score=0.62; hard target. |
>Single-strand DNA-specific exonuclease, C terminal domain (Length=202) QLFDVRGIKQVHKWNGLIPEENKKMICFHPSTLDKLNLPKDEVILLEESSTLDGLSTEKASLVLLDLPNSKPLLEELLKK GRPHRIYAHFFQEEDHFFSTMPTRDHFKWYYGFLTKRGSFDVNKHGDDLAKYKGWSKETIDFMSQVFFDLKFVTIDNGFI SLNPAKTKKDLSESRTYQKKQQQYELENELLYSSYQELKTWF |
| 2307 |
PF10128 (OpcA_G6PD_assem) |
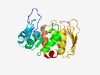 Estimated TM-score=0.62; very hard target. |
>Glucose-6-phosphate dehydrogenase subunit (Length=246) ANDASREHPCRVIVVARGARKAAPRLDAQIRVGGDAGASEVIVLRLYGELANEGASCVVPLLLPDTPVVAWWPNEAPAVP AQDPIGQLAQRRITDAAAEKNPIKALEQRRAGYTPGDTDLAWTRLTLWRAMLASAFDLPPYEKVTEAEVSGESDSPSTDL LAAWLAGCLRVPVKRAKAASGEGIVDVRLERRSGVVELSRPDGRVGTLTQPGQPERRVALQRRQVRDCLAEELRRLDPDE VYEAAL |
| 2308 |
PF10060 (DUF2298) |
 Estimated TM-score=0.62; easy target. |
>Uncharacterized membrane protein (DUF2298) (Length=477) LLGYVIWLPASLGWWRYDSAGLAGGLLVLIGIGLFLWWVGVRPERAAWRSLLPGEAIFLAGFAAMTIIRALNPDLWHPVW GGEKPMEQGFLNAILRSPQMPPYDPFYSNGYINYYYYGLYLMSVPIKLLGIDSAVGFNLALATLYGMLLAGAYELGTRLG GRRRYGIVALSLIGLMGNLTSLIPAGWSLGAQGLRLLLSGQAELLGDWFVGPSRVIPYTINEFPLFSFLYADLHPHLIAL PLALLAIVCGWHLILRPQRPLWLLSALTLGTLAVTNSWDFPTYGLLCGLAIIAGAIRRTGWRWQPLTLAVVQAVGLGLGS LLLFAPFFDRYWPMVRGIGLVTTGVTLPLDYLLIYGVPLIIVIPVIAGGLLQRWRSGFAGRRWALIASGIGLLMIIGAAV PELALRLSLFVVLMVTTILLMRRAAGAPAWYALLLCWVAWAVSLGVEIVYIRDHLDGSDWYRMNTVFKFGLHVWVLL |
| 2309 |
PF09969 (DUF2203) |
 Estimated TM-score=0.62; hard target. |
>Uncharacterized conserved protein (DUF2203) (Length=127) RYFTVKEANAVLPQIKVLVEEIVEKREEIYSLVSKYEELKELSEKSDLEELEYIYAKSKIQILDDEIKELVETIQNFGVM VKGVDPVLIDFPAKNGEEEILLCWKEDEDEIMYWHGLYEGFRGRKPI |
| 2310 |
PF09928 (DUF2160) |
 Estimated TM-score=0.62; hard target. |
>Predicted small integral membrane protein (DUF2160) (Length=88) WMAWTYPTAIFFITIALMLCGMTVWQIASPCVKRRGILPIATTRGDRLFIGLLSSAYIHLAWLGLTDFTLWVAFVFSLVW MVVVMRWG |
| 2311 |
PF09916 (DUF2145) |
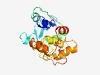 Estimated TM-score=0.62; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2145) (Length=199) LDASGASVVLLARAGQDLSKYGLRYSHLGWAYRTPEGPWRVAHKLNECGTAVAAVYRQGLGEFFLDDLWRYEAVWAVPAP EVQQRLLPLLQDRARTTALHQRPYSMVSYAWGTRYQQSNQWALETLAAAMEPASVHTRTQAQAWLRFKGYEPTALAIGPL ARLGGRMTAANIAFDDHPNDKRFADRIETVTVDSALAWL |
| 2312 |
PF09857 (YjhX_toxin) |
 Estimated TM-score=0.62; easy target. |
>Putative toxin of bacterial toxin-antitoxin pair (Length=85) MNISKFEQRTLHALARGGCIRVEKDDKGKIAEVACVTREGWVLTDCTLATFRRLRKRRLIASRDGSPYRITREGLHAVRA QLDNK |
| 2313 |
PF09652 (Cas_VVA1548) |
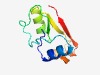 Estimated TM-score=0.62; hard target. |
>Putative CRISPR-associated protein (Cas_VVA1548) (Length=90) YFVSRHPAAIDWAAEEGIHVDAFVEHLDPDDIREGDTVIGSLPVNLAAQVCERGARYLHLSLELPPELRGRELTVDEMRA CGARIEAFRV |
| 2314 |
PF09343 (DUF2460) |
 Estimated TM-score=0.62; easy target. |
>Conserved hypothetical protein 2217 (DUF2460) (Length=204) HEVRFPDNISRGARGGPERRTQIVELASGDEERNASWANSRRRYDVAYGIRRADDLAAVVEFFEARNGRLYGFRFKDWAD YKSCLPSQTPSATDQAIGTGDGTTTAFQLVKRYTSGGQTWVRTITKPVVGSVTVAVDGVEQAGGWSVDTTTGLVTFTTAP ASGATITAGFEFDVPVRFDTDTLDVTLDLERLGSITSIPLLEIR |
| 2315 |
PF08894 (DUF1838) |
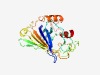 Estimated TM-score=0.62; very hard target. |
>Protein of unknown function (DUF1838) (Length=231) QSTFLIWTGKIYAFIPGEKRKLLFKIVGMSVSRCIATEEGKWDFTSRELTYYLNPETGEILNKWENPWTGETVTVMHVAN NPVQGEFKGKFPAQVEGDSTTFVFDIFPTYPNPLAADSKFAEYSPQQIYQAAELFKLTVPTADLENSEMTSVSQLKLSWD RIGQWLPWMKMGDRPGQLIYSATGSKVNGFTELPQLLQDEINTRVPLYKNAPKSFLDGEDMTSWLYFQQHF |
| 2316 |
