

Go to page (83 pages in total): [First page] << < 12 13 14 15 16 17 18 19 20 21 22 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 1601 |
PF09365 (DUF2461) |
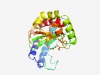 Estimated TM-score=0.66; easy target. |
>Conserved hypothetical protein (DUF2461) (Length=226) NTLLFLKDLKAHNQRSWLKSHDGEYRRSLKDWNTFVETITPKITEVDFTIPELPAKDLVFRIHRDIRFSKDPTPYKAHFS AAWSRTGKKGPYACYYIHLQPGSCFVGGGLWCPEASHLQKLRESIDERPRRWRRVLNDERFKKTFLPKSLQKGGEEAALK GFAETNKGNALKTKPKGYLVDHRDIELLKLRNFTISKKVDDKVFTEDDAQEQICEIIAAMHPFITF |
| 1602 |
PF08901 (DUF1847) |
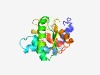 Estimated TM-score=0.66; hard target. |
>Protein of unknown function (DUF1847) (Length=159) CLTTNVPQKEIEKVNELYKNDSLVSKIAHTAAEIEGTYYGKLTRVEEIIAFAKRIGAKKIGIATCIGLMNEAKIFAKILK AKGLESYSVICKVGSIDKTEIGIAEEFKIQKGCHEALCNPILQARLLNREKTDLNVIVGLCVGHDSLFIKYSKAPVTTL |
| 1603 |
PF08855 (DUF1825) |
 Estimated TM-score=0.66; hard target. |
>Domain of unknown function (DUF1825) (Length=103) MGFFDSEIVQQEAKQLFEDYQSLIQLGGSYGKFDREGKKIFIDQMEAMMERYRIFMKRFELSDDFMAQMTVEQLNTQLSQ FGITPQQMFDQMHMTLERMKSEL |
| 1604 |
PF08813 (Phage_tail_3) |
 Estimated TM-score=0.66; easy target. |
>Phage tail tube protein, TTP (Length=166) SGWPELNERVVRAVNTASNTFALEGIDTTKVGRFSKGQGIGTATPVSTWVDLSQVTNVAKTGGEQQFYQWRYVEDRNSRQ RQRPTYKSAKFITLTLDYDPALAWYEALKEADAAKDAVVLRAKLPNNDELYYLVYPSFDSDPSMELDANMQNTATFSMMS DFTRYA |
| 1605 |
PF08520 (DUF1748) |
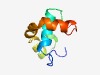 Estimated TM-score=0.66; hard target. |
>Fungal protein of unknown function (DUF1748) (Length=70) LGRLTHYAFDAVLFSAFLAGVKRSTGLTLNSDKITDSKDVKKWVDSYLGVGEWMMDQSVAVMGSSSWFER |
| 1606 |
PF08396 (Toxin_34) |
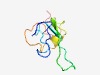 Estimated TM-score=0.66; easy target. |
>Spider toxin omega agatoxin/Tx1 family (Length=75) CIDIGGDCDGEKDDCQCCRRNGYCSCYSLFGYLKSGCKCVVGTSAEFQGICRRKARQCYNSDPDKCESHNKPKRR |
| 1607 |
PF08378 (NERD) |
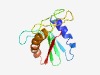 Estimated TM-score=0.66; easy target. |
>Nuclease-related domain (Length=111) KGKVGERRVKSVSSRLDPNQYTIFHDVTLPTTNGTTQIDHIIISRQGIFVIETKNYSGWIFGSEKSRFWTQVIYKAKNKF QNPLLQNYKHIKTLQKLLQLPIHYFHSVIVF |
| 1608 |
PF08168 (NUC205) |
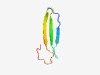 Estimated TM-score=0.66; hard target. |
>NUC205 domain (Length=44) EKSVIQSFSASVDRKFISLMSLSSDGCIYETLIPIHPSDPEKNQ |
| 1609 |
PF08067 (ROKNT) |
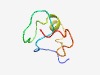 Estimated TM-score=0.66; hard target. |
>ROKNT (NUC014) domain (Length=43) METEQPEETFPNTETNGEFGKRPAEDMEEEQAFKRSRNTDEMV |
| 1610 |
PF07985 (SRR1) |
 Estimated TM-score=0.66; hard target. |
>SRR1 (Length=52) IRCLALGSPTDSNYARYQLALLLQLAKHLSSPLISLYDPIFTPGDKTLFESF |
| 1611 |
PF07712 (SURNod19) |
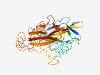 Estimated TM-score=0.66; easy target. |
>Stress up-regulated Nod 19 (Length=379) NVKSGVFLSPKFELGPGSVANKYDYDIDFPRGHIALKSFNAEVVDEAGNPVPLHETYLHHWVVARYYQPKYVTTHTKYDD HRILHPSDHIMVRNSGICQRNALGQYFGLGSETRGTATHVPDPFGIVVGNPAEIPEGYEEKWMVNIHAIDTRGVVDKMGC TECRCDLYNVTKDEYGKPLRSDYKGGLLCCYDHTQCRLREGFQGQKRSLYLRYTVKWVDWDKFIVPVKIYILDVTDTLKI SDDSGGMIPEHDCRVEYDVESCSTGDKDGNGCNHVKRTSLPMKNGGYVIYGVAHQHSGGIGSTLYGQDGRVICSSIPSYG KGKEAGNEANYIVGMSTCYPGPGSVKIIDGEIVTLESNYSSNRGHTGVMGLFYLLVAEQ |
| 1612 |
PF07585 (BBP7) |
 Estimated TM-score=0.66; easy target. |
>Putative beta barrel porin-7 (BBP7) (Length=362) WARLEMLLWTTRGMNVPALVTTSNTGTARNIAGVLGEGSTRVLYGDDTVNSYTRIGGRFTGGVWLDECRRLGIEGDYFGL ENETENYSQNTAGNAILARPFYDIVNGQETAQLVSFPNVISGSVNVDAVTKFQGSGIRFINNLCGAQGCCPSILNNCNTV PTSYQVQMLLGYRYLRLDDSLEITEDLTSLDTAAPGSFFVRDMFETDNQFHGGDIGFVYSTRRGCWSLDILSKVAIGSTY SRISIDGNTLITEGNQTTNNTGGLLAQRTNIGDYTANEFAMVPELGLTLGYQLNPCWRFTMGYTLLYWSRVARAGDQIDT DINPNLIPPEDPAVTTHLRPEFNLAYTDFWGTGLNLGLEATW |
