

Go to page (83 pages in total): [First page] << < 17 18 19 20 21 22 23 24 25 26 27 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 2101 |
PF11138 (DUF2911) |
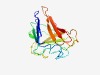 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF2911) (Length=145) SPSATAYQKIGTTDVTINYSRPGVKGRTIWGGLVPYNELWRTGANAATNIEFSTDVVVEGNKMPAGKYALFTIPSEKEWT VVINKNWEQGGTAEYAEADDVVRFKVTPKKVDHSHEWMFFTFFAQSKNSAKAILAWEKLMVPFTI |
| 2102 |
PF11079 (YqhG) |
 Estimated TM-score=0.63; easy target. |
>Bacterial protein YqhG of unknown function (Length=259) MQQHEIHQFLERYFRANECDILEADDGYIKVQLSIEMDKELMNRPFYWHYIERTGGTPAPMQLTLITKQTEKTNSLEGER LHFGSPRLHQIFRSAQKRGGFIRLYEEVDTYQQSHIPLHPWLGLNVKISYQCDRTKDQILSLGLHLISGTIIEQFHDRLQ KLKLTPKIPDYCFTISPLIKPKSGIGRLEQYIRRHIFQDDHTWAEKAKERWAEDLALLDHFYEGMEEKPECYYIEKEALE EQYKPRIIVSVINGGLFYL |
| 2103 |
PF11011 (DUF2849) |
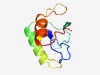 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF2849) (Length=85) KVVTANDLLDGDVIYLTADDRWTRDLSEAELITDEAHAQLRLLEAERQANKVVGAYLADAVAGENGPEPTHFREDFRRTG PSNYA |
| 2104 |
PF10983 (DUF2793) |
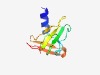 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF2793) (Length=87) HNEALRILDGLVQLSVLDRDLTTPPGSPAEGDRYIVGSGATGDWAGWDLNIALWTDGAWLRLPPRTGWRAWVEDEGLLLV YDGAGWV |
| 2105 |
PF10974 (DUF2804) |
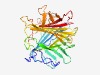 Estimated TM-score=0.63; easy target. |
>Protein of unknown function (DUF2804) (Length=324) DGSLREPGWSKSLVQTYDRKQIKAPRMRIKEWDYYLVLNEDFAGAFTLSDDGYIGLQSVSLLNFKEGWEHTETILNAFPM GKMQMPSDSRQGDIVYRDKRLHMEYTLEEGQRRIRCKFDNFYQGKTFRCDITLVQPKMDTMVIATPWKEKKTAFYYNQKI NCMPAKGKMEYDGKIYRFDPSTDFGTLDWGRGVWTYDNRWYWGSGNQMIDGKPFGFNIGYGFGDTTAASENMLFYDGKCH KLDDVTFHIPDGDYMKPWKFTSSDGRFEMNFEPVLDRAARTNALIIESDQHQVFGRMNGKAVLDDGTVIEVKDMMCFAED VHNR |
| 2106 |
PF10945 (CBP_BcsR) |
 Estimated TM-score=0.63; hard target. |
>Cellulose biosynthesis protein BcsR (Length=41) DQDDINALSDAFSLKAFRYIDIAREARLQQLLSRWPLLQEC |
| 2107 |
PF10942 (DUF2619) |
 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF2619) (Length=69) RLLSGSIEILAAMLMIKFNQVEKALVINSSLALVGPMILIATTTIGLVGITDKISFTKIIWILLGVACI |
| 2108 |
PF10920 (DUF2705) |
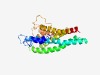 Estimated TM-score=0.63; easy target. |
>Protein of unknown function (DUF2705) (Length=229) VLLTIIMQGILISRFNLINTNFPFLDGIPITSSHALEYRYLLNWYLPIISMSFYFSGYLSDLTRIHGITFFVRTYSKSKW IIKQYLCMLMILLLFTIFQTVIFTLLGLGKSTFSTTNFIKSLSIYLLTLLTLFSIQILLELYIKTKISHLIINIYIVSSV LMTNQLYGSSKLLQYLFLPNYGMGFRNGLSQIPEFKIHTIDYLIGLFILLIIQTAVIYLSVNKIKKMDL |
| 2109 |
PF10902 (WYL_2) |
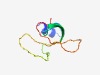 Estimated TM-score=0.63; hard target. |
>WYL_2, Sm-like SH3 beta-barrel fold (Length=69) ETLVDRLRKGIVSFVYRKKDGTERHACGTLYGIGHTIKGNKKHQQCKYTLAYYDVDCKGWRSFVIDNLV |
| 2110 |
PF10820 (DUF2543) |
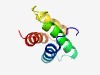 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF2543) (Length=81) MNNEIPLKYYDIADEYATECEKPVSEAERDPLAHYFQLLITRLMNNEEISEESQQEMAREAGINESRIDEIANFLNQWGN E |
| 2111 |
PF10807 (DUF2541) |
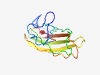 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF2541) (Length=132) SFKRMSAAALLSCLCLAAAGKAKAEQVVVLGQTTLAYSENESYMLKLANCDRQPRYTAVKLQALRGNADLKQVTLVYGNG EVDRLPIVKRLNQGNETGWIAVQGNKRCISRITIVGDTSNSSANRALLKVLG |
| 2112 |
PF10754 (DUF2569) |
 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF2569) (Length=139) YQRIGGWLLAPLAYLIVTLLSASLMLALYAMAIFTPESREYLVTNSQAFTLQWYFSVVTTLLMWIYTLWVIWLFCSRSQR FPKWFILWLLLTVLLALKAFAFSPISDDIALRTLGWPLLAAAIFVPYMKRSQRVKHTFI |
| 2113 |
PF10688 (Imp-YgjV) |
 Estimated TM-score=0.63; hard target. |
>Bacterial inner membrane protein (Length=157) WFAQAVGVLAFLVGITSFFNRDDRRFKLQLSGYSLTIGIHFFLMGANAAGSSALLSACRNLVSMRTRSLWVMWLFLSLTL VMGLSRYQHWVELLPIFGTSISTWALFRTRGLTTRCVMWCSTACWVTHNIWLGSIGGSLIEGSFLLMNGFNILRFRR |