PF08892 (YqcI_YcgG) |
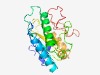 Estimated TM-score=0.62; hard target. |
>YqcI/YcgG family (Length=210) ADDADTFPCIPGRQAFLTDQLRIAFAGDPRENRTAEELAPLLAEYGKISRDTGKYASLVVLFDTPEDLAEHYSVEAYEEL FWRFLNRLSEQDEKEWPEDIPADPEHYKWEFCFDGEPYFILCATPGHEARKSRSFPFFMITFQPRWVFEELNGSTAFGRN MSRLIRSRLEAYDEAPIHPQLGWYGEKDNREWKQYFLRDDEKQVSKCPFS |
| 2317 |
PF08879 (WRC) |
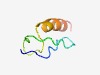 Estimated TM-score=0.62; easy target. |
>WRC (Length=41) PDDLRCKRTDGRDWRCKHSVVPGKTFCLSHLHRGQKRRVSS |
| 2318 |
PF08664 (YcbB) |
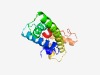 Estimated TM-score=0.62; hard target. |
>YcbB domain (Length=138) EFILNQLGIIAESGSKDLLSIVEYLYTRENTKTIEKDFPSLKEIFTKISEKKLGQSSSSIDIKKEIKASEQRVRRAVLQS LNYIASLGLTDYSNPKFEEYAAEFFDFTEVRKKMLELKNNTPLSQPIHINVKKFIKVL |
| 2319 |
PF08621 (RPAP1_N) |
 Estimated TM-score=0.62; hard target. |
>RPAP1-like, N-terminal (Length=44) QIDAENHARLQEMSPDEIAQAQAEIMEKMNPAIINALKKRGQDK |
| 2320 |
PF08592 (DUF1772) |
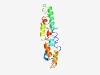 Estimated TM-score=0.62; hard target. |
>Domain of unknown function (DUF1772) (Length=150) GNIAALSLMAVPALRLSKANDGVSSTSLVKQWRYIYEAGKSQNPPIAAATAAAFIYLAWSVRRIAPRSQIPLYYGSAAVL TLGIVPFTLMVMSPTNNRLIQHSETMSMEGSAAPSRARDDEIDQLIAKWNTLNGVRSLLPLAGGILGLVA |
| 2321 |
PF08349 (DUF1722) |
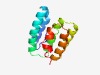 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF1722) (Length=117) KLLVMAHSPKHVGDLGKLVANGKRSHLDEVCAEYHRTLMEALKRKATVKKNTNVLQHLMGYFKKQLSSDEKQELSELIEH YHQGYLPLIVPVTVIRHYVRKYDQPYLKEQYYLNPHP |
| 2322 |
PF08179 (SspP) |
 Estimated TM-score=0.62; hard target. |
>Small acid-soluble spore protein P family (Length=43) MTNKNDGKDMRKNAPKGDNPGQPEPLSGSKKVKNRNHTRQKHN |
| 2323 |
PF08132 (AdoMetDC_leader) |
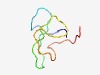 Estimated TM-score=0.62; very hard target. |
>S-adenosyl-l-methionine decarboxylase leader peptide (Length=51) MEFKSGKKQSSSSSSLQYEVPLDYTVEDVRPHGGIRKFQSATYSNCVRKPS |
| 2324 |
PF08044 (DUF1707) |
 Estimated TM-score=0.62; hard target. |
>Domain of unknown function (DUF1707) (Length=53) LRATNADRRQAADVLQMALREGRLDAVEFDSRSRAGESARYVDELDALVADLP |
| 2325 |
PF08012 (DUF1702) |
 Estimated TM-score=0.62; easy target. |
>Protein of unknown function (DUF1702) (Length=318) FGSLRRLLMTPSLAEVSFEGRGFPVTPSDATRRLEAIPQAVICGFEWGIDARDQWEVERRLSMVDPEQRGFAYEGATMAF TILDAMAGGRGHRTRDLLLGPGQPHIFLTYIGIGFAMARLPRPLWKKVMPDLGGSAYYPTMSWLAVDGYGFDRAYFDTRR WVDEQRVPASYPWEGSPDYFPRAVDQGIGRALWFIHGGQVPDVASAVGRFAGHRQADLWSGVGLASTFAGGADTEALATL RREAGEHRAELAQGVVFAVKARDFSGFVPPHTEAAAAVLADLSIENAVALADDTAVDPHGSGPVPQYELWRSRIRSHF |
| 2326 |
PF07843 (DUF1634) |
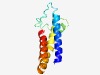 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF1634) (Length=109) IGNLLRIGVLFSAAVVFIGGIIYLFEYGNQKIHYHTFRGEPPELRSVTGVIHYALQGHGRGIIQLGLLLLVATPIARVIF SIYAFIREKDRLYVVVTLIVLTILLYSLI |
| 2327 |
PF07706 (TAT_ubiq) |
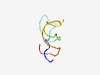 Estimated TM-score=0.62; very hard target. |
>Aminotransferase ubiquitination site (Length=40) MDPYMIQTNGNGNLASVLDVHVNIGGRSSVPGKMKSRKAR |
| 2328 |
PF07680 (DoxA) |
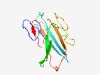 Estimated TM-score=0.62; hard target. |
>TQO small subunit DoxA (Length=130) FHGGVWGTLHNKSVKPKLEITEGKINNDTLSFDVFRTEGVDVYGSWVIAIEVLDQNNKSIVQYTQKELASLADQNISNYY VAKIKPGKHSLVVPLGAKAKMTFSNQVFSQLSNGTYTVKITDISGAFWTH |
| 2329 |
PF07639 (YTV) |
 Estimated TM-score=0.62; hard target. |
>YTV (Length=43) YTVCKPVYETKTCQIPYTVCKPVYETKTRQINYTVCKPVCETK |
| 2330 |
PF07157 (DNA_circ_N) |
