

Go to page (83 pages in total): [First page] << < 9 10 11 12 13 14 15 16 17 18 19 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 1301 |
PF10772 (DUF2597) |
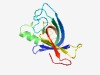 Estimated TM-score=0.68; easy target. |
>Protein of unknown function (DUF2597) (Length=134) MNFDITVGDMQVHVETVTLDITDNSAVSQTGGVPDGYVDGDVAASGEIEVDTTNFNLIVEAAKAAGSFRQLEPFDSLFYA KTPTDELRVEAFGCRLKISSLLNIDSKGGEKHKHKIPFDVTSPDFIRINGVPYL |
| 1302 |
PF10724 (DUF2516) |
 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF2516) (Length=91) NLQNGVLLILGVAAFGLCLWALVDCLRRPAGAFVAAGKRTKTFWTVIVAVATAIGFVSIFSILGIFGILAVVGASVYLAD VRPAVRQYGGG |
| 1303 |
PF10658 (DUF2484) |
 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF2484) (Length=77) MTLSLILACLWVVAANVLAMVPSRDNHWSRAYLLIAAGIPLVGYVTYENGPWWGLGVLFAGMSMLRWPVIYLGRWLR |
| 1304 |
PF10455 (BAR_2) |
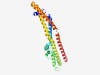 Estimated TM-score=0.68; trivial target. |
>Bin/amphiphysin/Rvs domain for vesicular trafficking (Length=270) LPSLAQSTQRMVQERLGQVTDISQLPREYTELEDKVDTIKLIYNHFLGVTAIYENGSYDYPKYINESVNEFSRSVASKLT ELTHATSASEAQNILVAPGPIKEPKTLNYALSKVALNSSECLNKMFPTEEQPLASALLQFSDVQAKIAQARIQQDTLIQT KFNKNLRERLSFEIGKADKCRKDVHSMRLRYDVARTNLANNKKPEKEASLRVQMETLEDQFAQVTEDATVCLQEVISHAN FSEDLKELAKAQAEYFETSAGLMKEFLSNS |
| 1305 |
PF10441 (Urb2) |
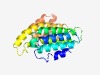 Estimated TM-score=0.68; easy target. |
>Urb2/Npa2 family (Length=212) RVCRILYTILEKKPQSMAQWNIESTLGAVCDLSSPKKASQTTVPYAWLCKLVEAIIRKHRLRLEGHHHMLMTTMQALLRN LIVSQAVSDAKDQPIQEAKAHCYARLVTLICEPTAGAVSRSQHPGSLDSATDAAKRSAGRHMYLVLMQYVKLQLEGDVAR PVREALEPAMNSVFDITPPEVRKILNDAMDGSGRAILREMYKRYTKFGKWSG |
| 1306 |
PF10302 (DUF2407) |
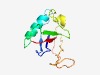 Estimated TM-score=0.68; easy target. |
>DUF2407 ubiquitin-like domain (Length=106) LLTVRFSASIPDLPLDIQNPDITTAAGLKQLIRTHLPPNLSSHRLRLIYAGRGLEDATPLSVSLKLPPSPSRTPVAQDDA TTAKGKGKAPVREQPRLYIHCSIGDI |
| 1307 |
PF10274 (ParcG) |
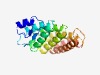 Estimated TM-score=0.68; easy target. |
>Parkin co-regulated protein (Length=184) TAFRKFYERGDFPIALEHDSKGNKIAWKVEIEKLDYHHYLPLFFDGLCEMTFPYEFFARQGIHDMLEHGGNKILPVIPQL IIPIKNALNLRNRQVICVTLKVLQHLVVSAEMVGEALVPYYRQILPILNIFKNMNVNSGDGIDYSQQKRENIGDLIQETL EAFERYGGEDAFINIKYMVPTYES |
| 1308 |
PF10084 (DUF2322) |
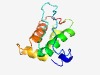 Estimated TM-score=0.68; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2322) (Length=98) FADNLKKLPGISHLAAINLLDADGNAVALLENKPGSAGSVAVYNHLAQTYGAITPQAAQKGLEIYAEHTDDARAHPGKHP NIDRLLQIIETGITLRIK |
| 1309 |
PF10067 (DUF2306) |
 Estimated TM-score=0.68; hard target. |
>Predicted membrane protein (DUF2306) (Length=147) FIHITTSVVALVIGPFTLSKKFREKNINSHRIVGRIYIVGILLGGISGLYLAFYATGGLVAKLGFGLLSVFWLTSAYQAL IRIKNKKIQDHRNWMIRNYSLTFAAVTLRIWLPLFVVLFGLEQFERSYAVISWLAWVPNLIIAELFI |
| 1310 |
PF09976 (TPR_21) |
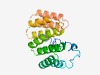 Estimated TM-score=0.68; easy target. |
>Tetratricopeptide repeat-like domain (Length=194) KAWWNKYGNLITWVLIIALGGFAAWNGWKTYQRKQAAQASVLYEEVQKAATAKDSAKVLRAAGDLKDKFGGTAYAQMGAL SAAKVAFESNDLKTAKAQLEWVVKQGKDEEYRSVAKIRLAGILLDEKAYDEGLKLLAGDFSPAFAADVADRRGDILVAQN KLEEARTAYRTALEKMDEKNPGRQLVQLKLDAIG |
| 1311 |
PF09964 (DUF2198) |
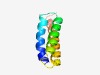 Estimated TM-score=0.68; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2198) (Length=72) WFLLALIVPALLVILFTRVTYNHYVGTLLTVALLVASYVKGYTDQLYEIVADIASLIVGFLYAKRMVEKQKQ |
| 1312 |
PF09852 (DUF2079) |
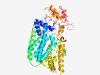 Estimated TM-score=0.68; easy target. |
>Predicted membrane protein (DUF2079) (Length=485) RYYSFYASYDQGIFNQVFWNSMHGHFFQSSLSSALSTNVVHQGQVSEVYYHRLGQHFTPALLLWLPIYVLFPYPITLTVL QVTLISVSGLVLYILARQHLQPQLSAMMSISFYGANAVIGPTLANFHDICQIPLFVFSLLLAMEKRWWSLFWILAVLTLA VREDSGVVLFGVGFYLILSKRYPKIGLAICTLSFSYMIALTNLIMPVFSEDISQRFMMERFGQYTEGDQASTLEIIWGMM SNPWRLIKEIFTPFFGTLKYLLGQWLPLAFVPAVAPAAWMIAGFPLLKLFSAKGLSVLSITIRYAMTVVPGLFYGAILWW GKRESEVKNSPVPSPKFLRFWVVCICLSLFFTFTSNPNRTFYFLVPDSVKPWVYIPANEQWQHVSQMRPLLDKIPDDASV AATTYIVPHLSSRRAILRFPRMQFRNDAKKIEKVQYIIVDLWRLNRYRVAFKSDRQRLETIVPRIEELYNSGEYGITDFG DGVVL |