| 2114 |
PF10607 (CLTH) |
 Estimated TM-score=0.63; easy target. |
>CTLH/CRA C-terminal to LisH motif domain (Length=144) FATMSKIRQSLEKGSVQEALSWCNENKKELRKMQSNLEFMLRCQQYIEMMRTGSQTKMIDAINHAKKYITPFNDTYPVEV SHMAGLLAYRPDTKIEPYASLYSASRWQKLAETFSEAYLKLLNLPMTPLLHIALSSGLSALKTP |
| 2115 |
PF10549 (ORF11CD3) |
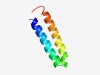 Estimated TM-score=0.63; hard target. |
>ORF11CD3 domain (Length=52) SVMEELNQACADMKRDKNIASVFATGLNEWKQVKAAHVSKIRTLVNEANMLL |
| 2116 |
PF10548 (P22_AR_C) |
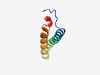 Estimated TM-score=0.63; hard target. |
>P22AR C-terminal domain (Length=73) KKYTFEFTEDELQSLVWAWFAFVRGIHTFRYIYPMFQKLGSNIAPEIYGQGFEYSHTAQSAHKILERITKEFD |
| 2117 |
PF10521 (Tti2) |
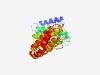 Estimated TM-score=0.63; easy target. |
>Tti2 family (Length=303) IVLALASFTSEADPWTTDESRTASTAILERFRTTSPSTPDASFWVVLETILKDKIRPLFTRTRNPAITAAGRKDMHPIPL PRFDTSILDPESKPWRSSDVYATTVFSWIVTQYQANDISRLESHFPLLVPPILALIDDETNPHKADGCTLLTHLLHPIQQ AKSDILKRTNLSSVFIDAIKPCLLSLPTITPESDSIRLLGTAYPTLFLLLKTSYLPSDKQTYTAELTKVLRENLISSFHH ISTMTPSSGTQSTLSSFPHPRLSTLLLNKIHDILFDLAVHTTKYLQEIIPLIYSTLSNPFGTA |
| 2118 |
PF10316 (7TM_GPCR_Srbc) |
 Estimated TM-score=0.63; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Srbc (Length=266) INTVGISAFILSLSNVLLNFYLIFSIFILKKVPKKADWTLIYCRFVVDMFYSLDTTTYLGYFLIRLVDPTIVIKNLAFYI AWPNFNLGTMRCWIVFFIGLDRVLATCAPISYHNNRSKIPTLAFFLFILSYTVFEYYILLVLCSFELDVPLDCTIFRCAV GTCYHDYWKWYQQIMYSFVGFLSIILFFRLFIWNSFTKNVVNKTISRATRISLLDSFIIFAFDFFPVFLTAQWPQYNTTT VGPLGSLCKSLGFVIESTIICHVLIG |
| 2119 |
PF10277 (Frag1) |
 Estimated TM-score=0.63; hard target. |
>Frag1/DRAM/Sfk1 family (Length=222) HWWMLPLASLIAWYGMLIAMLACWSFQGHPIYWFMDVRQFPVYISDIGATNLKPLFISCAGFQGLAFFLTLCCERYLRAK QKLIPNFNKKEVVSSYLSIFFCAIGELGILFVSIFDTHLFHTVHITMVGIFISFMFFSCFSLIFEYFSLGRNYYRKFNVK KYNYLMLSAITKLIWCIFAMGLAIGFGVCMRKRLRSQSAALEWTLSFWYGVFLMILIFDLYP |
| 2120 |
PF10020 (DUF2262) |
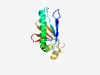 Estimated TM-score=0.63; very hard target. |
>Uncharacterized protein conserved in bacteria (DUF2262) (Length=142) DKVLGTLTLNREFDMFETNFSWNGAEIPLALEVNQESKSSWSRARTAAKKLIAEQEAWDKAMREFAAKELTELANDWLAD DDENEDAEPITEESFAQRITLSELTITSGGSFTAYYNDDDMFWGHVVEVSGSLKKGITDANI |
| 2121 |
PF09961 (DUF2195) |
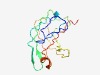 Estimated TM-score=0.63; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2195) (Length=121) ILHLSSGGVALAADTGRIIIRNDLAACVDMRVEKPPFEYSNGSNIVAINANFKLRKPIGACGCMSALARYASSVNERESR LFLQQGLFNLKKSDTKTLPLATEPALVKDGNIEITVGCARP |
| 2122 |
PF09951 (DUF2185) |
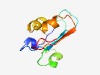 Estimated TM-score=0.63; very hard target. |
>Protein of unknown function (DUF2185) (Length=84) CIATDKITVEGFKIGYMYREKPNPKYPDSGWRFFQGEESDEYIKDVKNSGIYDLNTICNYDPSIIPLLKSPYGVCYYRDK MGTF |
| 2123 |
PF09885 (DUF2112) |
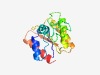 Estimated TM-score=0.63; easy target. |
>Uncharacterized protein conserved in archaea (DUF2112) (Length=143) MKVAIFPPNSLILADLVERKGHEPLILQKEIRKKVTDPEIDSPPFNITQEDPIKGLKYAAIEVPSGVRGRMSIFGPLIEE ADAAIIMEDAPFGFGCMGCSRTNELCVYYLRQKNIPILELKYPTSREETIELVNKINTFLDEL |
| 2124 |
PF09884 (DUF2111) |
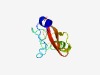 Estimated TM-score=0.63; easy target. |
>Uncharacterized protein conserved in archaea (DUF2111) (Length=82) IHELVNRLPLTMRSKEKRGLRIENGKVIDYNYTGPVLEKVLKTGKICKETPNEGPYKGIPVVVVPLKENGEVIAAIGVVD IT |
| 2125 |
PF09624 (DUF2393) |
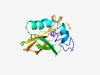 Estimated TM-score=0.63; easy target. |
>Protein of unknown function (DUF2393) (Length=143) FILFIIFALLLRKKLIVALFFIFLGFATLVLGPTFGYIQMHKYIFKNSTKLLSQKKLNFVQAVVVKGTITNESKFDFGEC KITAEVYKVTKNKYKNYLFKLKPFQKMSILEQNLPKGQMREFKIIIEPFRYQKDYNISLGASC |