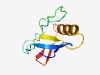 Estimated TM-score=0.62; easy target. |
>DNA circularisation protein N-terminus (Length=86) LQPASFRGVPFSIESDEGTFGRRVQTHEYPNRDKPYTEDLGRSTRRFTVSAYLIGDDYIDQRDRLIAAVDTPGPGTLVHP YYGEMA |
| 2331 |
PF07148 (MalM) |
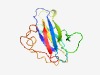 Estimated TM-score=0.62; very hard target. |
>Maltose operon periplasmic protein precursor (MalM) (Length=133) IQGAVAAFALPANQGSLEITLNSLIKDKQVYSPNVLVLDEQLRPAAFYPSHYFQYQRPGIMSVDRLEGTLKLTPALGQQQ IYLLVYTTRADLQSSTQMVDPAKAYAQGVGNAVPNIPDPMARHTETGTLTLKV |
| 2332 |
PF07103 (DUF1365) |
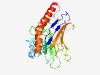 Estimated TM-score=0.62; very hard target. |
>Protein of unknown function (DUF1365) (Length=248) IYAGQVRHRRFQPKTHSFQYRLFMMLLDLAELDTVFKSRWLWSSRGFNLAWFRRKDHMGDPNKPLADEVRRLVREQTGRT PRGPIRLLTHLRYFGYCFNPVSFYYCYDASDTRVEAIVAEVNNTPWGEQHCYVLTESMNQGRDRHKRYTFDKDFHVSPFM QMAVHYDWRFSAPDKQLNVHMQNLKQGENQQQKVFDATLALSQQPVTGFNLAKVLIQYPFMTLKVTTGIYYQALKLWLKG VTFYEHPK |
| 2333 |
PF06989 (BAALC_N) |
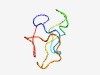 Estimated TM-score=0.62; very hard target. |
>BAALC N-terminus (Length=50) MGCGGSRADAIEPRYYESWTRETESTWLTYTDSDAPPSNAAPDSGPEAGG |
| 2334 |
PF06951 (PLA2G12) |
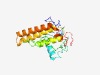 Estimated TM-score=0.62; easy target. |
>Group XII secretory phospholipase A2 precursor (PLA2G12) (Length=178) SLGHGLAQSDTSPDAEETYSDWGLRHIRGGFESVNSYFDSFLELLGGKNGVCQYRCRYGKAPMPRPGYKPQEPNGCSSYF LGLKMDLGIPAMTKCCNQLDVCYDTCGANKYRCDAKFRWCLHSICSDLKRSLGFVSNVEACDSLADTVFNTVWTLGCRPF MNSQRAACICVEEEKEEL |
| 2335 |
PF06926 (Rep_Org_C) |
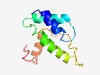 Estimated TM-score=0.62; hard target. |
>Putative replisome organiser protein C-terminus (Length=95) NSLLSKFLDTFINFSSKNRSKRAVATAEFIKLPSFQREQAVIGAMNYIQSYKNEHPDDETGQYSVNAVNFLSNMMFMDYQ EEVKADTGYDEDLGF |
| 2336 |
PF06850 (PHB_depo_C) |
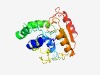 Estimated TM-score=0.62; easy target. |
>PHB de-polymerase C-terminus (Length=202) GGPIDTRRNPTAVNTLAEEKGIDWFRDNVIMQVPWPNPGFMRDVYPGFLQLSGFMGMNLDRHIIAHKDFFMHLVKNDGDN AEKHRDFYDEYLAVMDLTAEFYLQTVDTVFVRHALPKGEMKHRGEPVDPSAIRNVALLTVEGENDDISGVGQTQAAHNLC VNIPADMRAHYLQPAVGHYGVFNGSRFRSEIVPRIVDFISSY |
| 2337 |
PF06826 (Asp-Al_Ex) |
 Estimated TM-score=0.62; easy target. |
>Predicted Permease Membrane Region (Length=168) LALVAVIGLWIGHWKIRGVGLGIGGVLFGGIIVAHFTNQYGLKLDVHTLHFVQEFGLILFVYTIGIQVGPGFFSSLRKSG LKLNAFAILIIVLGSIAVVLVHKIADVPLDIALGIYSGSVTNTPALGAGQQILAELGVPQTTVTMGVSYAMAYPFGICGI LLAMWLIR |
| 2338 |
PF06772 (LtrA) |
 Estimated TM-score=0.62; hard target. |
>Bacterial low temperature requirement A protein (LtrA) (Length=372) VTRLELFLDLVFVYAFLNVTSVAAAHPDLFGIVRGLLLLALLWWCWASYAWLSNSIRLDRGIMPMVAFGLIAIIFVVGVA IQEAFVDRPGGLPGPLIFVLGYLAARAGALTVAMICARSDPVSRQRVRRGWLPLFGAAPLLLLAALLASRLIDVEIHPWA RLGLVLLAILIEYRGGVLTGAVGGRDIPVRHWVERHALIVMVAFGETIISVGVSRGIGSTEPITWPLVAAILLSMSIVGV LWWTYFDLARFAAELALEQVSGAARVRLARNAYTFLHLPMVGGLILLSLGLEKAVGAAGEPEPLPGLSLLALYGGVILYL AGLLAFEGRTMHIVGRGPLLGLVLLALGIPVAAQVPALAALGILAVLVAVLV |
| 2339 |
PF06635 (NolV) |
 Estimated TM-score=0.62; easy target. |
>Nodulation protein NolV (Length=207) MTADISVAPAAPQMRPLGPLIPASELEIWDNAAKACAAAERHQQHVRSWARAAYQRELARGHTEGLNAGAEEMAALISQA VAEVARRKAVLEQQLPQLVLEILSELLGAFDPGELLVMAVRHAIERQYSGAEVCLHVYPTQVDMLAREFAGWDGQDGRPR VRIKPDPTLSPRRCVLWSEYGNVDLGLDAQMRALRLGFGSLSEKGEL |
| 2340 |
PF06620 (DUF1150) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF1150) (Length=75) TPEALAHLGEGHIAYVKQVRSEDVPDLFPQAPKIAPGLKLFALHAADGTPIMLTDSREAAIANAWSNELQAVSVH |