| 1313 |
PF09834 (DUF2061) |
 Estimated TM-score=0.68; hard target. |
>Predicted membrane protein (DUF2061) (Length=52) MKKTLSFAVIHFTVAFTVAYILTGDILLGSLIAMVEPMVNTVAFYFHEKAWD |
| 1314 |
PF09820 (AAA-ATPase_like) |
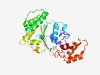 Estimated TM-score=0.68; easy target. |
>Predicted AAA-ATPase (Length=276) PVGIQSFEKIREGGYCYIDKTSLIYSLIKSGQYYFLSRPRRFGKSLLISTLEAYFLGKKELFKGLAMEGLEKDWTTHPVL HLDLNTQKYDTPEALNNVLEENLNYWESLYGASESEIGVARRFNGIIRRAAEKAGQKVVVLVDEYDKPMLQAIGNEPLLT DYRNTLKAFYGALKSNDKYLRFALLTGVTKFSKVSVFSDLNNLMDISLSNRFANICGITENELYRNLEEDIRLLAEANGM NETEARQTLKEWYDGYHFAENTEDIYNPFSLLNTLA |
| 1315 |
PF09799 (Transmemb_17) |
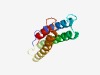 Estimated TM-score=0.68; hard target. |
>Predicted membrane protein (Length=107) ILMYLNSFYFGLYAAFKVCISVLKPIYLQYAENVLTREASILLAMCILETIRIILGRRSSLSDRAWQAIASVILTLPSLA IVIYLCCFQTYVLKLEIIMSALMIALQ |
| 1316 |
PF09605 (Trep_Strep) |
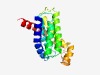 Estimated TM-score=0.68; easy target. |
>Hypothetical bacterial integral membrane protein (Trep_Strep) (Length=183) TKDLITTGIFTAIYFVVLFACGMLGFIPILFILLPFIAPIVTGIPFMLFLTKVKKFGMVTIMGLLLGLIMFLTGHTFIPI ITGLVFGLIADLIFKAGNYQNFKNSVLGYAVFSISVVGYMIPMWVMRDSYFEYIRSSMGTEYVNTVMALTPDWMLFVVIL VSFIAGIIGALLGKSLLEKHFKR |
| 1317 |
PF09527 (ATPase_gene1) |
 Estimated TM-score=0.68; easy target. |
>Putative F0F1-ATPase subunit Ca2+/Mg2+ transporter (Length=51) GNAATIGTHMVVSTFVGMGIGWYLDKWLGTKPWLLLVFLCFGIAAGFKNVY |
| 1318 |
PF09446 (VMA21) |
 Estimated TM-score=0.68; hard target. |
>VMA21-like domain (Length=63) AIKNLIRFTAAMFVLPLLTMFTTYHYIFRDYFHLPPDQAMLYAGMCGVAVVFIIIIVFIYVAY |
| 1319 |
PF09419 (PGP_phosphatase) |
 Estimated TM-score=0.68; easy target. |
>Mitochondrial PGP phosphatase (Length=171) VSATLNVFRLLVQPQLCLPHLTVSSFDKIPVPLFFSRYQTNHPQIKAIVLDKDNCFAKAHDDKVWPAYNEQWEQLRKAYP DASLLIVSNSAGTNDDKGHEQAKILEDRTGVTVLRHTTKKPGCHEEIMDYFRKKGIVEGPHQVAVVGDRLFTDMLMANMM GAWGIWVRDGV |
| 1320 |
PF09335 (SNARE_assoc) |
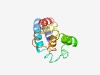 Estimated TM-score=0.68; hard target. |
>SNARE associated Golgi protein (Length=119) IPAIPLTMSAGLLFGSLTGTILVSISGTVAASVAFLIARYFARDRILKMVEGNKKFLAIDKAIGENGFRVVTLLRLSPLL PFSLGNYLYGLTSVKFVPYVLGSWLGMLPGTWAYVSAGA |
| 1321 |
PF08902 (DUF1848) |
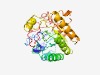 Estimated TM-score=0.68; hard target. |
>Domain of unknown function (DUF1848) (Length=262) ILSVSRRTDIPAFYSDWFFNRIKEGFVLVKNPFNSKQVSKVILNPKVIDCIVFWTKNPKKMMKRLDEIKEYNYYFQFTLN SYDKTLETNVPEKKYLVNTFIELSKRIGKDRVIWRYDPIILTDKFTKDYHYKWFEYLAKRLSPYTNKCVISFLDLYRKTE RNLKEINILPIDKDDMFELAEKFSKIASKYNVTIETCSEEIDLSKFNIKHGKCIDDRLISKVVDEKLSIDKDPNQREVCG CVKSIDIGAYNTCNHGCLYCYA |
| 1322 |
PF08891 (YfcL) |
 Estimated TM-score=0.68; hard target. |
>YfcL protein (Length=84) DFEARILAQIDEMVEHASDDELFAGGYLRGHLTLAVAETEQNGEHTLEALHSRVQESLERAISKGELSPPDQILVKTMWQ TLYQ |
| 1323 |
PF08889 (WbqC) |
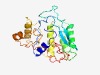 Estimated TM-score=0.68; hard target. |
>WbqC-like protein family (Length=215) QPSYLPWLGFFEQMTRSDKFVLLDDVQYTRRDWRNRNKVRVKEGWVWLTVPVQQKSRFSQSLLETRIDNSVSWRRKHLET LRQHYCKAPFFEKYFPRCQQIYEKDWEFLFDLCLETIQFLKEEMGIETPLLRSSEMKLSGEKTERLVSICRELGATHYLS GESGSDYISQEDFSNQGIELEYQNYEHPVYPQRYPGFVPHLSTIDLLFNCGEQSL |
| 1324 |
PF08873 (DUF1834) |
 Estimated TM-score=0.68; easy target. |
>Domain of unknown function (DUF1834) (Length=198) LIRTENAIIERLRQGLGQLVREVGSYGGELDESLPDAIRRFPAAWVTFGGISQTVAHGTSRQKFRATGQFVVMVADRSVR GEAAGRHGGAGPGEVGSYPLVYAVRRLLSGQDLGLPIDALLPGRVRSLSGVQTSAQALSVFACDFQTAWIEDALPRGHWP SPQHPAEPDRIFAQYAGRLEQPAPDWLRTGLDYHLTPD |
| 1325 |
PF08862 (DUF1829) |