| 2126 |
PF09582 (AnfO_nitrog) |
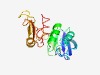 Estimated TM-score=0.63; easy target. |
>Iron only nitrogenase protein AnfO (AnfO_nitrog) (Length=189) EIAVYMGDNGETTSLYQNGKVMVYQRNQGCWRVSRERDFSLGDNFNIKALREIMDELIEFLGHCKTFVGLSITGIPYFVL EKSGCSIWEFEGNPLDFLDYILAKEEEEQRENSQQNSTILPTPVETFSGCYRISIKEIQEKNLGVSSKQVLLPFIRQGKF YSLEVLCNHVPPWLENEILLNQLESRTEV |
| 2127 |
PF08885 (GSCFA) |
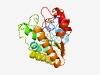 Estimated TM-score=0.63; easy target. |
>GSCFA family (Length=237) KIMMLGSCFAENIGEKLIQNKFQTLVNPFGTLYNPESIAHSLEFLIKKKKFTESDLFEQQGVWNSFYHHSRFSDTDKERC LANINTQIEKGSQFLKKADFLIITLGTAWVYQLKKSGTIVTNCHKVPTKEFERFRISKCDMVEKLRSVIKKIKNQNLNLK IIFTVSPVRHLKDGAIENQLSKASLLLTVHELVAEFEKVSYFPSYEIMMDDLRDYRFYAGDMVHPSGQAIDYIWEKF |
| 2128 |
PF08876 (DUF1836) |
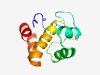 Estimated TM-score=0.63; easy target. |
>Domain of unknown function (DUF1836) (Length=100) PYWHELPDLDLYLDQVLLYVNQTTSSTEQLEQKSLTAAMINNYVKHKQLEKPVKKKYQKQQVARLIALTILKNVFSIQEI SQTLNLLLQSSNSETLYNHF |
| 2129 |
PF08723 (Gag_p15) |
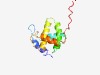 Estimated TM-score=0.63; trivial target. |
>Gag protein p15 (Length=123) GDPLTWSKALKKLEKVTVQGSQKLTTGNCNWALSLVDLFHDTNFVKEKDWQLRDVIPLLEDVTQTLSGQEREAFERTWWA ISAVKMGLQINNVVDGKASFQLLRAKYEKKTANKKQSEPSEEY |
| 2130 |
PF08636 (Pkr1) |
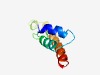 Estimated TM-score=0.63; hard target. |
>ER protein Pkr1 (Length=71) DFIKDLWESIFTPGPTPSLLIATNVSFAALQVVLFALLIATYSIHFVVLSGLCGGLWWAINWFAREVKEAQ |
| 2131 |
PF08629 (PDE8) |
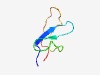 Estimated TM-score=0.63; very hard target. |
>PDE8 phosphodiesterase (Length=47) MGCAPSIHVSQSGVIYCRDSDESNSPHQTTTISQGTAATLHGLFIKT |
| 2132 |
PF08476 (VD10_N) |
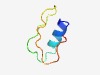 Estimated TM-score=0.63; hard target. |
>Viral D10 N-terminal (Length=42) LQYSHLMNYITQHNRKLSKTYTWTDDAQRVSATAFNNQRLRW |
| 2133 |
PF08410 (DUF1737) |
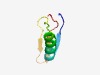 Estimated TM-score=0.63; hard target. |
>Domain of unknown function (DUF1737) (Length=53) MKAYRFLTADDTSAFCHKVTEALNKGWELHGSPSYAFDAANGVMRCGQAVVKD |
| 2134 |
PF08373 (RAP) |
 Estimated TM-score=0.63; easy target. |
>RAP domain (Length=55) IEVNGRHHFYRDTETLKTRQRLKEHILRCHHWHVINVNFYEWDALPTDADKAQFV |
| 2135 |
PF08346 (AntA) |
 Estimated TM-score=0.63; hard target. |
>AntA/AntB antirepressor (Length=68) VSARELHVFLEIKTRFDKWANRMFEYGFTENIDYQALVKNDQTLKREVLEDYALSMDCAKEISMIQRS |
| 2136 |
PF08177 (SspN) |
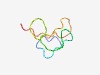 Estimated TM-score=0.63; very hard target. |
>Small acid-soluble spore protein N family (Length=46) MGNQKKRSQQFVPEHLGTQPRGFGGNKGKQMQDRSGQHPQVIQTKG |
| 2137 |
PF08167 (RIX1) |
 Estimated TM-score=0.63; easy target. |
>rRNA processing/ribosome biogenesis (Length=203) LGVLCRKLTSTPAVQLPHALPALTRHVVRCKDALSAPQDQKLKDNASQSSLLLHKIKTQITTLLNGRSREGRFAAVGLVK AVIDVGGWEMLRGSEPWLRSLLSILQKGDSFACKEMAVVTLTRIYMLLHPYPTLAREIATATLPAFITASLQIVKSDGRP LSIVETVCDAFAALIPLYPTTFRPQASQIPSAIRLYLAPTMSD |
| 2138 |
PF08052 (PyrBI_leader) |
 Estimated TM-score=0.63; hard target. |
>PyrBI operon leader peptide (Length=44) MVQCVRHFVLPRLKKDAGLPFFFPLITYSQPLNRGAFFCPGVRR |
| 2139 |
PF07879 (PHB_acc_N) |
 Estimated TM-score=0.63; hard target. |
>PHB/PHA accumulation regulator DNA-binding domain (Length=60) LIKKYPNRRLYDTKTSSYITLADVKQMVLKQEEFQVVDAKSGEDLTRQILLQIILEEESG |
| 2140 |
PF07842 (GCFC) |
 Estimated TM-score=0.63; hard target. |
>GC-rich sequence DNA-binding factor-like protein (Length=270) LEDCAQLFERLQGDFYEEYKAFGLADLAVAMVHPLLKEHFRDWEPLKDGIYGIEVIGKWRALLESGQMAHSQPDMNAMDP YHRMLWEVWLPFVRASVTRWQPRNCGPMVDFLESWAPILPIWILENILEQLIFPKLQREVESWNPLTDTVPIHSWIHPWL PLMQAQLEPLYPPIRSKLASALQKWHPSDSSAKLILQPWRDVFTPGSWEAFMVKNIVPKLGLCLSEFVVNPHQQHMDPFQ WVLDWEGMLSPSSLVGMLDKHFFPKWLQVL |