| 2341 |
PF06377 (Adipokin_hormo) |
 Estimated TM-score=0.62; hard target. |
>Adipokinetic hormone (Length=46) QLNFTPNWGKRAPEGESNRCKESVDTIMLIYKIIQNEAQKLVDCEK |
| 2342 |
PF06195 (DUF996) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF996) (Length=134) GLVGIILILISLNMFSKIFNDPQIFRNALISFILAIIGVFVIMLTIGITFFSFFTMMGSYHLHMFSSIGIGTIFSFIVTY LLLIISAHYLKSSFYLLSSYTGINLYKTGGFWYFIGTLLIIVVIGFLINLVAWV |
| 2343 |
PF06170 (DUF983) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF983) (Length=85) FDGYLSVKQSCDNCGEELHHHRADDAPPYFTIFIVGHVVVALAMWVEMAYVPPMWLHMAVWLPLTIIMSLTLLRPIKGAL VGLQW |
| 2344 |
PF06107 (DUF951) |
 Estimated TM-score=0.62; easy target. |
>Bacterial protein of unknown function (DUF951) (Length=54) YDLGDIVEMKKPHPCGTNRWEITRMGADIKIKCTGCGHRIMLPRREFEKRLKKV |
| 2345 |
PF06045 (Rhamnogal_lyase) |
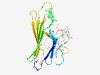 Estimated TM-score=0.62; easy target. |
>Rhamnogalacturonate lyase family (Length=219) ANVNKSSTYKLRVALASATFSELQVIMDNGIVRVNLSNPEGIVTGIQYNGLDNLLEVLNEESNRGYWDIVWDQGGEKRTE KGKKGKGTLDRLEATNFTVIVKNEEQVELSFTRTWNSSLEGKFAPLNIDKRFIMLAGSSGFYTYAIYEHLKEWPAFDLDN TRIAFKLRKDKFCYMAVADNRQRFMPLPDDRLPPRGEVLAYPEAVRLVDPLEPEFKGEV |
| 2346 |
PF06034 (DUF919) |
 Estimated TM-score=0.62; easy target. |
>Nucleopolyhedrovirus protein of unknown function (DUF919) (Length=62) SKDEDLRRQLNQILQAKRQLTIQMEHWERIKRITKDPKEVQDIESKLLKMRMDFLNFSTDKF |
| 2347 |
PF05857 (TraX) |
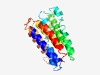 Estimated TM-score=0.62; hard target. |
>TraX protein (Length=226) SLDLVKWLAMLTMVIDHLRYLWPEATAWLFVVGRFAFPLFCLGIAANVSRSHPGDLYSENNFRYLSWILAFSLISEMPYR ALSEMSNTLNVMPTLMLGLIVAWSVHHSDRTGLVLGLFTLTVSIVLHDRLMYGAFGVLLPATMVLAVQRPWITWLIPAVF SVLANSRDGWIGAGPFGTWWAMATTFAAPLVGLWLLRQKITQHIWPVGRWGYYFYPGHLAVLALLR |
| 2348 |
PF05805 (L6_membrane) |
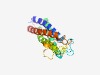 Estimated TM-score=0.62; hard target. |
>L6 membrane protein (Length=186) MCTGKCSLFIAISLYPLVLLSILCNILLFFPDWSTKSVKEGHITDEVKYMGGFIGGGVLVLIPALYIHLTGEQGCCANRL GMFFSVIFAAVGAAGAVYSFIVALLGLYNGPLCRTSSYKWERPFMDKNLEYVKNSNSWNDCLEPKNVVQFNMGLFITLMV ASVLQGLLCVSQVINGFVGCICGTCN |
| 2349 |
PF05659 (RPW8) |
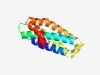 Estimated TM-score=0.62; easy target. |
>Arabidopsis broad-spectrum mildew resistance protein RPW8 (Length=136) SAAAGAGLELAFGSLLRIILEVRRKNVMFASTLQILIDTMEAIRPSIKRIDSFNTDFDRPDEIKGLRDLIRKGEDLVIKC SKVHKYNYVKKPYYTKKLIKLDESIRRYISTILPLYQTSDIKETLYEMRVLSTQIR |
| 2350 |
PF05425 (CopD) |
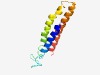 Estimated TM-score=0.62; hard target. |
>Copper resistance protein D (Length=108) ALERFSGIGSAIVAVLLLTGVINGWVLIGPDRLDSLWTSPYGRLLLLKVLLFGGMLVLAASNRFRLTPGLGKALHSGEET GAALGALRRSIALELALGIAILALVAWL |
| 2351 |
PF05415 (Peptidase_C36) |
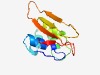 Estimated TM-score=0.62; easy target. |
>Beet necrotic yellow vein furovirus-type papain-like endopeptidase (Length=104) NLVSRPNNCLVVAISECLGVTLEKLDNLMQANAVTLDKYHAWLSKKSPSTWQDCRMFADALKVSMYVKVLSDKPYDLTYE VDGAGSSVTLHLVGKESDGHFIAA |
| 2352 |
PF05255 (UPF0220) |
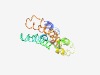 Estimated TM-score=0.62; hard target. |
>Uncharacterised protein family (UPF0220) (Length=162) EERLWKFRKPEWLNSAWSRNAGVYAAGGLFSLAFYVMLDSAVWSKTLNGSDTHVTFFDWLPLLFSSLGMLIINSVEKTRL SADSFSYSGNGVAWKARVVLFLGFAALAGGMAGGVTVFVLKFAVRGVAMPALSMGVENVVANALVMLSSVVLWVSQNMED EY |
| 2353 |
PF05212 (DUF707) |
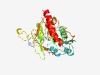 Estimated TM-score=0.62; easy target. |