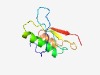 Estimated TM-score=0.68; easy target. |
>Domain of unknown function DUF1829 (Length=87) GKSGYVHNFDFVIPASKKSPERIIKTINNPNKNNTTSVLFSWSDTRENRTLSSKMYVFLNDTEKEVKPEIINAYIQYQIK PVLWSKR |
| 1326 |
PF08345 (YscJ_FliF_C) |
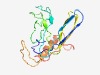 Estimated TM-score=0.68; easy target. |
>Flagellar M-ring protein C-terminal (Length=162) LEPMVGNNGVRAQVTADLDFTRTERTEETYNPDLPALRSEQVSEEQMGAGGGAVGVPGTLSNTPPGPQAEGAGMSQSNSS RRSTRNYELDKTISHTRLATGTIRQLSIAVVIDDKRVPGPDGTATREPWTEEELDKFTTLVREAVGFDARRGDSVQVINA AF |
| 1327 |
PF08318 (COG4) |
 Estimated TM-score=0.68; easy target. |
>COG4 transport protein (Length=322) LQQATMQLRHLITQKFDESVKKDDLASVERFFKIFPLLGMHDHGIEKFSQYICHKLNAKAQKELRTSMDTAKAEKRTVVA FADTLTILLENIARVVEVNQPIIETYYGKGRLHQMVRVLQEECDQEVKICLLEFNKGRHIQRRVAQINEYIKTGGSGGSS VAGHYRKPSGGSMDKLNPKDIDSLIGEITIMHSRAELYVKFIKRRVARDLDSSTLDEEKKRTALAGVEKCLKDSDLSRQM QELLGIYLLFERFFMEESVLKAIALDAHEPGQQCSSMIDDVFFIVRKCIRRANTTQSLDGICAVINNAATELENDFIAAI KE |
| 1328 |
PF08235 (LNS2) |
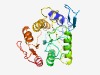 Estimated TM-score=0.68; easy target. |
>LNS2 (Lipin/Ned1/Smp2) (Length=229) KSLRLTSEQISKLVRQSRVKLQPGQNTISYSVTTQYQGTSKCVSHIYLWRYDDKIVISDIDGTITRSDVLGNILPMLGKD WSQMGVAKLFNKIHLNGYKFLYLSARAIGQSSLTRSYLKSIRQGELCLPDGPMLLTPSSLINAFHKEVIERKPEEFKINC LKDIASLFPATRPCPFYSGFGNKINDVWAYRAVGIPISRIFSINHRGELRSEISKTFQTSYNSLXLLVE |
| 1329 |
PF07947 (YhhN) |
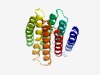 Estimated TM-score=0.68; hard target. |
>YhhN family (Length=191) QAFFKTCPIWYLVFFVIDMLQQTPPQIGNNFDDFRLWIAMGLLISSLGDVCLVWKDMLFIPGILFFGLAHTCYIVSFNLR GTNPHGNILLAEISIPIVIFCLTYILSGIDTWAMTILVALYTFVLLWMGYLSIRRYEHIQTPANLCGMVGGSCFLLSDFM LGVEKWQCCLPLPDGLCIMTYYTAQLGLACS |
| 1330 |
PF07865 (DUF1652) |
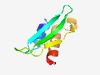 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF1652) (Length=67) LELRRIVETSFLPLHCQCTVGHDGAMTVRICDPASGRVDLLVTGIATRGLTSSRALSELIAELRYDL |
| 1331 |
PF07618 (DUF1580) |
 Estimated TM-score=0.68; easy target. |
>Protein of unknown function (DUF1580) (Length=57) PLLDAIEQETGHRTHPSTAMRWSLKPNRHGNVLESWMIGGRRMTSVEAVRRYIEANT |
| 1332 |
PF07592 (DDE_Tnp_ISAZ013) |
 Estimated TM-score=0.68; easy target. |
>Rhodopirellula transposase DDE domain (Length=308) ELDALVDPATRGDSQSPLRWTCKSTPRLAKELKERGHAVSRRTVCRLLAQLGYSLQSTRKTREGGKHPDRDAQFQHIASE VEQYQSAGDPVISVDTKKKELIGDFKNAGQEWQPQGQPEEVRVYDFIDSALGKVAPYGVYDLTANQGWVSVGIDHDTAEF AVESIRRWWNEMGKPLYPKARRLLITADCGGSNGYRVRLWRVQLQKLADELEIAIQVCHFPPGTSKWNKIEHRMFCHITQ NWRGRPLLSREVVVNLIGNTTTEAGLRIHAEVDGNTYEAGIKVSDQELATLAIERDEFHGEWNYKIIP |
| 1333 |
PF07532 (Big_4) |
 Estimated TM-score=0.68; hard target. |
>Bacterial Ig-like domain (group 4) (Length=57) DKVEVTTVAGVKPDLPYQVTAKLSDGSSKDVDVTWPEIDASKYAKAGTFTVEGTVEG |
| 1334 |
PF07387 (Seadorna_VP7) |
 Estimated TM-score=0.68; easy target. |
>Seadornavirus VP7 (Length=307) VQKNCFQVYKSQNVYRVRFCVEGKIHDVPLPGVNTHRDIIGTVKYFCELMGLNIVDNSLRLNWLNLSDMKNHGDNVTLAK VGSEYGDLFVKKLPTLPINIHSTNYVFGKYSVFARVHGIVKLLNDDNYDVGVILEKCNEIRVMTVNIIVMGLKSLMDSHR ESGGTLHGDCTVQNLMCDKFGMLKLVDPGNLMSQTVIWKNDTQYFGNTVDDEVCSFVMACLKTTANLRNIETSELTIPFI TDLYLLDTVLSDPNVIHGTRESGWLEVGSDFVSIVQMFVALANMVRPYNLDDVVSALKTQVTGENET |
| 1335 |
PF07198 (DUF1410) |
 Estimated TM-score=0.68; easy target. |
>Protein of unknown function (DUF1410) (Length=58) QYKNREVSSVFVDENGKEYIVSATVDANGKVSFDTSSLPKPGNYNLSKVVDKNTNEQI |
| 1336 |
PF07197 (DUF1409) |
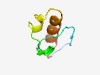 Estimated TM-score=0.68; easy target. |
>Protein of unknown function (DUF1409) (Length=48) RLDYPIDTLINNAGSIRSRIEEIQDRLPDDLIDAIAPAGYIESHRFPM |
| 1337 |
PF07158 (MatC_N) |
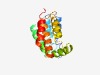 Estimated TM-score=0.68; easy target. |
>Dicarboxylate carrier protein MatC N-terminus (Length=149) MTPQIATILGLVIMFIVATALPINMGAVAFALAFIVGGVWVGLDGKEVLAGFPGDLFLTLVGITYLFAIAQKNGTIDLLV HWAVRAVRGHIVAIPWVMFVITGLLTAFGALGPAAVAIIGPVALRFAKQYRINPLMMGLLVIHGAQGGG |