| 2141 |
PF07810 (TMC) |
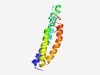 Estimated TM-score=0.63; easy target. |
>TMC domain (Length=111) CWETYVGQQLYKLILTDFAVQFVTTFFINLPRAFLARHTNSRCFKIIGEQDFYLPKHVLDIVYTQTIIWMGSFFCPFLPI IGTIFYFLIFYIKKFTCLTNCTPSPVVYKAS |
| 2142 |
PF07786 (DUF1624) |
 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF1624) (Length=222) RIVAIDIVRGIALIAMASYHFTWDLEFFGYTDPGLTAFGLWKIYARCIASTFLFLVGVSLYLAHGRRIRWNGFWKRFAMV AGAAIAISVVTRIATPDGFIFFGILHEIALASLLGLLFLRLPALLTLVVAALVIAAPVYLRLEAFDHPWLLWVGLSATSP RSNDYVPLFPWFGAVLLGIAAAKLASASGVRARLASLTPGRWANPLVFIGRHSLAFYLIHQP |
| 2143 |
PF07693 (KAP_NTPase) |
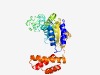 Estimated TM-score=0.63; easy target. |
>KAP family P-loop domain (Length=306) RKSIAEKVITLLRSDITVSPLVIDGSWGLGKTEFCQKLLSLMSTEETHHLIYIDAFKADHADEPLLTVLAKVLEVLPSQE EQQGLIQKAIPALRYGLKTGGKALVAHILRQDTDSIINGFDEEVKQVADKAIDASVESLLKDHVEAESSLQALQQALKSI AEQKPIVLFIDELDRCRPNFSVLMLETIKHTFDVEGVQFVLITNTNQLKASINHCYGPTIDAQRYLDKFIRFSFTLPHTT NENRHDVTMASVTHYKNLVAKSERLQGLSLDRDSDFWLVAQVINTNNISLREVETLVRHIEIYQAL |
| 2144 |
PF07659 (DUF1599) |
 Estimated TM-score=0.63; hard target. |
>Nucleotide modification associated domain 1 (Length=61) KNHDYGTSWRILRLPSITDQIFIKAQRIRTLQEKGVSKVGEGIEPEFVGIINYCVMALIQL |
| 2145 |
PF07637 (PSD5) |
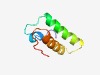 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF1595) (Length=62) CARELLKPFARRAWRRPVTPEEVERLVAFVALARQQGDTPEVGVKLALRAVLVSPHFLFRVE |
| 2146 |
PF07611 (DUF1574) |
 Estimated TM-score=0.63; easy target. |
>Protein of unknown function (DUF1574) (Length=354) LIRNRYLFVPFLVVFLTFCLDKLLILENIHTYFSKSLSDINYIQKNELYEDLKEYLSLKDRDKVLVYFGNSRGLLFDNEY IHKKYPGWVMFNFSVPGGKPDYVLQWMEEFEKDGVKPDFFLFDHSVEMFNSAASLKVDETLTNGLNVSFVLKHFSLFSTE DISTLIAKRMFRAYQYRPKLEVIIARAKNKDSFLYPYRDLRNNLMANLKSGKGSAMTPGTHQAVLPPELLKKSAYGDFHS YLVPFRFSENILSFTDQSLDLAKKLNVPSAVIWVRLSLPYMDHIRHLKVSVGENKEGTVYDDWYPRMVGYHKENGIPFWN MNDDPTYTCNEFSDAGHMSPSCYDEYTDYIFKNL |
| 2147 |
PF07593 (UnbV_ASPIC) |
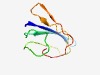 Estimated TM-score=0.63; hard target. |
>ASPIC and UnbV (Length=72) ATGARVTLTATINGQQVAQTRHVNTNPGGYGSQSSPLVHFGLGDALSVSGVTINWPSGQTWDTTGVFADQSL |
| 2148 |
PF07556 (DUF1538) |
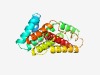 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF1538) (Length=211) IILVVAFFQIFIVQAVPENLTSIVIGLIIVAIGLALFIRGLELGIFPVGENLATDFAKKGSLFWLLLFAFTIGFATTVAE PALIAIADKASFISDGKIDSFFLRMTVAFSVGFAIALGTFRILVGHPIQYYIITGYILVVTLTFFSPIEIIGLAYDSGGV TTSTVTVPLVAALGIGLASSIKGRNPAIDGFGLIAFASLMPMIFVQGYGIF |
| 2149 |
PF07551 (DUF1534) |
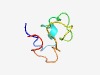 Estimated TM-score=0.63; very hard target. |
>Protein of unknown function (DUF1534) (Length=46) LSFRTLQRGNAVRDAPRHRSAPRRAFKIGRRASRTACDAERRTIVR |
| 2150 |
PF07539 (DRIM) |
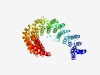 Estimated TM-score=0.63; easy target. |
>Down-regulated in metastasis (Length=602) WSMVDRKALIGVFSQFNNPRVLYQSQKVYDVLLGLMANGDIEIQKLALKAILAWKQDGIKPYQENLEYLLDEARFKNELT VLFQGDNQVQPEHRSDLMPVFLRLLYGRAISKKGAASGRHGLHATRLAVIRNLNIEDMGSFLSIALGELRDIRVVNASGV KEAVFAREALPVRRQVGLLNMVESIINELGTSVVPYMESLVNAVLYCLIEACRQLREDGEDADDAEATTNKSLYKVARTT AIKCLCKLFQNAQSFDWTPYQNAIVTEVISPRIEKLAAETTQGISGTWKLLATWSVLPKPAMFLSIDDRILPNVMECLGR GKAKDEVKVFALNIVRNLVKLAVAPAEESEFNELIKAELLDPNIDLILKQIGDMLRDQHEIGRDLLETAIETVVDLAPLV KSSKNVRDVVDITTYLLQQPSRRVSPKVKGSILLILKEFIVLDDVQSSKELKDKVYATLGPLFGYFKDTQNRQKLAEVLQ VFAAHEPSVQEIADICSDLNSYLEDRLDEPDYNRRLAAFNVISKDGESRFTIDQWIPLLHNMVYFICQEEEFGILATNSA DGLTKFIEAAEAGWEGSEHDRYVDLLSNVILPAIYSGAREQS |
| 2151 |
PF07483 (W_rich_C) |
 Estimated TM-score=0.63; easy target. |
>Tryptophan-rich Synechocystis species C-terminal domain (Length=105) TKLVKDTTNKLYAQIGNNNPIAIKIAATQITTNTYPGWQILAAETVNGINQVLLKNTSQNLLYIWNLDNNWNWQSSQGGW GLNSTPAFSQETNFQQDFNGDNQIG |