>Protein of unknown function (DUF707) (Length=290) TNPRGAERLPPGIVNAESDLFLRRLWGLPSEDLTFKPKYLVTFTVGYEQKKNIDAAVKKFSKDFTILLFHYDGRTTEWDE FEWSKQAIHVSVHKQTKWWYAKRFLHPDIVAPYDYIFMWDEDLGVENFNAEEYLKLVRKHGLEISQPALEPSKSTKAVCW NMTRRREHSEVHKEAVEPGKCKYPLLPPCAAFVEIMAPVFSRNAWRCVWHMIQNEFVHGWGLDFAFRKCVEPAHEKIGVV DTQWIVHQGIPSLGNQGETQTTGKPAWRAVKERCGMEWRMFQGRLTNAEK |
| 2354 |
PF05121 (GvpK) |
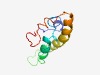 Estimated TM-score=0.62; hard target. |
>Gas vesicle protein K (Length=85) RINSDPESVERGLVSLVLTLVELLRQLMERQAIRRVDSGDLTDEQIERIGTTLMLLEEKMEELREHFGLEPEDLNIDLGP LGPLL |
| 2355 |
PF04910 (Tcf25) |
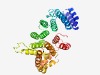 Estimated TM-score=0.62; hard target. |
>Transcriptional repressor TCF25 (Length=341) FTFEHHREYQQVQFKFLDAVESMDPNNIVLLLQMNPYHVDSLLQLSDVCRMQEDQEMARDLIERALYSLECAFHPVFSLT SGTCRLDYRRPENRAFFLALFKHLMFLEKRGCPRTALEFCKLILSLDPENDPLCVLLLIDFLALRAREYSFLTRIFQEWE SHRNLSQLPNFAFSVPLAYFFLSQQEERPELERSQARERAARLIQLALVMFPSVLMPLLDHCSVQPDARVASHSFFGLNA QISQPPALNQLTSLYVGRTHTLWKDPAVMAWLETNVHEVLRMVDTQDPLVEESEHKRKMRYQSAPRNIYRHVILSEMKEA TSALPLEVTSQPVMGFDPLPP |
| 2356 |
PF04854 (DUF624) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function, DUF624 (Length=77) LAYLNLLWMIFTIAGLVVFGLFPATAAMFSVTRKWVMKDTDIPIFKTFWQSYKSEFIKSNLLGLVLVLIGGILFFDL |
| 2357 |
PF04833 (COBRA) |
 Estimated TM-score=0.62; hard target. |
>COBRA-like protein (Length=164) YVAAVTMSNLQMYQPIMSPGWTIGWTWAKKEVIWSMVGAQATDQGDCSKFKGSIPHSCKRNPEIVDLLPGVPRDQQFTDC CKAGVMAAWGQDSSAAVSAFQVSVGLSGTSNMTVKPPKNFNLFGPGPGYTCSPATIVSPSVFLSPDRRRKSHAIMTWIVT CTYS |
| 2358 |
PF04827 (Plant_tran) |
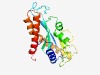 Estimated TM-score=0.62; very hard target. |
>Plant transposon protein (Length=204) EYLRKPNSNDVRRLLEMGEARGFPGMMGSIDCMHWQWKNCPKAWKGMFMSGHKGVPTLLLEAVASSDLWIWHAFFGVAGS NNDINVLDRSPLFDEVLEGRAPEISYTLNGNNYNFGYYLADGIYPEWATFDKTIPRPQGEKRKLFSKYQEGQRKDVERAF GVLQSRFAIVRGPARFWDKGDLARIMRACIILHNMIVEDERDTY |
| 2359 |
PF04475 (DUF555) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF555) (Length=101) NYKVVLEAAWIVKDVKSVDDAMSVAISEAGKRLNPRMNYVEVEVGATFCPFCGEPFDSVFVVANTGIVGLLLEMKVFNAE SKEHAERISKKEIGKALKDVP |
| 2360 |
PF04468 (PSP1) |
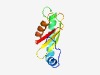 Estimated TM-score=0.62; hard target. |
>PSP1 C-terminal conserved region (Length=94) KQVLRFATPWESSTNLHNKFQDELKALHIAQLKLRSLNNSNSGGGLNIKILNAEFQFDRKKLTFYYICQERNDFRDLIKE LFKFYKTRIWLCAI |
| 2361 |
PF04341 (DUF485) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function, DUF485 (Length=87) QASPEFQELRRRLRSFVFPMTAFFLIWYIVYVLLSNYAHDFMSTPVWGNINVGLLLGLGQFVTTFAITGIYVRFANRELD PRAEALR |
| 2362 |
PF04312 (DUF460) |
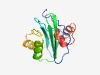 Estimated TM-score=0.62; easy target. |
>Protein of unknown function (DUF460) (Length=134) SREEDVQVTVTSVEKEAIEFIPLEKKKRDYIIVGVDPGTTTAVAILDLNGKLVEVSSSRVSSVPDIIEHISEMGRPVVIA TDVVPPPAAVEKIKRTFNAVLFTPNERIPVEEKVRLARPLGYSNDHERDALAAA |
| 2363 |
PF04290 (DctQ) |
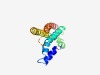 Estimated TM-score=0.62; hard target. |
>Tripartite ATP-independent periplasmic transporters, DctQ component (Length=131) TLILFLNVVLRYVFKSSASWAEEVIKYLMIWIAFIGGSICVRKGKHVSIDFFYEFLSVKNKKVLSVVVHLIATVFCGVMF FYGVKIIDFVMKSGQVSPALQIPMWIPYLAIPLGFILMFFRFIQDLIRIFK |
| 2364 |
PF04254 (DUF432) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF432) (Length=123) PVNLPKEITNFLLIRFKKPVFIEPKTSMEVNLTFPVEIGVFLAAKKRVDVIDIFSLQKPKYTLYGNPRMGIICRYWESDV YSDVPDVDGFREGVMRLRITNSSDEWIEVTNAVFDIFGMKIYY |
| 2365 |
PF04250 (DUF429) |