| 1613 |
PF07578 (LAB_N) |
 Estimated TM-score=0.66; trivial target. |
>Lipid A Biosynthesis N-terminal domain (Length=71) IGFLAQILFSSRLIIQWLYSEKQKRVITPTIFWTLSLLASFLLFVYGYLRDDFAIMLGQSLTYFIYIRNLQ |
| 1614 |
PF07423 (DUF1510) |
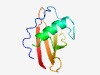 Estimated TM-score=0.66; hard target. |
>Domain of unknown function (DUF1510) (Length=93) IVNPDWKPVGTTQTGEHAAVYDESSVDWQEMLQAISYASGLDKNNMTVWRLGNNGGPNSAIGTVSQKGSDQKYRVAIEWV DGQGWKPVKVEEL |
| 1615 |
PF07298 (NnrU) |
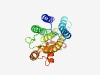 Estimated TM-score=0.66; easy target. |
>NnrU protein (Length=215) FALAYAVFFLSHSLPVRPPLRPWLQARLGASGFTIAYSALSLAALAWLIAAAGRAPYVALWDWAPWQVHVPLTFMGPVCL ILALSIGRPNPFSFGGARNDQFDPARPGIVRVWRHPLLLALALWAAAHVVPNGDLAHVILFGTFAAFALLGGRLIDRRKR REMGPEWQRMRDRVADAPFLCLPVSWQRLATRMAIGIGLYVALLWLHPWLFGVDP |
| 1616 |
PF06871 (TraH_2) |
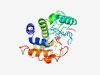 Estimated TM-score=0.66; easy target. |
>TraH_2 (Length=203) MLDAALIQQCADPSLKPAIVEQFVTAAGANDPFAVTVKSGGRLILVPKATTQDEAMAIIRQYVGQTVVRVGLTQFPAGVG VKDAADLNSDLVDPCENLRKGTAMFAKVLRIVAKWYGNPKSDDVFPQIFDDAVYAWKSGEFEGVSVFQADDPGKSVTIDN TLPANLDKASADIPPADANTESVEKGESVGSAEMRIDLSRIGG |
| 1617 |
PF06852 (DUF1248) |
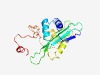 Estimated TM-score=0.66; easy target. |
>Protein of unknown function (DUF1248) (Length=178) RNVDIIVDPNENLIDEFMNGYGKERLNFERNDIDMWKKCFKDDYSLVFVCPKGSNTIIQTVHHINYHPIPSNNDTSHQYD GFMWIHPDHRGPDSMKMLEYVVKERLRCVTDSLVVQAFPQSMNFWKQMAGHRKYGHIQYVSYYKMEEMKVPEDLNTDGIL IKNATDVPDEDIVKYDRE |
| 1618 |
PF06713 (bPH_4) |
 Estimated TM-score=0.66; easy target. |
>Bacterial PH domain (Length=74) TGYKIENSKLEIQSGPYKKTINIDEINKVRNTKNPFTAPALSMDRIEISYGKYEFITISPKNKEEFIRQLLKVN |
| 1619 |
PF06672 (DUF1175) |
 Estimated TM-score=0.66; easy target. |
>Protein of unknown function (DUF1175) (Length=223) YFEADTSDDDEDGYPDCLVLSENDAEKFRNWFIWIGLSAIKNEPVLWNEHERDCAGLIRYCSREALKKHDGRWLERSNYS GPIFDDVEKYNYPDIPLIGDKIFRIISGKYTNPGEFSAFASARIMLENSLKFVSKDVEHALPGDIAVFFHPEDFEYPYHM MIYISDIYFLQNHRWFLYHTGPIGESAGEMRFVRYENLEKLDPSWSPREKNRYFLGFYRFKFL |
| 1620 |
PF06532 (NrsF) |
 Estimated TM-score=0.66; hard target. |
>Negative regulator of sigma F (Length=204) LSAGEGAVDRHALGKRFAIALLAGALGALLLMAAIFGVRPDLGEVARTPLFWAKVALPGSLAPLALWLSSRLARPGVGGG AAWVLLALPVLLVWIGGAVELAGVPADARTGLILGKTWRTCPLNIALLSVPTFIGVFWALRGLAPTRLRIAGAAGGLLAG ATATLAYCLHCPEMGIPFWGVWYLLGMLLPTALGAVLGPRLLRW |
| 1621 |
PF06338 (ComK) |
 Estimated TM-score=0.66; very hard target. |
>ComK protein (Length=149) DSYVINQTTMAILPVEEGKRVYSKVIERETSFYVELKPLQIIERSCRFFGSSYAGRKAGTYEVTGISHKPPIVIDSSNHL YFFPTYSSNRPQCGWISHKYIHTFQESSLGDTVVIFTNEQTVELDVSYKSFESQVHRTAYLRTKFQDRL |
| 1622 |
PF06287 (DUF1039) |
 Estimated TM-score=0.66; trivial target. |
>Protein of unknown function (DUF1039) (Length=65) IAGLNHNFCKESRAIMESLPFLVPDINVRLTCHALLLHGLGETQKAINLLKDSSLEEAIVLREIF |
| 1623 |
PF06099 (Phenol_hyd_sub) |
 Estimated TM-score=0.66; hard target. |
>Phenol hydroxylase subunit (Length=56) PACDTSLRFVRVMERRADGLVSFEFSIGWPELAVELMLPEAAFEAFCANNRVQRLD |
| 1624 |
PF06073 (DUF934) |
 Estimated TM-score=0.66; easy target. |
>Bacterial protein of unknown function (DUF934) (Length=107) GIWLDSNEEVEEIADDLEHFQVIALNFPKFTDGRHYSSARLLRERYGYKGEIRAIGDVLRDQLFYMRRCGFDAFAIRADR DPEDALQSLKDFSVRYQAAVDERLPLF |
| 1625 |
PF05990 (DUF900) |
 Estimated TM-score=0.66; easy target. |
>Alpha/beta hydrolase of unknown function (DUF900) (Length=226) AAAQHWINTHLPRTKRVMIFIHGFNNTFEDSVYRFAQIVHDSGADVAPIIFTWPSRASVFDYNYDKESTNYSRDALEHVL RVVANDPQVRDVTIMAHSMGSWLAMEALRQMAIRDRRVNAKITDVILASPDIDVDVFGKQFQAMGSHHPRFTLFTSRDDK ALALSRRISGNIDRLGQIDPSVEPYRSKLEKAGIVVIDLTQLKAGDRLNHSKFASSPEVVQLIGRR |
| 1626 |
PF05944 (Phage_term_smal) |