| 2152 |
PF07437 (YfaZ) |
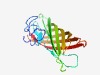 Estimated TM-score=0.63; easy target. |
>YfaZ precursor (Length=180) MKKIALAGLAGMLLVSASVNAMSISGQAGKEYTNIGVGFGTETTGLALSGNWTHNDDDGDVAGVGLGLNLPLGPLMATVG GKGVYTNPNYGDEGYAVAVGGGLQWKIGNSFRLFGEYYYSPDSLSSGIQSYEEANAGARYTIMRPVSIEAGYRYLNLSGK DGNRDNAVADGPYVGVNASF |
| 2153 |
PF07282 (OrfB_Zn_ribbon) |
 Estimated TM-score=0.63; easy target. |
>Putative transposase DNA-binding domain (Length=70) WRMFRQMLEYKCKWYGKKLIAVDPKNTSRICSKCGYNSGAKPLEIREWTCSKCQTKHDRDINAAVNILHK |
| 2154 |
PF07011 (Elf4) |
 Estimated TM-score=0.63; hard target. |
>Early Flowering 4 domain (Length=82) EVDGKVLQTFQKSFVQVQGILDQNRLLINEINQNHESKIPDNLSRNVGLIRELNNNIRRVVDLYADLSSSFTRSMEASSE GD |
| 2155 |
PF06793 (UPF0262) |
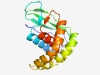 Estimated TM-score=0.63; very hard target. |
>Uncharacterised protein family (UPF0262) (Length=151) RLIEITLDESTVVRRSPDIEHERKVAIYDLLEGNSFRPEGSEVGPYRLHLSVEENRLVFDIRTGEDEQHAKVMLSLTPFR KIVKDYFLICESYYDAIKTAAPAQIEAIDMGRRGLHNEGSELLKARLDGKIAVDFDTARRLFTLICVLHIK |
| 2156 |
PF06750 (DiS_P_DiS) |
 Estimated TM-score=0.63; hard target. |
>Bacterial Peptidase A24 N-terminal domain (Length=84) IFGTIIGSFLNVCIHRIPKEESIITPPSYCPKCGKKIKWYDNIPIISYLILKGKCRNCKEKISIQYPLVELLTGFLTTGI VWKY |
| 2157 |
PF06743 (FAST_1) |
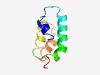 Estimated TM-score=0.63; hard target. |
>FAST kinase-like protein, subdomain 1 (Length=68) ATLLPFAVLDYDPPQGDEFFETCIQRFKPHLSSFDPHLLVLLAYSLAVADCFPEELIRTIFNVDFLSK |
| 2158 |
PF06649 (DUF1161) |
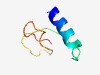 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF1161) (Length=47) CGELKDEIAAKLTAAGISPSVLAIVDKDWVGAGKIVGSCENGTKRIV |
| 2159 |
PF06543 (Lac_bphage_repr) |
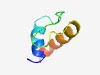 Estimated TM-score=0.63; hard target. |
>Lactococcus bacteriophage repressor (Length=48) NHQEPIDLAPLVDESKVDWDEWVSFDGKPLTDEVKTALKLILGKRLED |
| 2160 |
PF06541 (ABC_trans_CmpB) |
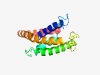 Estimated TM-score=0.63; hard target. |
>Putative ABC-transporter type IV (Length=154) WCFFLFSFLGWCSEVVFAAVRERRFVNRGFLNGPLCPIYGSGVVAIDFFLRPFGSSLPVQLVGAMLLGSVLEYAAGFLLE KLFHQKWWDYSDTPHNLHGYICLKFSVVWGLAGALIVRYVMPLAHRLFGQIPRQVGWVLLVVLGSLFVADFLVT |
| 2161 |
PF06306 (CgtA) |
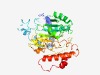 Estimated TM-score=0.63; easy target. |
>Beta-1,4-N-acetylgalactosaminyltransferase (CgtA) (Length=347) MLFQSYFVKIICLFIPFRKIRHKIKKTFLLKNIQRDKIDSYLPKKTLVQINKYNNEDLIKLNKAIIGEGHKGYFNYDEKS KDPKSPLNPWAFIRVKNEAITLKASLESILPAIQRGVIGYNDCTDGSEEIILEFCKQYPSFIPIKYPYEIQIQNPKSEEN KLYSYYNYVASFIPKDEWLIKIDVDHIYDAKKLYKSFYIPKNKYDVVSYSRVDIHYFNDNFFLCKDNNGNILKEPGDCLL INNYNLKWKEVLIDRINNNWKKATKQSFSSNIHSLEQLKYKHRILFHTELNNYHFPFLKKHRAQDIYKYNWISIEEFKKF YLQNINHKIEPSMISKETLKKIFLTLF |
| 2162 |
PF05939 (Phage_min_tail) |
 Estimated TM-score=0.63; hard target. |
>Phage minor tail protein (Length=99) PDMEVNSQPSVREVRFGDGYSQRMAAGLNADLKTYRVTLSVTREEARHLEAFLAEHGGWKAFLWKPPYAYRQIKVTCAGW SARVGMLRVEFSAEFKQVV |
| 2163 |
PF05804 (KAP) |
 Estimated TM-score=0.63; easy target. |
>Kinesin-associated protein (KAP) (Length=708) VKGGSIDVHPTEKALVVNYELEATILGEMGDPMLGERKECQKIIRLRSLNSTTDIAALAREVVEKCKLIHPSKLPEVEQL LYYLQNRKETTQKAGAKKGELTGKLKDPPPYEGTELNEVANINDIEDYIELLYEDNPEKVRGTALILQLARNPDNLEELQ SNETVLGALARVLREEWKHSVELATNIVYIFFCFSSFSQFHSIVAHFKIGALVMGVVEHELKKHLSWNEELLKKKKTAEE NPASKRDFEKSTKKYHGLLKKQEQLLRVSFYLLLNLSEDPKVELKMKNKNIIRLLIKTLERDNAELLILVVSFLKKLGIY VENKNEMAEQQIIERLAKLVPCDHEDLLNITLRLLLNLSFDTDLRGKMIKLGMLPKFVELLANDNHRLVVLCVLYHVSQD DKAKSMFTYTDCIPMIMKMMLESPTERLEIELVALGINLACNKHNAQLICEGNGLRLLMRRALKFRDPLLLKMIRNISQH DGPSKAQFIEYISDVARIIKESNDEELVVEALGIMANLTIPDLDYEMIINEFGLVPWIKEKLEPGAAEDDLVLEVVMLVG TVATDDSCAAMLAKSGIIQSLIELLNAKQEDDEIVCQIVYVFYQMVFHEATREVIIKLTQAPAYLIDLMHDKNSEIRKVC DNTLDIISEFDEEWARKIKLEKFRWHNSQWLEMVESQQIDDGEGGLMYGDESYSPYIQESDILDRPDL |