| 1338 |
PF07000 (DUF1308) |
 Estimated TM-score=0.68; easy target. |
>Protein of unknown function (DUF1308) (Length=163) LNLDVTAMLAYVSGLTNGRCNFKFKEPVLTQQAEWERAKPVKPVLDQLFEGKSLFCCESAVKDFQSIVSTLGGPGEVSRA NELLARITVIPDMTSLRAQAQLKLGGKIKQRSLAVFGTGDAVRAVTVSANAGFVRAAKCQGVEFAVFVHESRALTEGKEH DAR |
| 1339 |
PF06993 (DUF1304) |
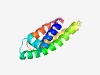 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF1304) (Length=109) GLVAVEHIYILYLEMFAWTKPSTAKSFGLTPEFAQQTKSLAANQGLYNGFLAAGLIWGLVHPNPDFGLQIQLFFVCCVVV AAIYGGLTAKRSILFVQGGPAIIALVALL |
| 1340 |
PF06974 (DUF1298) |
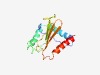 Estimated TM-score=0.68; easy target. |
>Protein of unknown function (DUF1298) (Length=145) WGNALGFIILPFFIGVHKDPLDYVRKAKKVVDRKKSSLEVVFTHLAAEVILKLFGLKAAAAIFHRMISHTTISFSNMIGP VEQVEFCGHPVVFIAPSGYGPPEALTVNFQSYVNTMMVNLAVDEAQFPDCHELLDDFSESLRQIK |
| 1341 |
PF06848 (Disaggr_repeat) |
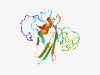 Estimated TM-score=0.68; easy target. |
>Disaggregatase related repeat (Length=180) DNGLRTSSPTSAFPTINYIDIGRNSASCRDLMMFDLSNYKTTDVISKATLSLYWYYPAGKTRTSDTVVQIYRPAADWDPR YVSWTNRASSTKWTTAGGNWFDKNNVAQGATPYASLTFAGSKVPDNKYYDFDVTQLVQEYVSGKYKNTGFFLKAKTESGN YIAFYSSEWTNAAQRPKLTV |
| 1342 |
PF06685 (DUF1186) |
 Estimated TM-score=0.68; easy target. |
>Protein of unknown function (DUF1186) (Length=253) QVSNALFELTINSGIFPREALTKAIELREQVTPRLLGYLETTEKYAYEQRDNDNLMLHIYALYLLAQFREQRAYPIIINF FSLPGKISLEITGDVVTEDLHRILASVYDGDDTLLKNLIEDEQVNEYVRDAGIRTFVTLVVCGQKSRDEVIAYLKTLFGK LERKPSFTWNSLVYVCMDLYPEEVDAEIRQVAKEGLTEEVLLTKEINILGLHEEALKKSYALGKEKTLEKLQNDDHYTLI DDTIRELKDWSCF |
| 1343 |
PF06348 (DUF1059) |
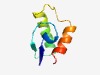 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF1059) (Length=57) KYTFKCADVGMDCGFEIVNAGSEDELLEALKSHAKMSHGLTSIPPDLVNKIKQNIKK |
| 1344 |
PF06282 (DUF1036) |
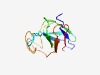 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF1036) (Length=114) AEAGLRICNDTAQPQMVAVGYLRDGGWVSLGWWKIEPQGCAVTVDGDLQNRYYYYRAEIDAGPFNGNPEFTFCTDPKVFE ILGDDNCEARGYEAETFAEIDTGPSATDFTFRLT |
| 1345 |
PF06168 (DUF981) |
 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF981) (Length=204) MVFIDDLALMLFTLPLISILIAYTTIQSYLGFRKEGYQAIKKSLDDSIIPAGILGLVTLIISLWGEFTWTLPGSYNILFY DVYLLMGIIILSYSMAVYMGKRLQTTGIFALFVGLITIYYGVIGFNLGLTQEPLALLGLYVTYGLAGILGYPVTVFLDRL KEQPSERRNLGSWIIVFVVFWIIVIVAGILGAVIGVETIPAHLA |
| 1346 |
PF06159 (DUF974) |
 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF974) (Length=234) LTLPQNFGNIFLGETFSSYISVHNDSNQVVKDILVKADLQTSSQRLNLSASSAAVAELKPDCCIDDVIHHEVKEIGTHIL VCAVSYTTQTGEKMYFRKFFKFQVLKPLDVKTKFYNAESDLSSVTDEVFLEAQIQNITTSPMFMEKVSLEPSMMYNVAEL NTVDTAGESESTFGSRTYLQPMDTRQYLYCLKPKQEFAEKAGVIKGVTVIGKLDIVWKTNLGERGRLQTSQLQR |
| 1347 |
PF05691 (Raffinose_syn) |
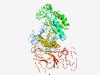 Estimated TM-score=0.68; easy target. |
>Raffinose synthase or seed imbibition protein Sip1 (Length=737) LSDGKLTVFGNSILHHVHHNVVLTPASSDNANANGAFIGVTSDHKGSTTLFPLGKLEGLRFMCVFRYKFWWMTQLMGASG KDIPFETQFLLVEVPGHPDGSRDEAQPSVIYAVFLPIIEGDFRAVLQGNEHNEIEICLESGDPGVDKFEGSHLVFVSAGT DPYDVITNSVKTVEKHLQTFSHREEKKMPDILNWFGWCTWDAFYTNVTAEGLKQGLESLDKGGTPPKFVIIDDGWQSVGM DPTGIEYRSDYSANFAERLIHIKENHKFQKDGKEGHRVDDPALGLGYVVSEMKERHNLKYVYVWHAITGYWGGVKPGIVE MEPYEPKLQYPIPSPGVQSNEYCEVLQSITRNGVGLVNPEKVSDFYNDLHSYLASAGIDGVKVDAQSILETLGAGHGGRV KLTRKFYQALEASISSNFHDNGIIACMSHNTDTLYSAKKTAVMRASDDFFPRDPASHTIHIASVAYNTVFIGEFMQPDWD MFHSLHPMAEYHGAARAVGGCAIYVSDRPGHHDFDLLKKLVLPDGSVLRAKLPGRPTRDCLFSDPVRDGKSLLKIWNVNE FTGVIGVFNCQGAGWCGVRKKMVSHEDQPGAGNAIISGVIRARDVEYLRQVAVAEAEDGCTWTGDTILYSHRAGEVAYVP RNASVCVTLKPREYEVYTVVPVKVALNGAKFAPIGLIKMFNSGGAIKELKYDDQGTSIRISMRVEGCGVFGAYASTQPQT TLLDSHQVEFEYEGGSD |
| 1348 |
PF05589 (DUF768) |