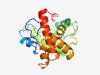 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF429) (Length=207) PCPPGWLVQAAKVQGATFAPERPRVYANYLEILDERPAYEMILLNAPVGYRDLPGDPPRLCDAEARRLLGARGATIHNAP TRAVLDGTMTWQEGGLDAVTATLLPRYRQVAQEMSPFRQRVVYEGNPELSFYQLNRDTSMRRSKRLAEGRDERVEVLDAH IPGGIGNALDNPPEGVALRHQLDAFALLWTARRVWGHGARRIPREPE |
| 2366 |
PF04233 (Phage_Mu_F) |
 Estimated TM-score=0.62; very hard target. |
>Phage Mu protein F like protein (Length=108) GVLDDLAQNIQRGGTFEEWQKNYTAQGLPRHYLETVYRTACLRAYNAGKWVQFRAQKENRPILRYSAINDSRTRPHHKAL HGFMAPVDDAVWRTLAAPNGFNCRCTLM |
| 2367 |
PF04186 (FxsA) |
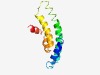 Estimated TM-score=0.62; hard target. |
>FxsA cytoplasmic membrane protein (Length=108) LFILIVPALEIGLLIWAGNWIGPWWVILLIILTGVIGAWLAKQQGLETIRNFQNSMSSGQMPQDTLLDGACILVGGAVLL TPGFITDAIGFLLLIPPTREPIKRLIKK |
| 2368 |
PF04168 (Alpha-E) |
 Estimated TM-score=0.62; hard target. |
>A predicted alpha-helical domain with a conserved ER motif. (Length=311) MLSRVADSIYWLNRYVERAENIARFADVNFNLMLDSPTGITQQWEPLVKITGDLPLFQARYGEATAENVIQFLTFDTEYP NSIISCLRSARENARSIQEIISSEMWQQVNQFYMMVQEAANQQTMSQLLEFLAEVKMAGHLFAGVMDATMTHNEGWHFGQ MGRFLERADKTSRILDVKYFILLPSVQYVGTALDEIQWMALLKSASAYEMYRKRKQHRITPTGVAEFLIMDREFPRSIHF CLLQAESSLHEITGTPSNTWKTPVERALGRSRAELDYLTIEEIIKKGLHEFMDDLQQQINDVGHKIFESFF |
| 2369 |
PF04118 (Dopey_N) |
 Estimated TM-score=0.62; easy target. |
>Dopey, N-terminal (Length=332) DKAYRRYASGVERSLSLFDTALQEWADYISFLGRLLKSLQAHPSTITTVPSRAIVAKRLAQCLNPSLPSGVHQKALEVYA YIFTLIGKDSLSRDLPLYFPGLASTLSFASLSVRTPFLDLLEKHMLKLDAGALRPALKAIILSVLPGLEEETSEEFERTL KLLDEFKHSVRSTRGDDLGHDHNSADEYFWQCFFLAAITSNTRRLGALAYLTRNLPKLGKAVALDSTKSAGDESSPEEDK DSSTSRLADLVTSPEPGLLLRCFAAGLADEQLLIQRGFLDLLVTHLPLQSHVLQNRVKAEDLELLITAAAGVVARRDMSL NRRLWTWLLGPE |
| 2370 |
PF04024 (PspC) |
 Estimated TM-score=0.62; hard target. |
>PspC domain (Length=56) RRVYRSRKNRMLGGVAGGIAEHLGVDPTLIRLIWILSIFTGVGVIAYIIAWIIIPE |
| 2371 |
PF03994 (DUF350) |
 Estimated TM-score=0.62; hard target. |
>Domain of Unknown Function (DUF350) (Length=55) FALVGIFFIYLAKRLDDWRTREFDDDRHIDDGNVAVGLRRAGLYLGIAIAMAGAL |
| 2372 |
PF03773 (ArsP_1) |
 Estimated TM-score=0.62; hard target. |
>Predicted permease (Length=299) HLGSAVNFFIYDTIKIFILLATLIFVISFIRTYIPPNKVKETLEKRHRYTGNFIAALVGIITPFCSCSAVPLFIGFVEAG VPLGATFSFLISSPMINEIAIILLLGLFGWQITAFYIVSGFIIAVLGGILIGKLKMETELEDYVYETLEKMRALGVADVE LPKPTLRERYVIAKNEMKDILRRVSPYIVIAIAIGGWIHGYLPEDFLLQYAGADNIFAVPMAVIIGVPLYSNAAGTIPLI SALIEKGMAAGTALALMMSITALSLPEMIILRKVMKPKLLATFIAILAVSITLTGYIFN |
| 2373 |
PF03601 (Cons_hypoth698) |
 Estimated TM-score=0.62; easy target. |
>Conserved hypothetical protein 698 (Length=318) PGILLLIGVGLLGKYAQIWWKDLAKAQHWTVPDIEYVLWAIVIGLLITNTVGLHRIFRPGVQTYEFWLKIGIVALGARFV LGDVLKLGGTSLVQIVVDMTVAGAIILLAARWFGLSGKLGSLLAIGTSICGVSAIIAAKGAIRARNSDVSYAIASILALG AVALFTLPAIGHGLGLSDYEFGLWAGLAVDNTAETVATGRLYSEHAGDIATVVKSTRNALIGFVVLGFALYWAARGEAET LAPGIKAKAAFVWDKFPKFVLGFLAVSTIVTLGWLTKGQVNNLVNVSKWAFLLTFAGVGLNTNFREIARSGWRPLVVA |
| 2374 |
PF03564 (DUF1759) |
 Estimated TM-score=0.62; hard target. |
>Protein of unknown function (DUF1759) (Length=145) FDGAYEKWLAFRDMFNSLIHSNLTLPKVQQFHYLRSSLKGEAANIVQSLEVCDESYDAAWSLLEDRYDNKRLIINNHLKG ITNITPLLKESSSGLRSLLDNLSKNLRALKVLKQPVEYWDTIIVFLVVQNLDPTSRREWEKEVSS |
| 2375 |
PF03436 (DUF281) |
 Estimated TM-score=0.62; hard target. |
>Domain of unknown function (DUF281) (Length=53) CIETKATCTRNDGMICTDIKMFGTTATGQVSITDSTTTTKVSATLSCANDGTY |
| 2376 |
PF03016 (Exostosin) |
 Estimated TM-score=0.62; hard target. |