| 2164 |
PF05675 (DUF817) |
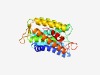 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF817) (Length=237) ELLLFGFKQGWACLFGGLLLALLLGTHLFYPEGAALHRYDFLTLAALLIQTGMLALRLETWREARVILIFHLVGTVMELF KTQAGSWTYPEASVLRIGAVPLFSGFMYAAVGSYIARVWRIFDFRFTGYPPAWMTWVLAAAIYINFFTHHYTIDIRWGLF AATAAIFWRTRVHFRNWRAHRWMPLLVGFGLVALFIWFAENIATFANAWNYPGQEKGWRMVSLAKFGSWYLLMLISF |
| 2165 |
PF05605 (zf-Di19) |
 Estimated TM-score=0.63; easy target. |
>Drought induced 19 protein (Di19), zinc-binding (Length=56) TFKCPVCFTVFVQANKLQQHIFAVHGQEDKIYDCSQCPQKFFFQTELQNHTLSQHA |
| 2166 |
PF05509 (TraY) |
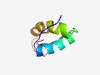 Estimated TM-score=0.63; easy target. |
>TraY domain (Length=48) VMARFDEDTNKKLIAAKNKSNRSKSHEVYLRLKAHLFEFPDFYSSEVE |
| 2167 |
PF05449 (Phage_holin_3_7) |
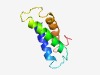 Estimated TM-score=0.63; easy target. |
>Putative 3TM holin, Phage_holin_3 (Length=79) NVATCSAIVLRLMLFRKPGARHRWWASWLAYLIILAYASVPFRYFFDFYVHTHWASVIINLIICAAVFRARGNVAQLFQ |
| 2168 |
PF05421 (DUF751) |
 Estimated TM-score=0.63; hard target. |
>Protein of unknown function (DUF751) (Length=60) FFDNISRYPRYFITIILGIFFAFFEWIKPLFKNPLTAIAVVGIFIGGFVFLFFTLRAMLG |
| 2169 |
PF05154 (TM2) |
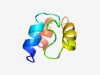 Estimated TM-score=0.63; hard target. |
>TM2 domain (Length=49) RKSLVEAYLLWLALGLFGAHHFYLNRPLFGIMYLTTFGLCGCGWLIDLF |
| 2170 |
PF05145 (AbrB) |
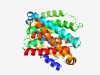 Estimated TM-score=0.63; easy target. |
>Transition state regulatory protein AbrB (Length=325) WLLGPMTFTLVGSRVVKVVRPSWPSWIRDTALILIGYSIGLTFTMTTLRQIGHQLPTMALMTFILILLCGLLAWIITRLS GIPFPTVLMGSIPGGLTQMVVLAEDTKGIDLTVVAFLQVVRLLMIIFCVPLLVYSPLFGGDSGTTAVSNTVSAVVEHAAT VAGTGWGVVVLYAVVCVAAALLANKIHFPSAYMLGPMIITAILHLAGVEGPALSAWLLSAAQVMIGTYVGLLLKPEKLQN KVRVAILAVTSSTLLIASTIGFSYLLTQLHAISSTTAFLSLAPGGMDQMGIIAHEVNADISIVTGYQIFRTWFIYFLIPP ILRML |
| 2171 |
PF05107 (Cas_Cas7) |
 Estimated TM-score=0.63; easy target. |
>CRISPR-associated protein Cas7 (Length=268) NRYEFVILFDVENGNPNGDPDAGNMPRIDAETGYGIVTDVCLKRKIRNYVEIVKEDCEGYKIYVKEGAVLNKQHETAYIG LGIDPKDEKAAKQNRDSLRDFMCKNFFDVRTFGAVMTTKVNCGQVRGPVQINFARSIDPIVPQEITITRMAVTTEKDAES KTTEMGRKHIIPYALYRTEGYVSANLAKKTTGFSEDDLDLLWESIINMFEHDHSAARGKMAVRKLIVFKHDSELGNAPAH KLFELINVKRKDSERPARSYSDYIVDID |
| 2172 |
PF05004 (IFRD) |
 Estimated TM-score=0.63; easy target. |
>Interferon-related developmental regulator (IFRD) (Length=310) EYDTAEETASVTSSHISEPDSIEEAIASHGLDDVDETSGQDAFEEKIRDFIDGTTQKSAAGRKHCFEGIRAAFSKKYIFD FVDSRKITIADCLERSIKKGKGDEQVLAAKLLNTLWIQLGAGPECEEVFAALRPVLMSKVLDKTASPAARASCASALGLG TFIAAAEFETVCSCMNALEDIFSASYLKGDGSSPTHKPGVSTLHTAALNGWMLLLSITPTSRIKHIVETHLPKLPDLLSS DDVNLRIVAGEAIAMLYELAREDDEEFEGDDIDELVVKIRNLATDSNKHRAKKDLRQQRSSFRDIIQTIE |
| 2173 |
PF04922 (DIE2_ALG10) |
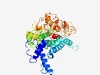 Estimated TM-score=0.63; easy target. |
>DIE2/ALG10 family (Length=397) REPYMDEVFHVPQAHKYCHGRFTEWDPMITTLPGLYLVSVGVIKPVVWLADLTGDVVCSTAMLRFINLLFNCGNLYLLYL LISKLHSREKMRSTSRRVLSALSLSTFPVLYFFNFLYYTDAGSTFFVLFTYLMTLYGCHKAAALLGVCSVLFRQTNIIWV AFCAGTLVATKMDEAWRTEHSKKREEKSPPTQIPLSLSGVRRLMIFMLEFFSSAYHMKAVLQVAWPYAAVALGFLGFVVL NDGIVVGDRSSHEACLHFSQLFYFFSFSLFFSLPVSLCYQRVMRFLQALKKQPLFFLCVTTVSLLMVWKFTFVHKYLLAD NRHYPFYVWKNVFQRHELVRYLLVPVYTFAWWNFADSFKSRSLFWSQAFLVCLLVATVPQKLLEFRYFIVPYLMYRL |
| 2174 |
PF04572 (Gb3_synth) |
 Estimated TM-score=0.63; hard target. |
>Alpha 1,4-glycosyltransferase conserved region (Length=127) GHGHELAEMCVRDLLANFNGKDWGNNGPGVITRVLQKYCDARTTVRMTRERCRHFTVYPPKAFYAIGFDDYKHFFEESYL EHALATLNQSIVVHVWNKFSKSHPVIVGSRVAYGVLADQYCPKVYRS |
| 2175 |
PF04459 (DUF512) |