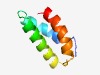 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF768) (Length=64) MSTRGINFLDQWIANNIPETTKADVISVDELTHKLIADAKALGIKRGEIDEEVDSLYRTILDAI |
| 1349 |
PF05232 (BTP) |
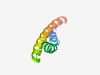 Estimated TM-score=0.68; hard target. |
>Chlorhexidine efflux transporter (Length=64) RTLKERILHAVGFEAVALAIVAPTAAWLMDKPLFQMGALAIMLSTVAMLWNIIYNAGFDRLWPR |
| 1350 |
PF05128 (DUF697) |
 Estimated TM-score=0.68; easy target. |
>Domain of unknown function (DUF697) (Length=158) LQSQRLGEEARRLIDTQRRRQAEKVVERFQWIGAGVIAVTPLPVVDVLATAAVNAQMVVEIGKIYGCELNLERGQELAMS LAKTLAGLGIVEGAIKLISTALQLTVATFLIGKAIQGVSAAYLTRIAGKSFIEYFRQNQDWGDGGMTEVVQRQFQLVK |
| 1351 |
PF05125 (Phage_cap_P2) |
 Estimated TM-score=0.68; easy target. |
>Phage major capsid protein, P2 family (Length=333) KFNAYMSRVAELNGVETGDMNKKFSVEPSVAQKLMTRVQESSAFLTRINIVPVPEMKGEKIGVGVSGSIASTTDTAGGDE RETADFSALDSQGYECAQVNFDFHIRYNTLDLWARYDDFQTRLRDAIIQRQALDRIMIGFNGTRRAKTSSRAQNPMLQDV AVGWLQKYRNEAPDRVMSKVTDEEGTVISAKIRVGKDGDYANLDALVMDATNNLIEPWYQEDPELVVIVGRQLLADKYFP IVNQSQPNTEQLAADVIVSQKRIGNLPAVRVPYFPPNAMMVTRLDNLSIYWQEGTQRRHIEEVPKRDRIENYESANEDYV VEDYAAGCLIENI |
| 1352 |
PF05117 (DUF695) |
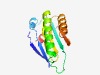 Estimated TM-score=0.68; hard target. |
>Family of unknown function (DUF695) (Length=132) DWASYFCRVEDKPASIRLNLALHDCAPIQNYKHRIWFSIKLLNPDENGFTTREEFPVICTIEDAIAEALEKIGAIFVGAL KTNGTFDMYSYSKNVENLDEVIHGVMKNYSDYNYATDTKEDTQWDTYFNFLY |
| 1353 |
PF04982 (HPP) |
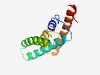 Estimated TM-score=0.68; hard target. |
>HPP family (Length=127) LIGSFGAMSILVFGAVEAPLAQPRNAILGNTLSAFIGVAIAKVLHTAEDVDEYMWLACALSVSLSMVAMQLTDTVHPPGG ATALIAVTSPQDIQNLGFFYAVFPVFVGSCILVGVSVIFNNIQRQFP |
| 1354 |
PF04666 (Glyco_transf_54) |
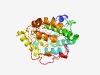 Estimated TM-score=0.68; easy target. |
>N-Acetylglucosaminyltransferase-IV (GnT-IV) conserved region (Length=289) TNRLAESLKLPTAFHFLPHLLDDSSSLQPSYIESKKRQGVSIVLGIPTVKRDKQSYLVETIDNLIANMDEEEQNDTMIVV YVGETDLEYVKLVAHQIRTRFASYIDSGFLEIIAPAASYYPNMNNLRQTLNDSQERVRWRSKQNLDFAFLMAYAQPKGAF YLQLEDDILTKKGFVTIMKNFALEKTAKKEHANWFVLDFCQLGFIGKMFKSADLPWLITFFQMFFNDKPVDWLLYHLIYT KVCSVEKDGKSCKQEMSKLWIHYKPSLFQHIGTQSSLRGKVQKLKDKQF |
| 1355 |
PF04664 (OGFr_N) |
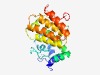 Estimated TM-score=0.68; hard target. |
>Opioid growth factor receptor (OGFr) conserved region (Length=206) KGDMTNLCFYKNEICFQPNGLYIEEILHNWKDNYVVLEENHSYIQWLFPLREPGVNRHAKPLTLKEVEAFKSSQEVKERL VRAYELMLGFYGIQLEDRDTGAVCCAQNFQPRFQNLNSHRHNNLRITRILKSLGELGLEHYQAPLICFFLEETLVQHQLP SVRQSVLDYFMFAVRCRHQRRELVYFAWEHFRPRCQFVCWPRDKLQ |
| 1356 |
PF04504 (DUF573) |
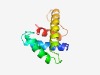 Estimated TM-score=0.68; easy target. |
>Protein of unknown function, DUF573 (Length=94) KMIWTRNNELIILAGIVDYESEKELSYNDDWDALYNFIKGSIELKFSKKQLIDKIRGLKKRFMDNMVKSEDGKGPSFTNT EDDEIFKLSMIIWG |
| 1357 |
PF04492 (Phage_rep_O) |
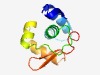 Estimated TM-score=0.68; easy target. |
>Bacteriophage replication protein O (Length=94) VADLDDGYTRIANELLEAVMAADLTARQLKVVLAVIRKTYGFGKKFDRITNTQIAMMTGIHHTHVCKAKNEMIAMNIIIS NGQTIGVNKVISEW |
| 1358 |
PF04343 (DUF488) |
 Estimated TM-score=0.68; hard target. |
>Protein of unknown function, DUF488 (Length=125) LSELIALLRENKVSCLIDIRAYPGSRTNPQFNKKNLESEIPKADIEYFHMHALGGRRHNSKGVTSSPNTFWQNKAFQNYA DYALSDAFQEALSGLKNITHKKTCAIMCAEAVWWRCHRRIVAYYL |
| 1359 |
PF04294 (VanW) |
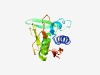 Estimated TM-score=0.68; hard target. |
>VanW like protein (Length=130) NRVNNIKIAAEELDGTIIEPGEIFSFNETVGRRTKEKGYKEATIFVDGEKSKGVGGGICQVSTTLYNAALEAGLKIVERH RHSREVSYVEKGKDAAVAYNSKDLRFKNTKDYPIEIKVTVTEDEIRVSIY |
| 1360 |
PF04066 (MrpF_PhaF) |
 Estimated TM-score=0.68; trivial target. |
>Multiple resistance and pH regulation protein F (MrpF / PhaF) (Length=52) PDRILALDTMYINCIALIILYGIWQGSNLYFEAALLIAMLGFISTVAVCKYL |
| 1361 |
PF04064 (DUF384) |