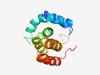 Estimated TM-score=0.66; hard target. |
>Phage small terminase subunit (Length=130) LLQLTTDRARLKQIQSEQGKAELKRELLPAYQPYVEGVLSAGQGAQDEVLTTVMVWRMDAGDYTGALDIADYVLRHGLVM PDRFARTTGCLVAEEIANAALKAQKAGGAFDLAVLERTQTLTQEQDMPDE |
| 1627 |
PF05876 (Terminase_GpA) |
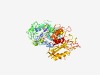 Estimated TM-score=0.66; easy target. |
>Phage terminase large subunit (GpA) (Length=566) LPQKASPEPGPWRNDRTPYLVEIMDCMSPDSPTEEVAFMKGTQVGGTESGNNWFGYVVHHSPGPMMIVQPTLNLAKRFSK QRIAAMIEEMPILQERIKDSRSRDSGNTTLMKDFTGGVLVIAGANSAADLRSMPCKYLFFDEVDAYPHDLDGEGDPIDLA EKRATNFARRKNLKVSTPTTKGFSRIEASYDAGTREHYHVACPHCGEYQWLKWAGIKWDKADDGAHLPETAHYVCEHNGC IIAEHHKTAMLAGGMWIADNPAAGPRRRSFHLSALYSPLGWASWASIVERYLKAKNALAAGDDTKMKVFVNTDLAETWEE QGDKVQTSDLQARAEPYRLRTVPMAALLLTAGVDVQGNRLEVKVKAWAPGEESWTVDWVTLYGDPSEDDVWNQLDAYLAR PLTHQSGTAMAISATAIDTGGHHTQRVYTYCRNRGYRNVIAVKGMSTAGKPIIGKPTDVEVNHHGTKIKKGVKLWPVGTD TAKALIYGRMRLTQYGPGYMHHSTDLPAEYYDQLTAERLVTRYSKGHALMSWVKPNGKRNEALDCEVYALAAAYFLGLQR YKEQDW |
| 1628 |
PF05585 (DUF1758) |
 Estimated TM-score=0.66; easy target. |
>Putative peptidase (DUF1758) (Length=164) VNVRNAKGDLVACRLLLDTGSELSYISERCIQTLGLARTPSRILVTGISSTKADTTRGCSTISIQSRISQDQLVVQAHVL GKITSSLERQTIDASVLRVFNDLQLADSQFSTNAPIDILLGSEHVWSVITGRKIYDDKGKLIAISSIFGWVITSLLSSRA CSAT |
| 1629 |
PF05575 (V_cholerae_RfbT) |
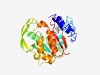 Estimated TM-score=0.66; easy target. |
>Vibrio cholerae RfbT protein (Length=286) MKHLIKNYVQKLIKTELDAIQSKSVHDNRNFIYNGEFLILESEFGWHCFPRVQLNHALSYKNPNFDLGMRHWIVNHCKHD TTYIDIGANVGTFCGIAARHITQGKIIAIEPLTEMENSIRMNVQLNNPLVEFHHFGCAIGENEGENIFEVYEFDNRVSSL YFQKNTDIADKVKNSQVLVRKLSSLDISPTNSVVIKIDAEGAEIEILNQIYEFTEKHNGIEYYICFEFAMGHIQRSNRTF DEIFNIINSKFGSKAYFIHPLSSAEHPEFNKATQDINGNICFKYVS |
| 1630 |
PF05420 (BCSC_C) |
 Estimated TM-score=0.66; easy target. |
>Cellulose synthase operon protein C C-terminus (BCSC_C) (Length=360) SQLTEAKAPMSFSFVPFDTSRVTLGVTPVTLDAGSSSGSANNRFGTGALHGELSPLAQAQQNASNTPASPSQYIAKESSG SQSESGVELNAALSGDNYKVDVGSTPLGQDLTSLVGGAQWAPKLGDFTTLTVTAERRAVNDSLLSYVGARDAYTGSKWGQ VTKNGGSVNLSYDDGDAGFYAGGGYYSYLGDHVTSNKAVNANVGLYIRPYRYDDRELKTGVNVGYMNFDKNLSYFSYGQG GYFSPQNYVSVSFPVEYTQKYDDWDLKLSGSVGYQSYTQDESPYFPEDSNLQNQLEQLVADGKAQEAFYSGGSKNGIGWN AGIGGNYHLNKNMQIGGQVGYDTFGDYNESKAQLYFRYLM |
| 1631 |
PF05413 (Peptidase_C34) |
 Estimated TM-score=0.66; trivial target. |
>Putative closterovirus papain-like endopeptidase (Length=92) KVKFVKGRFDCLFSSIAEVIHKKPEEVMAFVPHILDRCISNRGCSIDDARAICEAYEIKIECEGDCGLVECGTVGLSVGR LLLRGNHFRVAS |
| 1632 |
PF05219 (DREV) |
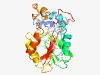 Estimated TM-score=0.66; easy target. |
>DREV methyltransferase (Length=262) WYVCNREKLCESLQAVFVQSYLDQGTQIFLNNSIEKSGWLFIQLYHSFVSSVFSLFMSRTSINGLLGRGSMFVFSPDQFQ KLLKINPDWKTHRLLDLGAGDGEVTKIMSPHFEEIYATELSETMIWQLQKKKYRVLGINEWQNTGFQYDVISCLNLLDRC DQPLTLLKDIRSVLEPTRGRVILALVLPFHPYVENVGGKWEKPSEILEIKGQNWEEQVNSLPEVFRKAGFVIEAFTRLPY LCEGDMYNDYYVLDDAVFVLKP |
| 1633 |
PF05206 (TRM13) |
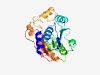 Estimated TM-score=0.66; easy target. |
>Methyltransferase TRM13 (Length=245) QASSLLGILENDHQLTDVTSYVEFGAGKGQLAYYLATALQDQQLSQSQVVLIDRMSLRHKKDNKLANKEVVQRIRADIAD LKLSALPELKKTQRTVAISKHLCGAATDLTLRCLLSDENASSDYVLIALCCHHRCSWRSYVGRKFLQEAGIGPREFAILT KMVSWAVCGTGLSRERRKAMETAAFPPTETNTQRLNRQEREQIGQQCKRVLDYGRLEYLRSHGYQAELKFYVSRDVTLEN VVLLA |
| 1634 |
PF05176 (ATP-synt_10) |
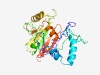 Estimated TM-score=0.66; easy target. |
>ATP10 protein (Length=254) TLKEASKPALEKSRPITKPIGLESPVLLNHKLGDTYSLTNIKQELFSSEAKERRQKKLDYEIRHSPFYETKSFRNTNGKI FTPPISFFKKDKALYFPDFISSTLDGQSRSLYEILADKVSIVRLYSTVSGEQCSHSYFKVDGKDYAQGDYGVFEENHPQS QIIDINIPQNWMKGFLLGLSKSNIKKTISPQRHNKYFILPDHIFQYSLREKLMCDNMCSGYIYLLDHNGKLRWATSGYAN DEELALMWKCVRGL |