>Exostosin family (Length=285) EKTLKVYVYKEGAKPIFHQPQVVLSGIYASEGWFMKHMKENKHFVTNNPKQAHLFYIPFSSRMLEEKLYVVGSGSRDNLV QHLKDYIHLIAERYDSWNRTGGSDHFLVACHDWATDETKKSMNSCIRALCNSDVKKEGFELGKDVSLPETYVRSPQNPLR ELGGKPPSQRSFLAFFAGQMHGDVRRILLHYWENKDPDMKIFGKLKKSKNNRNYIQYMKSSKYCICAKGYEVNSPRVVEA IFHECVPVIISDNFVPPFFEILNWESFAVFVLEKDIPNLKKILVS |
| 2377 |
PF02791 (DDT) |
 Estimated TM-score=0.62; easy target. |
>DDT domain (Length=56) HLMNVIAIYEVLRNFGTVLRLSPFRFEDFCAALVSQEQCTLLAETHVALLKAVLRE |
| 2378 |
PF02652 (Lactate_perm) |
 Estimated TM-score=0.62; easy target. |
>L-lactate permease (Length=521) LSALVALSPIVLFFISLIVFKLKGYSAGFLSLLLSIIIALFVYKMPAQMVSASFFYGFLYGLWPIAWIVIAAIFLYNLSV KSGYFEILKESILTLTPDHRILVILIGFCFGSFLEGAIGFGGPVAITAAILVGLGLNPLYAAGLCLIANTAPVAFGAVGI PITAMASVVGIPELEISQMVGRVLPIFSIGIPFFIVFLMDGFRGIRETFPAVAVTGFSFAIAQFLSSNYLGPQLPDIISA LVSLIATTLFLKFWQPKRIFTSNGKEPTINTEKHHICKVVVAWMPFVLLTITIIIWTQPWFKALFKEGGALAFSSFAFEF NSISQKIFKTVPIVTEATNFPVVFKFPLILTTGTSIFLAALLSVFLLRVKISDAIGVFGATLKEMRLPILTIGVVLAFAY VANYSGMSATLALALADTGHVFTFFSPVVGWLGVFLTGSDTSSNLLFGSLQMLIATQLGLPEVLFLAANTSGGVVGKMIS PQSIAIACAAVGLVGKESELFRFTVKYSIALAIIMGIVFTL |
| 2379 |
PF02067 (Metallothio_5) |
 Estimated TM-score=0.62; hard target. |
>Metallothionein family 5 (Length=41) MVCKGCGTNCQCSSQKCGDNCACNKDCQCVCKNGPKDQCCS |
| 2380 |
PF01688 (Herpes_gI) |
 Estimated TM-score=0.62; easy target. |
>Alphaherpesvirus glycoprotein I (Length=157) LVLVGLWVCATSLVVRGPTVSLVSNSFVDAGALGPDGVVEEDLLILGELRFVGDQVPHTTYYDGVVELWHYPMGHKCPRV VHVVTVTACPRRPAVAFALCRATDSTHSPAYPTLELNLAQQPLLRVRRATRDYAGVYVLRVWVGDAPNASLFVLGMA |
| 2381 |
PF01492 (Gemini_C4) |
 Estimated TM-score=0.62; hard target. |
>Geminivirus C4 protein (Length=84) MGNLISTSCFNSKEKFRSQISDYSTWYPQPGQHISIRTFRELNPAPTSSPTSTRTETQLNGGNSRSTVEVLEEVNRQLTT HMPR |
| 2382 |
PF18798 (LPD3) |
 Estimated TM-score=0.61; very hard target. |
>Large polyvalent protein-associated domain 3 (Length=116) GKKALRSKVLEHLTETLKGKWVKNLFLDRDIEIRKRGIKKTLANSANPLKLKALAQLEEIIKTAKGSKDYIQDNFKSDKK PNVLRYFHLENHVLVDNRPLKINVVIEEDTNGLLHY |
| 2383 |
PF18735 (HEPN_RiboL-PSP) |
 Estimated TM-score=0.61; hard target. |
>RiboL-PSP-HEPN (Length=191) DDFVWRKKEISDLKALIETRSFTTSKHNALLRSGVALLYAHWEGYIRTAATSYLEFVARQKLTYGELAINFVAIAMKAKL NEANDTNKATVFTAVTNLILTQVHQTSSIPYEDIISTASNLSSAILREITCLLGLDYSFYQTKEVIINEQLLKRRNAIAH GEYLSLDRDEYQQLHDEILAMMENFRTQVEN |
| 2384 |
PF18734 (HEPN_AbiU2) |
 Estimated TM-score=0.61; hard target. |
>AbiU2 (Length=205) EYFKKYHEKLIWETQNTRAHLELWERLERYKSSHLKELNQAPHFFTFTIKAHLDDALLTLSRIVDRRLRHDPLSIWKFLN FVEQNYDMFSTEAFCQRMMQKPNYDEHWTKSHEPITIKEINEDRQRLTSLENVISNIEKWRDKVIAHIDLKVVTRNKVIS KEYPLKLQQLQEVIDTLFRILNRYSSAFESTSYTEQFVGEDDIQY |
| 2385 |
PF18723 (aGPT-Pplase1) |
 Estimated TM-score=0.61; hard target. |
>alpha-glutamyl/putrescinyl thymine pyrophosphorylase clade 1 (Length=278) TEVFDTYWKFAATRQKVYERRIAGLPGPWTDDSIISEHRFTNCFRASDRVSQFLINNVIYEGAQDPEEVVFRILLFKFFN KISTWELLQSELGAPTREKFSIVEYDRVLSAAMSAGKKIYSAAYVVPPPKMGEVRKHKNHLRLLERMLDDSISDRLQDCA EMQDAFGVLRSYPAMGNFLAYQFLIDINYSEVLDFDEMDFVVPGPGARDGLRKCFGDRSRGIEAEIIRYMCDSQDEHFAR LGLEFRGLQGARSLQLVDCQNLFCEVDKYARVAHPDIE |
| 2386 |
PF18722 (MazG_C) |
 Estimated TM-score=0.61; very hard target. |
>MazG C-terminal domain (Length=188) VPLFDESFPEHEQLPREFEIEFIERGGRENGHVVQRLCGVYIGDRLTDNSNEADDYRFHDVFHLAYAAYLGWSPVLRALL KRKRKSNPSKDENEDGARAMIIEEGIATWIFDHAKKHDCYETVEVGSLDYGLLKQIQSMVRGYEVDQCKLWQWELAIIEG FKVFRQLRDNRGGTVKVDLTNGSLTYVA |
| 2387 |
PF18549 (NitrOD1) |
 Estimated TM-score=0.61; hard target. |
>Nitrosopumilus output domain 1 (Length=68) SNAVLIRYKLKGDKQYTTCVVTRIQYENFKILPVVKECEIVQRYVGITGDQIEHANQTLGEAIRKEIK |
| 2388 |
PF18165 (pP_pnuc_1) |