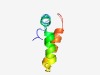 Estimated TM-score=0.68; trivial target. |
>Domain of unknown function (DUF384) (Length=54) AIMQLCATKKCREYIKNSGAYYVLRELHKSESCRPVVVACENLVDILISDEPEP |
| 1362 |
PF04019 (DUF359) |
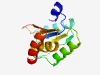 Estimated TM-score=0.68; hard target. |
>Protein of unknown function (DUF359) (Length=123) TTFHLLEAGIIPDICIVDNRTKRKPVSRDVSARNRDKVYEEVSVDNPAGIITDELIKTLCEAFASEKLLRIFVRGEEDLA TLPVILMAPLGSVVLYGQPDEGVVFVEVTEEKKEEIRALFEKL |
| 1363 |
PF03791 (KNOX2) |
 Estimated TM-score=0.68; hard target. |
>KNOX2 domain (Length=46) YKDPELDQFMEAYYDMLIKYKEELTRPIQEAMEFMRRIESQLSTLT |
| 1364 |
PF03698 (UPF0180) |
 Estimated TM-score=0.68; hard target. |
>Uncharacterised protein family (UPF0180) (Length=76) KIGVEQSLTDVQQALQERGYDVVQLNQEQDAAGCDCCVITGQDRNVMGMQDTTTQASVIDAHGRTADEICQEVEQR |
| 1365 |
PF03142 (Chitin_synth_2) |
 Estimated TM-score=0.68; easy target. |
>Chitin synthase (Length=525) SGFIHDSVVPQPPSDWMPFGFPLAHTICLVTAYSEGELGLRTTLDSVAMTDYPNSHKVILVICDGIIKGKGESKSTPEYV LDMMKDHTIPVEDVEAFSYVAVASGSKRHNMAKIYAGFYDYGTQSNIPLDKQQRVPMMVVVKCGTPDEMVKSKPGNRGKR DSQIILMSFLQKVMFDERMTELEYEMFNGLWKVTGISPDFYEIVLMVDADTKVFPDSLTHMISAMVKDPEIMGLCGETKI ANKRDSWVSAIQVFEYFISHHLAKSFESVFGGVTCLPGCFCMYRIKAPKGAQNYWVPILANPDVVEHYSENVVDTLHKKN LLLLGEDRYLTTLMLRTFPKRKQVFVPQAVCKTTVPDSFMVLLSQRRRWINSTIHNLMELVLVRDLCGTFCFSMQFVIFI ELIGTLVLPAAIAFTFYVVIISIINQPPQIIPLVLLGLILGLPAILIIITAHSWSYVLWMLIYLLSLPVWNFVLPAYAFW KFDDFSWGDTRKTAGEKTKKAGIEYEGEFDSSKITMKRWAEFERD |
| 1366 |
PF03108 (DBD_Tnp_Mut) |
 Estimated TM-score=0.68; trivial target. |
>MuDR family transposase (Length=65) DLRVGLCFKDIDGLKKAVDWCSFRGQRSCVMREAEKDEYMFECARWKCKWTLEAARMEKHGLIEI |
| 1367 |
PF02681 (DUF212) |
 Estimated TM-score=0.68; easy target. |
>Divergent PAP2 family (Length=131) ANKVLIIALSASLVAQLLKVIIETIRFKKFKFQVFFETGGMPSSHSALVSALATGVGLTQGWDSSLFAIACVFAVIVMYD ATGIRRAAGQQAQVLNQIVVEVFEDAAPDPLKELLGHTPVQVLVGAIVGIL |
| 1368 |
PF02498 (Bro-N) |
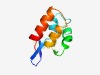 Estimated TM-score=0.68; easy target. |
>BRO family, N-terminal domain (Length=88) VRVQLDEHKRRWFNANDICAALELLNPRAALAQHVDAENVSKRAAIDTIGRTKHVNYLNESGVYALLIGSTKEAAKRFRQ WLISEALP |
| 1369 |
PF01831 (Peptidase_C16) |
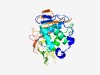 Estimated TM-score=0.68; easy target. |
>Peptidase C16 family (Length=249) AFDAVCSEALSAFYAVPSDETHFKVCGFYSPAIERTNCWLRSTLIVMQSLPLEFKDLEMQKLWLSYKAGYDQCFVDKLVK SVPKSIILPQGGYVADFAYFFLSQCSFKAYANWRCLECDMELKLQGLDAMFFYGDVVSHMCKCGNSMTLLSADIPYTLHF GVRDDKFCAFYTPRKVFRAACAVDVNDCHSMAVVEGKQIDGKVVTKFIGDKFDFMVGYGMTFSMSPFELAQLYGSCITPN VCFVKGDVI |
| 1370 |
PF01679 (Pmp3) |
 Estimated TM-score=0.68; hard target. |
>Proteolipid membrane potential modulator (Length=45) DIPKILLAILLPPIGVFLERGCDNHLIICILLTILGYIPGIIYAL |
| 1371 |
PF00811 (Ependymin) |
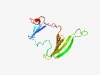 Estimated TM-score=0.68; trivial target. |
>Ependymin (Length=126) NKTFTYDVLLLYREATMYEIHQHNRTCKKMPLKVQFYPLAIPKDATLLGQAVVGSSSGPGQGLLVNSWMGDLPDKSGKYS SVVTEFGCIPVITNYQTKEYGWVMMSFFDNVIGIEDPNLLNVPSFC |
| 1372 |
PF18848 (baeRF_family6) |
 Estimated TM-score=0.67; easy target. |
>Bacterial archaeo-eukaryotic release factor family 6 (Length=150) YTYYLLALNRDSMKLYFVDNKEISEVALPDDAPIDVKTALGDELTGGNLNYSSQGSQTGSKEGVAYHGVNAKDEEVEIDW VNYYQAVDTFFKDTFNNAENYPLYLYALPENQTMFKKYAKTPYYRADAAISGSPAQLDIQKIQADLAEIT |
| 1373 |
PF17426 (Putative_G5P) |
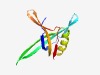 Estimated TM-score=0.67; hard target. |
>Putative Gamma DNA binding protein G5P (Length=108) MRTGFYIVGKLLGQKSTSFTHRETGEVKYKHTIGIQLQDPDGFGGYNTFTQDLRIDERSVNQGLSSSIERLKGKNVMVLV FPREWAMEGGRKGIIYHFDENSVIEEIK |
| 1374 |
PF17314 (DUF5360) |
 Estimated TM-score=0.67; hard target. |
>Family of unknown function (DUF5360) (Length=125) LKPFFLVTDIGFIIYWLITFFHLIPEEVAFKDYDNPIIIAWNWSFFPLDIFISITGLYSIYLYNKNNPAWRNWALISLVL TFCSGLQAIAFWAFSKDFDITWWAANLYLMIYPLFFIKDFIAEKN |