| 1635 |
PF05042 (Caleosin) |
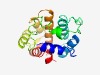 Estimated TM-score=0.66; hard target. |
>Caleosin related protein (Length=170) DQNVLRKHVAFFDRNNDGVIYPWETFQGFRAIGSGIILSSVAAVFINVGLSSKTRPGKKFPDLLFPIEIKNINMAKHGSD SGSYDTHGRFVASKFEEIFINYARTNSDALTACEVDEFVKGNREPKDYGGWIGSLTEWKILYYLAKDKNGFLQKDTIRGV YDGSLFEKMA |
| 1636 |
PF05030 (SSXT) |
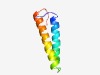 Estimated TM-score=0.66; hard target. |
>SSXT protein (N-terminal region) (Length=60) QPPSTATIQKILDENCSLIQSIQAYQSEGKHQECMSHQQALHRNLVYLANLADSTQNIPQ |
| 1637 |
PF04967 (HTH_10) |
 Estimated TM-score=0.66; easy target. |
>HTH DNA binding domain (Length=52) LTEKQASVLRAAYHAGYFEWPRGSTAEELAESMDVSSPTLHNHLRRAQQKLL |
| 1638 |
PF04575 (DUF560) |
 Estimated TM-score=0.66; easy target. |
>Protein of unknown function (DUF560) (Length=289) WSFQGGLTYLNDPNINNAPNAGTTYGNWTAPKKESAQGVGFHFEADKKWSWGNGFFHEFRLNGNGKYYWNNKKYNEYSLR ESLGLGYQNAKQKITLLPFFEQMWYAGGSSQTESTKRYSNAGGVNARWQYWITPQWQSAISYEYAEQRYTRRQHLDGNYH FIAPSLYFYPNSRQYWFVGANFNRTSPRDLDDSFIRRGVSIGWGQEWKNGLSTQLSASYARKQYRAPMPIFQITQRNSEY GVQASIWHRALHFKGITPRLTWQYNKVKSNHSFYSYDKNRVYIEFSHTF |
| 1639 |
PF04535 (DUF588) |
 Estimated TM-score=0.66; hard target. |
>Domain of unknown function (DUF588) (Length=146) KGVAIMDFILRLGAIAAALGAAATMGTSDQTLPFFTQFFQFEASYDSFTTFQFFVITMALVGGYLVLSLPFSVVAIVRPH AVGPRLFLIILDTVFLTLATASGASAAAIVYLAHNGDQDTNWLAICNQFGDFCAQTSSAVVSSLVA |
| 1640 |
PF04188 (Mannosyl_trans2) |
 Estimated TM-score=0.66; easy target. |
>Mannosyltransferase (PIG-V) (Length=434) LLRYAVTSRLLILTLIIIWRSLLSPYDTSASINPQCLSSSNSSNPPPAALFPRVGSAIERSIVWDGVYFTRVAECGYEYE QTYAFLPLLPLCASLLSKTVFAPLVRVIGYRAVLGLSGYVLNNIAFVFAALYLYRLSVVVLKDNEVALRASILFCFNPAS IFYSSIYSESLYAFFSFGGLYHFMNGAYNYTTFWFALSSCARSNGVLNAGYICFHAMLQVYEAFFSKKRAYLVTLKIILA AGLRSVCIFIPFISFQAYGYFNICLDHSVGEMRPWCKARLPLLYNYIQSHYWGVGFLRYFQVKQLPNFLLASPILSLALC SIVYYVKLWPEVFLSLGLQAPPSKSETSDGLHYDPVVVLPFVLHLSFMVATAFFVMHVQVATRFLSASPLVYWFASLIMG SPGIGKKWGYFIWAYCAAYIFLGSLLFSNFYPFT |
| 1641 |
PF04134 (DUF393) |
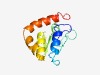 Estimated TM-score=0.66; hard target. |
>Protein of unknown function, DUF393 (Length=107) LYFDGECPLCAREIKFLQGRAAKERLLFVDISSGAFDARALGLTFEQMQFSLHARFADGRWLTGLDATLWSWRAAGLGFW ATPLTWRVLRPLLEFGYRLFCRLRPHL |
| 1642 |
PF04083 (Abhydro_lipase) |
 Estimated TM-score=0.66; trivial target. |
>Partial alpha/beta-hydrolase lipase region (Length=62) VTEMIESYGYPVESHVAQTADGYLLTMHRIPYGRAPGAGPEPNKPVVFLQHGLLSSSADWVV |
| 1643 |
PF04070 (DUF378) |
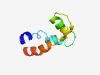 Estimated TM-score=0.66; hard target. |
>Domain of unknown function (DUF378) (Length=58) MDKTALTLVIIGALNWGLIGLFNFDLVATLFGGQTSWLSRIVYSLVGIAGLYAISLLF |
| 1644 |
PF04031 (Las1) |
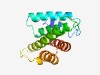 Estimated TM-score=0.66; hard target. |
>Las1-like (Length=155) TPWRDRAELLAVRRQFYPDAAPQAQLKPGHAGQHDAVARVSLWMQRGGCPHLVESTALLVAAILSDAEGGGSSAGTYAAR AAYAAAFSRFVTGLLDGHQDKQRKLSMYSVAKTIGLPATFVELRHQATHEQLPSLAKLRAAARRALDWIWEYYWR |
| 1645 |
PF03891 (DUF333) |
 Estimated TM-score=0.66; hard target. |
>Domain of unknown function (DUF333) (Length=45) AANPASVFCEEQGGEVVIKDENGGQVGYCKLSDGRLIEEWTFMNE |
| 1646 |
PF03769 (Attacin_C) |
 Estimated TM-score=0.66; hard target. |
>Attacin, C-terminal region (Length=114) GSGRTTTVAGQANIFKNEKAEMNASAYHTRNRTHDQFGGGMDISTATGHSASLGVHHVPVFNMTSVNASGSANLYSSPDG KLNVAATANAMRHTSGPFRGKSDMGYGLNMQYKF |
| 1647 |
PF03650 (MPC) |
 Estimated TM-score=0.66; easy target. |
>Mitochondrial pyruvate carriers (Length=100) PAWNHPAGPKTVFFWAPTIKWALVGAGLADLARPAHKLSPYQNTAICATGAIWTRYCLVITPINYYLSSVNFFVMCSGLV QLCRIAHYRYTNPDWEQQQK |
| 1648 |
PF03626 (COX4_pro) |