 Estimated TM-score=0.61; hard target. |
>Predicted pPIWI-associating nuclease (Length=139) QLLSNQFELDLFEASIASLNDRTNKLRFNNFAYSIRELSRHFLYNLAPEERVKNCVWFSPETADGKPTRNQRIKYAVQGG IDDTFLDRWGFEVDELKDMIKDVKETINTLSKYTHINPEVFNISDSEIERMSQDVLNSF |
| 2389 |
PF17486 (Cys_Knot_tox) |
 Estimated TM-score=0.61; trivial target. |
>Cystine knot toxins (Length=70) NSGYGIPHIVEKLPNGQWCRTPGDDCSESKQCCKPEDTATYAHGCSQQWSGQRGELVKMCYICNKESSMC |
| 2390 |
PF17434 (DUF5413) |
 Estimated TM-score=0.61; hard target. |
>Family of unknown function (DUF5413) (Length=133) MKRYLIFVAIGPALGGFCLLLVTTYQSGYWTETNAAEVAKLFKVFAISLQYSYLFGFLPALMMCAIDDILFHVRRIGPVL RMLLVGAIAFLLAAFNYSSHGADYGVAQFILYGLVGFIPATISSWLVHKFAEE |
| 2391 |
PF17306 (DUF5355) |
 Estimated TM-score=0.61; easy target. |
>Family of unknown function (DUF5355) (Length=326) IEEEIAIALINVAVFYQDIGIETLYRAYESSQASNNLWTTSGTYLKRGLGLICFLGKNFQINTANDCQKMQVLNVLNQLS LEFQLLQQLGIVVLALSKLRSKISKDAVADLEPQELEELGKSSVFYAKLCIGSYSTASQCQGGRIVDALFMNYLQSLTYL FLSINQYNNDECGIAIGMLQESIKKLLNIVPNSQLKELDILSSTDITKKRDLIKMSFKRKIHGSTLKNQRIFEKKVPFSS KAYMMPLLKSSLDDFVIPLTILLRYRYQTTNENYSFKTVETDVSKLKELFPRGKSSDIEGTVWSFQDGHLTFADSNNATH NCGNYF |
| 2392 |
PF17293 (Arm-DNA-bind_5) |
 Estimated TM-score=0.61; hard target. |
>Arm DNA-binding domain (Length=85) LCYKSKTLANGEHPLMIRVCKDGKKKYVSLGISINPKYWDFNKNKPKPNCPNRQEIEKLIINKSKTYNDQIIELKAEDKD FTAKT |
| 2393 |
PF17274 (DUF5339) |
 Estimated TM-score=0.61; hard target. |
>Family of unknown function (DUF5339) (Length=68) TETCNAYFAEVDSLIAKASENPQAKTQLDAMKGQLEEGKKQVAALPKDQQDKACQQGIDAMKQMKAAL |
| 2394 |
PF17248 (DUF5317) |
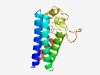 Estimated TM-score=0.61; hard target. |
>Family of unknown function (DUF5317) (Length=149) NMKLRGGWLFPLLLAVQFIVFFLQERIEWIAKMSNVIFLLVYITGLAFLWLNRHQPGFPVIFAGVLLNFIVMAVNGGRMP VSIEAAQFLGREYIEALKAGLYGKHQAITSDTLLPFLGDIIPLSPPYPRQQVISIGDVVMNVGAFLFIQ |
| 2395 |
PF17112 (Tom6) |
 Estimated TM-score=0.61; hard target. |
>Mitochondrial import receptor subunit Tom6, fungal (Length=45) PKQPKSKFEQFKETPAYTVLLNGTLFVAGICFIQSPLMEMLAPQL |
| 2396 |
PF16947 (Ferredoxin_N) |
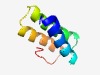 Estimated TM-score=0.61; hard target. |
>N-terminal region of 4Fe-4S ferredoxin iron-sulfur binding (Length=65) HPLGESWQDDLEEMLDDTEYDTELGMEMGRDAMRVTKGELSEAEFHERYHDDVVAEFGEDDRPIA |
| 2397 |
PF16887 (DUF5081) |
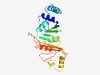 Estimated TM-score=0.61; easy target. |
>Domain of unknown function (DUF5081) (Length=230) IINENSFSVSELFLLAEACGTSSVFGLPDKKVQLLLNSTMMQDAGETLQEKGILSEDETLTEGGYYVLSALNSYINSSQY TRINNCMVGFLPDDQENLVLLVEVEANKKYQLKIIDKLVLLKNLFDESPLLQREPLEDETEFILKKMRNSQRRIAEKLEL KTNLLNIEIFKGELYKEEVDSAEYHEQWLMYQDQGELYAVDFNEPKYYRASQYWLMKFVFDSLAIPYGAA |
| 2398 |
PF16711 (SCAB-ABD) |
 Estimated TM-score=0.61; easy target. |
>Actin-binding domain of plant-specific actin-binding protein (Length=43) LKDIVAQETQQLSDQHKRLSVRDLASKFDKNLSAAAKLSNEAK |
| 2399 |
PF16506 (DiSB-ORF2_chro) |
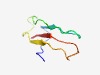 Estimated TM-score=0.61; hard target. |
>Putative virion glycoprotein of insect viruses (Length=51) SPYMFDRSCLNVYRTNDYLFGECTLPPNCSEPSVVKLDKTFYGQETVVCHS |
| 2400 |
PF16271 (DUF4924) |
 Estimated TM-score=0.61; hard target. |
>Domain of unknown function (DUF4924) (Length=178) MYIAQELRKKSIAEYLLYMWQVEDLIRAYGCSLTRIRHEYIDRFQYTDEQKEEEIDWFGNLVRMMNEEGCREKGHLQINK IIVQDMTDLHNQLLQSPKFPFYNAEYYKVLPFIVELRNKGDKEVNEIETCLDALYGVMLLRLQKKMISPDTEHAIKEITT FIGMLSDYYQKDKTEGLK |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218