| 1375 |
PF17269 (DUF5335) |
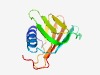 Estimated TM-score=0.67; hard target. |
>Family of unknown function (DUF5335) (Length=107) RKLERSEWESYFDQLSKKLGAKEVQIEIINEDIGDQVETWWQPLTGLSYDPKDDEFEVAAERHDHLIHKPVEIYVDEDVD GVKTVEVIQEDGTKHIIKLRSPEALPE |
| 1376 |
PF17166 (DUF5126) |
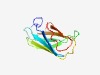 Estimated TM-score=0.67; easy target. |
>Domain of unknown function (DUF5126) (Length=102) LTPSYIEAYNTLETNETFGGIAISMQNPSASDLAIEVITPDSLNQWTTVQTNYTAAKNINITARGYKPEERKFGVIVRDR WDNATDTVFTTVTPWEEYELDK |
| 1377 |
PF17157 (GAPES4) |
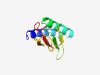 Estimated TM-score=0.67; easy target. |
>Gammaproteobacterial periplasmic sensor domain (Length=97) IQYKFSHRVQAVATAIDTHLVSNDFSALRPQITELMMSADIVRVDLLHGDKQVYTLARNGSYRPVGTSDLFRELSVPLIK HPGMSLRLVYQDPMGNY |
| 1378 |
PF17064 (QVR) |
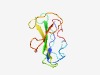 Estimated TM-score=0.67; easy target. |
>Sleepless protein (Length=92) RLWCYECNSWSDARCKDPFNESSGLQEPALLRRCEGCCVKIVTDKGTKQEAIRRTCTSNLQINLFLVDHVCMKESEGKGT MCFCESEACNSV |
| 1379 |
PF17013 (Acetyltransf_15) |
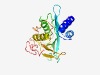 Estimated TM-score=0.67; easy target. |
>Putative acetyl-transferase (Length=214) QGIKTLDMVKDKYLEVHKAFSTKTVEDWQILQSCNNFEIFEITCDKEFIGSVYLKRCKYYSKIKSLFKTYMKEEEAVEIY SLYINDKYKGKGYSRKLLNYALNAVKNHYNLNNKFIIGLHLNEEDKYMNVSFSLYYSMNFIKSTFLESGPTDYAPIFHKS RYFIDPLKLVKNPSLCKNKGIYFGMFTRYNEFLPFNTIRNKEILKVGEQLRNIL |
| 1380 |
PF16459 (Phage_TAC_13) |
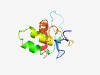 Estimated TM-score=0.67; easy target. |
>Phage tail assembly chaperone, TAC (Length=104) DSLKEMGAFTGAPVEKEITWKQGKKELTATVYVRRLSYRTAVSDIQAMKNNSDAVAGRIAASICDEKGKPIFTPEDITGD ADPDRGPLDGNLTMALLSAIGEVN |
| 1381 |
PF16379 (DUF4989) |
 Estimated TM-score=0.67; easy target. |
>Domain of unknown function (DUF4989) (Length=293) CQDDEVVDLVKYPVNQPAITIDDAEGASKATLTAVYKSDGTLELDGPVTRTYTFHFAASPEDATVTFDIINTNIPKENVE ISATKVVLPAGSTDASVTVTLKDEDFSFAASNYDATTYELGVKASVEGYKIGTEPIQSKVVIEKEAYTASCSVVGESGNN VTFERAFSQGAIVNPDPISYTFKMKLDKPARKDVKVKLATTGLDEQFMNDITVTPAEITIAAGELESAEVTWTITDDFLL TTAGEESHTLVVTASAESEDPVVKVNSKENFLTFNVDKVLRNFKYLSVIGSNW |
| 1382 |
PF16376 (fragilysinNterm) |
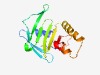 Estimated TM-score=0.67; trivial target. |
>N-terminal domain of fragilysin (Length=144) SIDLQSVSYTDLATQLNDVSDFGKMIILKDNGFNRQVHVSMDKRTKIQLDNENVRLFNGRDKDSTSFILGDEFAVLRFYR NGESISYIAYKEAQMMNEIAEFYAAPFKKTRAINEKEAFECIYDSRTRSAGKDLVSVKINIDKA |
| 1383 |
PF16342 (DUF4972) |
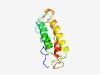 Estimated TM-score=0.67; easy target. |
>Domain of unknown function (DUF4972) (Length=127) MKNNYLYNWVVIVFMMLQMLAFTSCSDDDDNSGWPTETDKTEIFIERFSTLVTNLTALRDGATYGELKDNYPVSSKAMLD DEIVYLEETIAKLKEGNKKLADSEADRIIREANQIEKNFRATRRTED |
| 1384 |
PF16324 (DUF4960) |
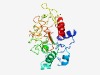 Estimated TM-score=0.67; easy target. |
>Domain of unknown function (DUF4960) (Length=255) YVGLASTIDGLGYEERAAADWMLKNVANSQYVSFEDIRNGNVDLGECKVMWWHLHIDGGIDNMDKFDAAAPAALQALPAV KDFYNNGGCLLLTRFATYYAAKLGATADGNNPNNCWGQVEESGEITSGPWSFFVDGHENHPIYQGLVTTIDGKQGVYTCD AGYRITNSTCQWHIGSDWGGYPTLDDWRNLHGGLDLGYGGDGAVVVWEYPANAAGGRVLCIGSGAYDWYSYGVDASGDQY HGNVAKMTLNAINYL |
| 1385 |
PF16289 (PIN_12) |
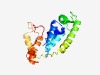 Estimated TM-score=0.67; easy target. |
>PIN domain (Length=169) VILDTNIIFSDFHLKGAKIKNLCESVKSTGDSIHIPAVVVDESINKYKEKAQECKSKIDRGISDFKRLTGKDIGADPCSD EFILKETEEYVEKFKKRLQELGIKIIPYPSTPHQELVKRDLSRKKPFQETGKGYRDALIWESVKNICEKYLYSSEIPKII FVNKNHKDF |
| 1386 |
PF16277 (DUF4926) |
 Estimated TM-score=0.67; easy target. |
>Domain of unknown function (DUF4926) (Length=57) LLDVVALTEDLPAQNLYRGQVGTIVEILDDGVFEVEFTDDEGQTYAMATLRADQLMV |
| 1387 |
PF16155 (DUF4863) |
 Estimated TM-score=0.67; easy target. |