 Estimated TM-score=0.66; hard target. |
>Prokaryotic Cytochrome C oxidase subunit IV (Length=74) ITGFILSVILTVIPFWLVMSPSLPKPTILMIIVGFALVQILVQLGYFLHMNTKSDEGWNMISFVFTVLIIAIVV |
| 1649 |
PF03591 (AzlC) |
 Estimated TM-score=0.66; hard target. |
>AzlC protein (Length=140) PILLGYLPIGLAFGVLANNAGISPVNSLLMSLLVFAGASQFIAIGLIEKGITTFSIILTTFLVNSRHILMSASLSPYFQK INSWKSSLLSFGITDETFALNSVELKEDQTKNHYYIAGVHLTAYIAWVSSTAIGAYLGNY |
| 1650 |
PF03196 (DUF261) |
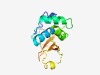 Estimated TM-score=0.66; easy target. |
>Protein of unknown function, DUF261 (Length=135) KIKQDNRTLRPEIQKWGCYFLCLHYYTSLFKQCEFNVYEINAAYYRFIGLGYIKSNCFIINPCMILNYYGIRSSVRYESL NYLGAANEFEISEVKIDKVNGYHFIATKNKEILYDSLDLKPRGKIFKVTSKRIFK |
| 1651 |
PF03077 (VacA2) |
 Estimated TM-score=0.66; hard target. |
>Putative vacuolating cytotoxin (Length=60) GYIGFITGVFKARDIFITGAVGSGNEWKTGGGAILVFESSNELTTNGAYFQNNRAGTQNS |
| 1652 |
PF02592 (Vut_1) |
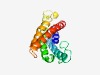 Estimated TM-score=0.66; easy target. |
>Putative vitamin uptake transporter (Length=159) IAVLCYPITFAVTDIISEVYGKRTAQKVVWTGFFASLILVVYSQIAVIYPPASIFGNNEAFAKVFGSTPRIVFASILAYI ISQSHDVWAFHFWKRVTKGSHLWIRNNLSTMVSQFIDTLVFITIAFLGTVPITVLVKMIFSQYAVKLIIALIDTPFVYL |
| 1653 |
PF02456 (Adeno_IVa2) |
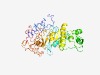 Estimated TM-score=0.66; easy target. |
>Adenovirus IVa2 protein (Length=372) LDRDAVEHVTELWDRLELLGQTLKSMPTADGLKPLKNFASLQELLSLGGERLLAHLVRENMQVRDMLNEVAPLLRDDGSC SSLNYQLQPVIGVIYGPTGCGKSQLLRNLLSSQLISPTPETVFFIAPQVDMIPPSELKAWEMQICEGNYAPGPDGTIIPQ SGTLRPRFVKMAYDDLILEHNYDVSDPRNIFAQAAARGPIAIIMDECMENLGGHKGVSKFFHAFPSKLHDKFPKCTGYTV LVVLHNMNPRRDMAGNIANLKIQSKMHLISPRMHPSQLNRFVNTYTKGLPLAISLLLKDIFRHHAQRSCYDWIIYNTTPQ HEALQWCYLHPRDGLMPMYLNIQSHLYHVLEKIHRTLNDRDRWSRAYRARKT |
| 1654 |
PF02001 (DUF134) |
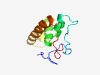 Estimated TM-score=0.66; easy target. |
>Protein of unknown function DUF134 (Length=99) MSRRGRPKIPRFISEEPKFRIFKPHGVSLTEVDKVILSVDELEAIRLVDYLDYTQEEASKLMGISRRVLWSLLTEGRKKI ADALINGKAIVIEGGEYKI |
| 1655 |
PF01733 (Nucleoside_tran) |
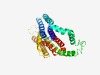 Estimated TM-score=0.66; easy target. |
>Nucleoside transporter (Length=312) MIKIVLINSFGAILQASLFGLAGVLPANYTAPIMSGQGLAGFFTSVAMICAVASGSKLSESAFGYFITACAVVILAILCY LALPWMEFYRHYLQLNLAGPAEQETKLDLISEGEEPRGGREESGVPGPNSLPANRNQSIKAILKSIWVLALSVCFIFTVT IGLFPAVTAEVESSIAGTSPWKNCYFIPVACFLNFNVFDWLGRSLTAICMWPGQDSRWLPVLVACRVVFIPLLMLCNVKQ HHYLPSLFKHDVWFITFMAAFAFSNGYLASLCMCFGPKKVKPAEAETAGNIMSFFLCLGLALGAVLSFLLRA |
| 1656 |
PF18867 (HEPN-like_int) |
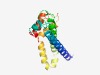 Estimated TM-score=0.65; easy target. |
>HEPN-like integron domain (Length=151) DKNTLLKMYFYFQYTAIEIQSAVNLLYMLEQFIEGKPYKEMIANEMLVLAPSQGSLNAYVTLSRVAFHNLIINIFKLGEL IEKKQGILTHLPEFNKSVNEFRKIFFTQDLRLYRNTYVAHHSDKNKDKSDNFLNYEELIQTFCKIIGVDLS |
| 1657 |
PF18789 (DarA_C) |
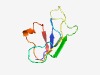 Estimated TM-score=0.65; very hard target. |
>Defence against restriction A C-terminal (Length=68) GLYKYYSTQRPVDIGTFPKPPYNKPDEIFNYDQRVPVENGSFLAWGYLTYTRPLTEKQASDYELRPAP |
| 1658 |
PF18788 (DarA_N) |
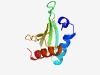 Estimated TM-score=0.65; hard target. |
>Defence against restriction A N-terminal (Length=100) FSFQDMATGDKASKIIERQFSRKGANVVQSDVTTSVKRAAGVSYREMMLTFADSQTVTLRVKQTGDIFQVLLNKKPLPIK NQDDHEAAVNEIVKLMDAGR |
| 1659 |
PF18763 (ddrB-ParB) |
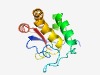 Estimated TM-score=0.65; very hard target. |
>ddrB-like ParB superfamily domain (Length=132) KGAEVKTAFKVVDASSLIISNNLDGTINPAFPEELQPRDRTRLSSKLQVNRIASNLRPAQLTDSGMSSHGAPIVGPDNVV ESGNGRSMGIWRAYEQGQADEYRQYLIDHAKEFGLNPDEISQMSMPVLVRER |
| 1660 |
PF18728 (HEPN_AbiV) |
 Estimated TM-score=0.65; easy target. |