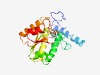 Estimated TM-score=0.63; very hard target. |
>Protein of unknown function (DUF512) (Length=216) VPAGITKYREGLYPLELFTKEEAGQVIDMIESRQKKYYEEFGLHFIHASDEWYILAGREFPEEERYDGYIQLENGVGMMR LLINEFQEALEQLRRSQEYEQMKKSFSRTVTIATGKLTYQTISKFAETLMEEFPGLTVHVYAIRNDFFGETITVSGLITG QDLIGQLKEKKESGVKLGDTLLIPGNMLRSGEQVFLDDLTVEDARRALEMEVTAVE |
| 2176 |
PF04428 (Choline_kin_N) |
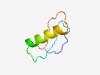 Estimated TM-score=0.63; hard target. |
>Choline kinase N terminus (Length=50) SSAGEEDLPPPNVNFFLDNRLPTEYFKQDILMLIHTLKIPKWKYVESTMA |
| 2177 |
PF04387 (PTPLA) |
 Estimated TM-score=0.63; hard target. |
>Protein tyrosine phosphatase-like protein, PTPLA (Length=162) QTAAIMEILHSIVGLVRSPVSSTLPQITGRLFMIWGILRSFPEIHTHIFVTSLLISWCITEVTRYSFYGMKESFGFTPSW LLWLRYSTFIVCFPVGMVSEVGLVYIVVPFMKASEKYCLRMPNKWNFSIKYFYASVFFMVLYAPVYLHLFHYLIVQRKKA LA |
| 2178 |
PF04178 (Got1) |
 Estimated TM-score=0.63; hard target. |
>Got1/Sft2-like family (Length=113) GVFLVFIAVTVFLPVMVFAPQKFAICFTIGCALIIASFFALKGYRTQFAHMTSKERLPITLGFIGSMLGTIYVSMVLHSY VLSVFFSILQVVSLSYYAISYFPGGSAGVKFFT |
| 2179 |
PF03993 (DUF349) |
 Estimated TM-score=0.63; hard target. |
>Domain of Unknown Function (DUF349) (Length=71) VRATFDTAITTARTRLTEHRDAVAQAKADNLKQLEEICTAFEAILGAEDIAKIETPGRELQTAWSKVGMVD |
| 2180 |
PF03969 (AFG1_ATPase) |
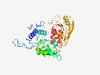 Estimated TM-score=0.63; hard target. |
>AFG1-like ATPase (Length=364) MTPKQRYEKDLQRTDFYSDEAQARAVDALDNLYHQWLNYLNQPVVRPSVWQKLFGKKPTVTQPPQGLYMWGGVGRGKTYL MDTFFEALPTEKKLRVHFHRFMYRVHDELRTLSDVSDPLEIVADRFCAEAKIICFDEFFVSDITDAMILGTLFEALFRRG IVLVATSNIAPKDLYRNGLQRARFLPAIALVETHCQILNVDSGVDYRLRTLQQAEIYHYPLDSKATDNLQRYFQQLISSE QTPETQIDVNHRMITVEAAGDGVLYATFAQLCQTARSQNDYIELSKIYHTVLLADVKQMNKSSDDAARRFIALVDEFYER HVKLIISAEVPLTDLYTDGLLEFEFKRCQSRLIEMQSHDYLAKE |
| 2181 |
PF03732 (Retrotrans_gag) |
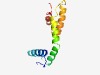 Estimated TM-score=0.63; easy target. |
>Retrotransposon gag protein (Length=100) KLAVIEFTDYAIIWWDQLVTNRRRNNERPVETWGELKALMRRRFVPSHFYRDLYQKLQNLTQGSRSVEDYHKEMEVAMIR ANVEEDREATMARFLSGLNR |
| 2182 |
PF03703 (bPH_2) |
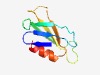 Estimated TM-score=0.63; easy target. |
>Bacterial PH domain (Length=75) CKVTDRRIEFQGGYLNRVSKQIPLDRVQDVAVNEGCCQRMFGVKSIDIQTAGSAKPTAELTLNAPKDAEMVRDLI |
| 2183 |
PF03661 (UPF0121) |
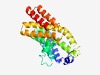 Estimated TM-score=0.63; hard target. |
>Uncharacterised protein family (UPF0121) (Length=237) QPRGVDALKQHVILNKIDTALWASRVLTIIFAIGYVIPIFGNPTNAYYKVLMANAATSALRLHQRMPAPQFSRAYLQQLL LEDSCHYLLYSLIFLYAYPVLLIIFPVTLFAVLHSASYSLTLLDTLGQNSWWGARLLISLVEFQTRNILRLAAFAEIFIM PLAVVLVFLGKAGIMTPLVYYQFLVLRYSSRRNPYTRNVFYELRLVTDNFANGARTPPFVRKVLQSSVSFVSRLAPP |
| 2184 |
PF03596 (Cad) |
 Estimated TM-score=0.63; hard target. |
>Cadmium resistance transporter (Length=192) YIATALDLLVILLMFFARAKTRKEYRDIYIGQYVGSVALIVISLFFAFVLNYVPEKWILGLLGLIPIYLGIKVAIYGDSD GEERAKKELNEKGLSKLVGTIAIITIASCGADNIGVFVPYFVTLSVTNLLITLFVFLILIFFLVFTAQKLANIPGVGEIV EKFGRWIMAVIYIALGLFIIIENDTIQTILGF |
| 2185 |
PF03235 (DUF262) |
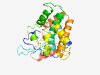 Estimated TM-score=0.63; hard target. |
>Protein of unknown function DUF262 (Length=230) IKEIFGNADSLYQIPRYQRPYSWGDEQLEKLWEDLVDAKENEPNYFLGSIITAKPEDKNNYLDIVDGQQRLTTLTILLCV YRDLYPEINNDILEEDPFAIDGKTVSNSIKFNDRFQRLRLRTHSNHQSDFDKLILSENATVDSKEIEKPTKQDLRKDEPK FKFKNSAFFFREKLTSIGEKEAGELLNYLFNKIKIIRIDCKSVSFAIKLFQVLNDRGLDLSNSDLIKSFL |
| 2186 |
PF03101 (FAR1) |
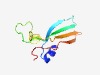 Estimated TM-score=0.63; easy target. |
>FAR1 DNA-binding domain (Length=85) LYNDYATRIGFSIRKEKLRFAKAVLRQREFVCSKEGFQRDSDLSKIKKFKKLQTRTGCKASIRFTVEDGEWKVTHFNPNH NHELA |
| 2187 |
PF02393 (US22) |
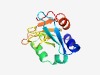 Estimated TM-score=0.63; easy target. |