>Domain of unknown function (DUF4863) (Length=153) RCREFLSEVQDLTPGKDLEERLNREYGPGNKFYEDFCTYIKQGLVEGWVAQTEIDGPKYRRGKIALPSEETRYFSITTVY MESKEEYTGQYHAHPYGEINCVIQLDATAELKGMQGWQGAGWTSPGPGTHHYPEVRKGALVALFFLPAGRISY |
| 1388 |
PF15982 (TMEM135_C_rich) |
 Estimated TM-score=0.67; easy target. |
>N-terminal cysteine-rich region of Transmembrane protein 135 (Length=134) PHNCYEIGHTWSPSCVQSAADVTRSALEVSFKIYAPLYLIAAVLRRRKKDYYFKRLIPEIFWSTSFLTTNGGLFIVFFCI LRRLLGGFYSWSVGFGSALPASYIAILLERKSRRGLLTIYMANLATETLFRMAV |
| 1389 |
PF15943 (YdaS_antitoxin) |
 Estimated TM-score=0.67; trivial target. |
>Putative antitoxin of bacterial toxin-antitoxin system, YdaS/YdaT (Length=65) IREACRIVGGQAVLARLLGVSPPTVNQWVSGARQVPAERCPAIEKATGGAVTCEELRPDVDWAYL |
| 1390 |
PF15580 (Imm53) |
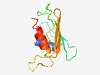 Estimated TM-score=0.67; very hard target. |
>Immunity protein 53 (Length=96) LDVLEDWALLYSTKKEQVKDLIHIYNIDNPGWVIEIDLKETILDGISIEWEIIEGSKDGWHTGDWHGIAVVDAAFDGSGG PRKLRFLLHYFKALVE |
| 1391 |
PF14995 (TMEM107) |
 Estimated TM-score=0.67; hard target. |
>Transmembrane protein (Length=117) LVPARFLTLTAHLVIVITIFWSRDNNVRACLPLDFSQDEYRSQDTQLVVALSVTLGLFAIELAGFFSGVSMFNSTQSLLS IAAHASASVALLFFLFEQWSCAIYWWIFGFCRLVSLT |
| 1392 |
PF14898 (DUF4491) |
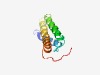 Estimated TM-score=0.67; hard target. |
>Domain of unknown function (DUF4491) (Length=92) NFEGLIIGIATFLIIGLFHPVVIKAEYYWGTKCWWIFLVLGVAGTIAALFIESIFWSAICGVFAFSSFWTIKEVFEQEER VKKGWFPRNPKR |
| 1393 |
PF14897 (EpsG) |
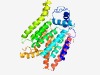 Estimated TM-score=0.67; easy target. |
>EpsG family (Length=356) KIYLIVTFGVLILVASLRDPSVGTDLAGHYAKRFNMIGDYSWSQIPTFSTTIGYEIGYCYFTRFLHVFSSDVQFFIFVTS FLMCAAFGYFIYKESTDVILSTELMLFTCFYYRFMTMMRQGLAISIILVAYTLMNNSERRLKDYIKFALLILLASTFHSS AILCTLMILFDRLRFTKKQIIIGVGVMVVFYLFYMNFYTAALNFFGTGNNYERYLTSASEGVGHINFQTIYNFILAFVPF LLGYYVLVWEKKTEKHLFKGDESMYCLAKNESFLLYMVLIASTFNLLIFRMNILNRFSLYFTPFVLILCPYAISSMKLKS NKPIVRACIYFMFGFYFVWLTITKAAEFYGVVPYRF |
| 1394 |
PF14737 (DUF4470) |
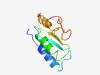 Estimated TM-score=0.67; hard target. |
>Domain of unknown function (DUF4470) (Length=97) LWGTVPAVDTINIAKNEGTSPVPPTLALAYVVSGGLRNVVRTINSLPSDFSGEVTVLLNDKDPIISIRSLMIFSILARLE DVSRAADVALHFWASAY |
| 1395 |
PF14462 (Prok-E2_E) |
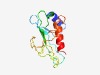 Estimated TM-score=0.67; easy target. |
>Prokaryotic E2 family E (Length=121) LPSNDRQYLENRGLPFEEVVDASQKGVILREFQLPLGRFDTEQADILILLPSGYPDAPPDMFYLLPWVKLVQGAKYPKAA DQPHQFNGQKWQRWSRHNNEWRPGTDGIWTMLKRIENALEV |
| 1396 |
PF14428 (SCP1201-deam) |
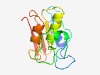 Estimated TM-score=0.67; easy target. |
>SCP1.201-like deaminase (Length=135) APVPSWIRQTGQDLPTRPDDHGPTHGQAFDSTGRPLSAEPWRSGRNIASTSDLRPIPGLKGFPWTLTDHVESRAAQQMRR PGAPREVSLVVNKEPCTDDPYGCDRILRHIIPAGSRLTIYVRDPDAPAGVRTVGQ |
| 1397 |
PF14399 (BtrH_N) |
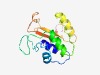 Estimated TM-score=0.67; easy target. |
>Butirosin biosynthesis protein H, N-terminal (Length=132) HCENGVISNLLNHRGVHISEPMAFGIGSGLFFVYLPFIKVNFAPGFSYRPMPGNIFKKLAKRLGIGVKHQRFSSPEKAKA VLDEKLAQGIPVGLQVGVYHLTYFPDEYRFHFNAHNLVVYGKEAGEYLISDP |
| 1398 |
PF14338 (Mrr_N) |
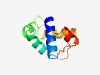 Estimated TM-score=0.67; easy target. |
>Mrr N-terminal domain (Length=86) YQEFMLPFLRTLGDGQEHKLREIYKLLASESNLTEEEMEQKLPSGKQLVFHNRIGWARTYLKKAGLIDAPQRGVFTITQR GRDLLK |
| 1399 |
PF14297 (DUF4373) |
 Estimated TM-score=0.67; hard target. |
>Domain of unknown function (DUF4373) (Length=93) YFPLDVDIDQDDKVQLIEAEYGIIGFAIIIKLLMKIYKEGYYYEWTEKEQLLFSKKVNVDINQVNDIINACLKWGLFDKT LYDKYKILTSKGI |
| 1400 |
PF14129 (DUF4296) |
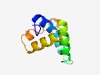 Estimated TM-score=0.67; hard target. |
>Domain of unknown function (DUF4296) (Length=83) PETVIPDAKMENVLYDFHIAKAMGEEVPYSDSYKRVLYIESVYKKHGITQADFDSSMVWYARHPDALTKVYEKVNQRLKA ERD |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218