>AbiV (Length=170) ARTFWKALMDNASALISDAHALLELGSFGRARSLTVLAQEELGKALWIYESFEQSWSTGSEKAREVPRLSADGRRHTVKY MEAFVFGQELAAFWGDYNSYEFPKDQSQEGWKAHFATKRREAEAAGKRANNEKMAGFYVDMDRPGDTVLSPADVSSGTIA EDLQTAAKVV |
| 1661 |
PF18291 (HU-HIG) |
 Estimated TM-score=0.65; easy target. |
>HU domain fused to wHTH, Ig, or Glycine-rich motif (Length=120) MSAMYDLIEKPNPKGDDEKQLLYPRIVSSGTISTEKLIERITGASSLTPGDVKSAITEISDVMAEYLKDGYTVELGDIGY FSPSIKGRGVSNKKEIRSTSIYFDNVNFRASKKFRQNLYG |
| 1662 |
PF18157 (MID_pPIWI_RE) |
 Estimated TM-score=0.65; easy target. |
>MID domain of pPIWI_RE (Length=136) EWRTPELLVRLHCRRLTEGLGDDLALPEGRRTKAMVTAAVAERRDRAARWMAAERPGTAPSLALVELDRAADFTTSDHDP KFALRLGFAKAGLLTQFVAVPKKTAGYDSTRNIGHRAEKAWDDGLRQLGVRVLPEH |
| 1663 |
PF17761 (DUF1016_N) |
 Estimated TM-score=0.65; hard target. |
>DUF1016 N-terminal domain (Length=137) DLKTRIRQAQVKAALAVNKELVLLYWQIGRDILQRQQEEGWGSKVITRLAKDLKREFPDMKGFSRTNLMYMRSFAEAYPD EQIVQQLVGQIPWGHNVRILDHVKDPQERIWYAQETIENGWSRNILELQIESKLYQR |
| 1664 |
PF17618 (SL4P) |
 Estimated TM-score=0.65; easy target. |
>Uncharacterized Strongylid L4 protein (Length=87) EDRKTITILLYRYDVPLINITYRNRKDLFRTFKRELRKHQLPIGEVSWADYENGDRSVIRNADDLFGAVSHCNSVKMYCR PKNADNP |
| 1665 |
PF17569 (DUF5475) |
 Estimated TM-score=0.65; easy target. |
>Family of unknown function (DUF5475) (Length=81) MSILKVVEACDLANTFLKLGYLFRAKTCLDIALENLELLRQKTNIKEVAVMLSKKTTECLQLKRKIDKKIAQRVLIKIYN I |
| 1666 |
PF17492 (D_CNTX) |
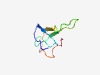 Estimated TM-score=0.65; easy target. |
>Delta Ctenitoxins (Length=53) ATCAGQDKPCKETCDCCGERGECVCALSYEGKYRCICRQGNFLIAWHKLASCK |
| 1667 |
PF17470 (Gp45_2) |
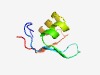 Estimated TM-score=0.65; hard target. |
>Phage gene product 45.2 (Length=57) NHLSMPEIKHWVIIDSFTGQEKILPETQLLEAFGQERWQKITSGIDDYFTAYEYNFE |
| 1668 |
PF17441 (DUF5419) |
 Estimated TM-score=0.65; hard target. |
>Family of unknown function (DUF5419) (Length=54) NFTRWLRRLDIELDKITGGIGLTRNDFADWRYAVAFTNGIAPRQAAIDMLAEDH |
| 1669 |
PF17196 (DUF5133) |
 Estimated TM-score=0.65; hard target. |
>Protein of unknown function (DUF5133) (Length=64) MLMAHPTVLRNLIEQYDTLRILHAEEGSPEVRRRMEDIAYTLCVSTGTRDVDAALIAARHQLPG |
| 1670 |
PF17172 (GST_N_4) |
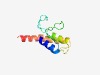 Estimated TM-score=0.65; easy target. |
>Glutathione S-transferase N-terminal domain (Length=92) PYVMKVHTYLRMTQIPYKIDGKDPFGEKGKAPWVSLNGQHISDSEFIIAFLNREFNKDLNKEHPVEQLALGTAVRVMLED QFSWGMALERFV |
| 1671 |
PF17133 (DUF5108) |
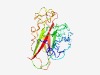 Estimated TM-score=0.65; easy target. |
>Domain of unknown function (DUF5108) (Length=221) FDFCDYPEVSSYIAAKGKGQLYQQVSRDDDDTYTALTKFIGRQEIVPQVSSYKIEMGPSGTLASDWSYLSYCTKGANTSG NWTKLMNNDALVVNIGYNGTLDLTIPPILAGKYRITLYYAYDPSMKFIGERGEGSQAGLTNFRLDSKRLAGGDNKRIYDG KTGDVQDCFNTPLTSNLSGEETAFEFTTTASHTLNVVVVDPAASSHNKFRMYFDYILFEPV |
| 1672 |
PF17014 (Mad3_BUB1_I_2) |
 Estimated TM-score=0.65; trivial target. |
>Putative Mad3/BUB1 like region 1 protein (Length=127) LLAYIDIPYFRNNPTYIERWRTYYQRYKDPQILLLMYHKQISTFYHWIYLELSQHFLKKSQPQIAHFILHEALKNNVYDA TRIKEAIEKIPPFEKKYSKGDMLGLLNTRNIQALGKTWNCSNETFFY |
| 1673 |
PF17003 (Actin_micro) |
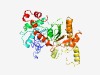 Estimated TM-score=0.65; easy target. |
>Putative actin-like family (Length=361) IEQITTPNKNVQSYFMRTDASYYVNEKYRMAGYRTDEPIIHVLVYGRRLIVDNMAETVDIEGFFDIVNISIDRFGSLEEG LGWLGNRIVAVTERLRIDRKRSFVSVVFHDLLSRTELLGLCNLFINLLSFKGVLVTPYSLSQAVCVVSPSCVVINMYERY STVSFVEDFWVVDAMCVSGESSKGFVLHDSVDFVDEFNKVRIFEEKNVYWCDLCDYKGECLDIMRQHFNEAHAGNVECHE GCDDHFKMHILMYDESGDLCSRIYARLGYVLTKEKIKKIGSKVMVIKHNECEVPEDELSACFERFGVHAEITVYEEPMDE AILGVMERFSNLDCMKEVWMTDKEWNSVGLRVLKEKVLFVI |
| 1674 |
PF16462 (Phage_TAC_14) |