>US22 like (Length=115) SGVHLQRYVRATAGRWLPLCWPPLHGIMLGDTQYFGVVRDHKTYRRFSCLRQAGRLYFIGLVSVYECVPDANTAPEIWVS GHGHAFAYLPGEDKVYVLGLSFGEFFENGLFAVYS |
| 2188 |
PF01809 (YidD) |
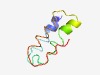 Estimated TM-score=0.63; hard target. |
>Putative membrane protein insertion efficiency factor (Length=65) ILTIGIKIYRKLISPLFPGCCRYTPTCSAYGLEALRVHGALYGSWLTFKRVLRCNPWGGSGYDPV |
| 2189 |
PF01684 (ET) |
 Estimated TM-score=0.63; easy target. |
>ET module (Length=77) CFVGIVTNYNSTSIGTEVFCNGLCGSISTSAGGYNFTTYNCFPTSFCSSAGLNESCTTVYSDRDLTGCCCGTDNCNV |
| 2190 |
PF01585 (G-patch) |
 Estimated TM-score=0.63; hard target. |
>G-patch domain (Length=44) TKGFGSKMMAKMGFVEGGGLGKDGQGMAQPIEVVQRPKSLGLGV |
| 2191 |
PF01578 (Cytochrom_C_asm) |
 Estimated TM-score=0.63; hard target. |
>Cytochrome C assembly protein (Length=226) LSNLYESLMFLTWGLLFSAIYLEYKTNLYLIGAIVSPISLFIVSFSTLSLPQDMQKAAPLVPALKSNWLMMHVSVMMLSY STLIIGSLLAILYLVLIKAQQKKHSLKDFAFANLEFTFPKSTNSTNFNLLETLDNLSYRTIGFGFPLLTIGIIAGAVWAN EAWGTYWSWDPKETWALITWLVFAAYLHARITKSWTGERPAYLAALGFVVVWICYLGVNFLGKGLH |
| 2192 |
PF18861 (PTP_tm) |
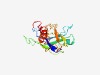 Estimated TM-score=0.62; hard target. |
>Transmembrane domain of protein tyrosine phosphatase, receptor type J (Length=184) PSPDGSPNITSVSHNSVKVKFSGFEASHGPIKAYAVILTTGEAVHPSADVLRYTYEDFKKGTSDTYVTYLITTEEQKRSQ GLSEVLKYEINVGNESTTHGYYNGKLEPLGSYRACVAGFTNITFNPHSDGLIDGNESYVSFSRYSDAVFLPQDPGVICGA VFGCIFGALVIVAVGGFIFWRKKR |
| 2193 |
PF18840 (LPD25) |
 Estimated TM-score=0.62; hard target. |
>Large polyvalent protein associated domain 25 (Length=99) NQQPTVTINFSENGELEQGDVLPYGLAERTFERLDREARAERDDPDRGRYYKTDFTVTYMMEGQLYEYNGRQDFGDGDGP LSDHIRGHAEYYRNDPQYQ |
| 2194 |
PF18739 (HEPN_Apea) |
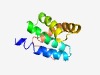 Estimated TM-score=0.62; hard target. |
>Apea-like HEPN (Length=98) ISPFEREILLAINWYGTAVDMTDPVLKFLNYAIVLEVLLSKQEKDSDRTITDKLAESVAFLLGKKYENRVEIKRDIKRLY GVRSSIVHGGKDFVQDRE |
| 2195 |
PF18736 (pEK499_p136) |
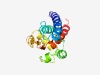 Estimated TM-score=0.62; very hard target. |
>HEPN pEK499 p136 (Length=153) MGNYYDFETEFVQRTLALIDQYNIMIEGWPFPEQYNYTLTLNCLLGLIVLPKERAFTFIPSTRLTQELKAEMGLHQSQLP GPEMNLRELIHKMRNSVAHFCVQIESVTDEHLVNRIVFKEARGERDGYATFSAPELLPFLKYYASLLLNNMGH |
| 2196 |
PF18545 (HalOD1) |
 Estimated TM-score=0.62; hard target. |
>Halobacterial output domain 1 (Length=73) STPPPVAVIKTVAVALDRKTTALEPLYESVDPDALDALLQSNGSSATPDDLSVTFTVADRQVTVHGSGEVVVR |
| 2197 |
PF18167 (Sa_NUDIX) |
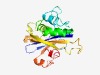 Estimated TM-score=0.62; easy target. |
>SMODS-associated NUDIX domain (Length=201) IRISFAYLFRIKVDGKYFLVQNSRTKKYQPVGGAYKFTEQEEAYLRDNIPVENDDRIPGNRITKRDYRLLVKNKYLRKFI RRFNKTQYRENIENLSREFMEEIFSTGILDKNLFGVLSYKYCGRHMTNVELSNVFCHYELLLADIVEVQLSDVQEDLFRN LMKTECDKYHFVTADEIKTLGVKFGTNELADNIANHTPKIL |
| 2198 |
PF18166 (pP_pnuc_2) |
 Estimated TM-score=0.62; hard target. |
>Predicted pPIWI-associating nuclease (Length=121) NHCRAQLARALEAQIHDEVVALAISETIGAIDELATHHCVDEIDVEEVTVTKVGSEFIHLRASGTIGVELQWGSNSDLRR GEGATGNDSFPFSCELQSHVEDPAELEAIEDTLCVDTSSWW |
| 2199 |
PF18163 (LD_cluster2) |
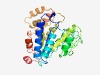 Estimated TM-score=0.62; hard target. |
>SLOG cluster2 (Length=270) LLHKLIGISISDSPDLARLGFSHFHLKRALVEISRHLLAQGASVAYGGDLRPDGFTQDLIEMVKAYNKQALELGEKIQNF LAWPLHLGQTVQFLAQHKNEITIRPIALPQELKQDFSIDEQKYLEPNSTVNQYIWSRCLTAMREQMAQQIDARIILGGKV VNYKGAFPGIAEEAALAISHHKPLFVLGAFGGCAKAVGEAMLGESPEVLTQAYQTAQSEDYAKMIQFYNQRANQGTSHKE INYTTLVETFQNTGFQGLNNGLTELENQNL |
| 2200 |
PF17546 (Defb50) |
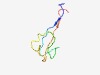 Estimated TM-score=0.62; hard target. |
>Beta defensin 50 (Length=50) HPGTFHVRIKCMPKMTAVFGDNCSFYSSMGDLCNNTKSVCCMVPVRMDNI |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218