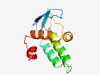 Estimated TM-score=0.65; easy target. |
>Phage tail assembly chaperone protein, TAC (Length=113) TLKAALLTPRPQIKAVALFGTQINLRRMTALELLELEEKAETLSDAGNGREASRLNIQMVLDCLVDDKGKPIDKADLPTT DELMAIHDNATLIEAIQTVKRHAIGTLEEAEKN |
| 1675 |
PF16238 (DUF4897) |
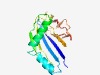 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4897) (Length=158) FEIVYYRTKMEYDYNGSAVFTTTAGLFFKDKNKQASYVQQYQQVSLDTFRKYFEDVSKKIGREIEVVSMESSINERAGIL EIVEVAKLNNAAVVTDGVVDTQMKDITLSASGDSEIVIVIPKDAVVVSVEPTPTKIVENQIYWQPIGSRMAFPRVVFK |
| 1676 |
PF16235 (DUF4894) |
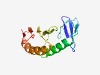 Estimated TM-score=0.65; easy target. |
>Domain of unknown function (DUF4894) (Length=120) IIFYKNRFWWISEKGFLLHTASIDDIVKQAFITDVNVNSLIVDANDVNIIQRIEDVLKNPYIAEIKLSNKSAILLKGVVL YFNEWDDLIRHSSKLEKLLKTMEPRTEYFLSSSGLVYKIR |
| 1677 |
PF16152 (DUF4860) |
 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4860) (Length=97) YQTSTPLAYLTEKVRQNDVADSISIEELSGISCLALRGSSDDISYTTYLYTYDGWLRELLVRDGTKASPETGKKIIEASD FSIEELSNGLYRLTITD |
| 1678 |
PF16116 (DUF4832) |
 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4832) (Length=205) RIGHHNDCFVASYNDVGTYPIDAVEQWKDYVWGDTRYTPHGGETCAPSSYAHSSKAIPEMEKLHTSYLFSGWHPAVYQGW KDQGKFDEISRRLGYRLVLDNAGWTKRFRPGQTASVELNLRNVGFAAMFNERPVYIVLDGAGGRFNFPLPDADPRWWAPG EATNVTAQFQVPANVPPGDYKLALWLPDDAESIRNRPEYAVRLAN |
| 1679 |
PF16090 (DUF4819) |
 Estimated TM-score=0.65; easy target. |
>Domain of unknown function (DUF4819) (Length=86) YDVIGDASPSLGQVTLDTKVCIRCPIANNHVDATKVFVKGTVCKILTKPNRFVVKISREDDQSESYVVKRADLRLVQPPW ADELEE |
| 1680 |
PF15920 (WHAMM-JMY_N) |
 Estimated TM-score=0.65; very hard target. |
>N-terminal of Junction-mediating and WASP homolog-associated (Length=50) QPDSLEGWVAVRENAFAEPEPHKLRFLVGWNEIESKFAVTCHNRTLQEQQ |
| 1681 |
PF15649 (Tox-REase-7) |
 Estimated TM-score=0.65; hard target. |
>Restriction endonuclease fold toxin 7 (Length=88) GNEGERRAGIDPATGKQRIYPQNPVGKPKYRIPDGRNDVTKQITEVKNVNDIKSKDSKQIIDEANWAAQNGYTMTLVTDH RTVLSQDV |
| 1682 |
PF15639 (Tox-MPTase3) |
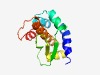 Estimated TM-score=0.65; hard target. |
>Metallopeptidase toxin 3 (Length=132) RKYPKLAYYVKVNLPQVVHVPAIVQALKKIANLNDVKLRDALIWGKGPQIKVTYLVDAYGEFSPNQQSNELRIDTSLVTD FEKGRGKRMARAGNVYLVGITLLHELTHWGDDQNGIDRPGEEGEEFEKLVYG |
| 1683 |
PF15631 (Imm-NTF2-2) |
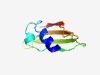 Estimated TM-score=0.65; easy target. |
>NTF2 fold immunity protein (Length=67) TAIKIAEAVWLPIYGEKIYSQKPYMVSLKDDSIWIVTGTLAENKRGGVAYIEIQKKDCKILKVTHGK |
| 1684 |
PF14952 (zf-tcix) |
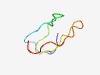 Estimated TM-score=0.65; easy target. |
>Putative treble-clef, zinc-finger, Zn-binding (Length=40) DLGKPTLRGVRKCPKCGTYNGTRGLSCKNKSCDVVFKETS |
| 1685 |
PF14877 (mIF3) |
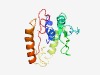 Estimated TM-score=0.65; easy target. |
>Mitochondrial translation initiation factor (Length=175) KKILITWSTGTDRAKEAANSVVSEIFKKNHKGNIKVVDPTTHRIEASNIRYFAKGIDLDKVGLSIVNVEQIDNENQIPLV KIVESRVALKKYSDFLAKKKEKELMELGVLNKSYKNLVTDKKEDNLKHIKISWQIESDDLKRQKAHEIVSLLKKGNKVTL YLDDKNNINSNNWLE |
| 1686 |
PF14783 (BBS2_Mid) |
 Estimated TM-score=0.65; easy target. |
>Ciliary BBSome complex subunit 2, middle region (Length=102) AMITFDFDKDGENELLIGCDDSFIRVLKNNQFIAEIAETGPILCLSPVNEVRFAYGLANGTIGIYEEGIRLWRVKSKQNA LALQWSGETLACCWSNGRIDWR |
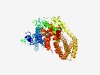
| 1687 |
PF14745 (WASH-7_N) |
 Estimated TM-score=0.65; easy target. |
>WASH complex subunit 7, N-terminal (Length=567) GMFMEDYATQLREIEDALDDSIGDSWDFTLDPIALQLLPYEQTSLLQLIKTDNKIFNKIITVLASLCSEMKALRHEAESK FFNALIFYGEGEPEKGLAEGEAQVQIGRMMPLIQELSCFVNRTNEVVKNVIQQFACLFSVVRNGPKLLDVSDVHFQVVYE HLGDLLGVLITLDEIIENQGTLKDHWILYKRMLKSVHHDPGKFGIAAEKIRPFEKLIMKLEGQLLDGRIFQDCVEQPFDT PTVYVSKNPNLAEEFAVSVKEYLMELEARIGEPNEMDQRYKFVGLCGLYILHFQIFRVIDKKVFKSVWDMHKKIPAVHLI GNIVWFPTEFLLSKMPQVDKLLDRKAKDSVTSTRTSWLAQRQQLLTKDVQNFHTTVMAWMVEMESAMTKANLMEDLNNRC VLFIQGVLHAYNISHLVRTTMNLHVAMSKPMTKSCLLSLCRLIELLKAIEHTFHRRTMLIAESVNHITQHLSFICLNSIA VAKKRITSDKRYSERRLDVLSALVLAETALNGPGTKERFLITQLALALGTKMKTFKDDELQNLAATMKKMEVICDLREKV RQACDCS |
| 1688 |
PF14421 (LmjF365940-deam) |
 Estimated TM-score=0.65; easy target. |
>A distinct subfamily of CDD/CDA-like deaminases (Length=187) VARAVDEIKAFRDQFVQGEHLGEVGQFWLRKSRKPVLAVLLVEKRTSSGDVQVVVHRGMNCEVSMPTGSLCAERNAIGSA LANDPTLLRQSLKMIAVLSVITTAVEASPKTGRKPKRPRTISCDASVPSIQAALDAVDASAATEKDLNPLAPCGACNEWL LKIAEANPSFKIVTFDSIDCDSVYIHQ |
| 1689 |
PF14394 (DUF4423) |
 Estimated TM-score=0.65; easy target. |
>Domain of unknown function (DUF4423) (Length=166) DVHKVRIIEGDSFRYFDSWKNPVLRELAPAMPGAKPLALARACRPKITAAEVSESLSFLIKANLLQKDEDGNYVQTEKSV TTGPMDVTPVAVRGMHRQMGEFALDAIEGVPQKERHFSGLTLGITQSAYDEIVQEIAEFRKRIIAIATKNDAADEVYRLN VQFFPM |
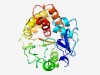
| 1690 |
PF14393 (DUF4422) |
 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4422) (Length=242) KIFVSHRIDLESEIIDNPLYYPVRCGAVFDNKESEIPGDNTLENISEKRDTFCELTVQYWAWKNAEADYYGLCHYRRFQS FADKQFEETDIYKHVVEQQIDPEFIYRHNLNETAMRKVIEKYDVISLVPMDLNDLEKGLTVYKSLENNAYVYNMMGVDLF IKIFKETYPEYSEDIDEYFNGHIWRAFNCYILKKELFFDYSERLFNVLFQLEKKLDISKFNQEQKRLAGYMGECMFAIYY NH |
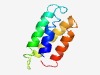
| 1691 |
PF14357 (DUF4404) |
 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4404) (Length=83) LQQQLQQLRDQLAQEPPLDAEERASLIELMQEIELRLAAEAAAAPDATLVDGVNLAVERFEASHPTLAGTLRNIMQSLAN MGI |
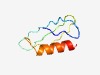
| 1692 |
PF14354 (Lar_restr_allev) |
 Estimated TM-score=0.65; hard target. |
>Restriction alleviation protein Lar (Length=62) ELKPCPFCGSDNVGTERHYGFADKDYEEWVNCYNCDASGSHTCWFDDVGVADSEAIKLWNRR |
| 1693 |
PF14330 (DUF4387) |
 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4387) (Length=97) LVDVASVIRSKNSGPFELTFDVIFKDFETYNKVKDANIFNEKMFCELYHIKAEDIINIIHFDPAKAIKVTIVRPIPSGSL GETDVYGAQQHTPLMKM |
| 1694 |
PF14326 (DUF4384) |
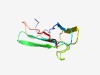 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4384) (Length=79) TYNIGESITFYFKSNRDCYVHLLDIGTSGKVTLIYPNQFATDNFIKGGQTYKLPAPETFQFVASGPEGTEMVKAIATLK |
| 1695 |
PF14299 (PP2) |
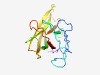 Estimated TM-score=0.65; easy target. |
>Phloem protein 2 (Length=152) KNCFMVYARGVSMTWAENNSYWQWLTEKDAPSDDAPVEMAELLNVCWLDVHGMFDVSKLSLGTTYEVAFVILMRSSGDGW HIPVNIQLTLPDGTKQEHTENLMEKPRRDWIEIKAGELSTTKHKEGEMEVSMFEHGEHWKSGLVVKGVLIRP |
| 1696 |
PF14261 (DUF4351) |
 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4351) (Length=59) REGQREGEVTLIVRQLTRRLGEINPALQQQIRGLSTLALEELGEVLLDFSAVTDLVAWL |
| 1697 |
PF14216 (DUF4326) |
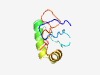 Estimated TM-score=0.65; hard target. |
>Domain of unknown function (DUF4326) (Length=76) VVHCKRDEYDVYIGRPSKWGNPFVVGRDGDRETVIQKYREWIMEQDELLADLHELRGKRLGCWCAPKACHGDVLAE |
| 1698 |
PF14169 (YdjO) |
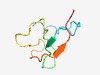 Estimated TM-score=0.65; hard target. |
>Cold-inducible protein YdjO (Length=59) MNYRKKPLEEVPEENTAVWSCTNDDCNGWMRDNFTFEDAPACPLCHAPMEQGMRMLPQL |
| 1699 |
PF14168 (YjzC) |
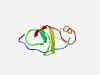 Estimated TM-score=0.65; very hard target. |
>YjzC-like protein (Length=55) MGQQRHFQPGDKAPNNGVYIEIGETGDNVKNPKKIKLKAGDTFPETSNHNRHWTY |
| 1700 |
PF14143 (YrhC) |
 Estimated TM-score=0.65; hard target. |
>YrhC-like protein (Length=72) KKWSGLVTDYKQYAFILLAVSTFLYIGTIIPNQHIEASPKMLMMGIDCVFLIAAFLCVRRAIFFQKKLQELD |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218