

Go to page (83 pages in total): [First page] << < 6 7 8 9 10 11 12 13 14 15 16 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 1001 |
PF14088 (DUF4268) |
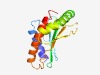 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF4268) (Length=140) IRKRYWTYALVQIHEAHGNPGSFSNVNPSTDNWINGFFGIGGFYLCCVANFDSARSEVVFGRADKDENKAAFDALYQHKT EIESKLGTELQWNRGDDIKSSKVFIQLDGVSIENEDDWPRMAKFHAEWSKKFYDVIVPYI |
| 1002 |
PF14084 (DUF4264) |
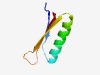 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF4264) (Length=52) KLELIATKSFEPYSDMYRIVDFLNKSLKEKRVMFGLTKDKENGKMIITIYEV |
| 1003 |
PF13973 (DUF4222) |
 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF4222) (Length=50) PQPNHRYKDAHGALVTVERVEHNRVTFYRDGYQHPCVQPIERFMKEFREV |
| 1004 |
PF13809 (Tubulin_2) |
 Estimated TM-score=0.70; easy target. |
>Tubulin like (Length=353) ICIGIGGTGRDILMRIRRLIVDSYGDLEKLPIVSFVHIDTDKSASQISGLRTGSTYHGVDLSFKEAEKVSATMSSRDVDN FTQELEKHSTSERTGPYDHIRQWFPPQLNRNIKAIEEGAKGIRPVGRLAFFHNYRKIQKAIEIAEKKTRGNEGILLKMGL RVDGGVNIFIVGSLCGGTGSGMFLDVAYSLRHSYGQQNTKIVGYLVISPELYGNNTNMKANTYAALKELNHYTTPGTKFE ACYDIQDLVIVQSERPPYDYAYLVSNETTGGYKILEQRKLCNVIAHKIALDFSGELAPIVKGMRDNFLQDLIQYDNHPRP NIQRYLTFGMAAIYFPRDKIVQIALNRISLNLV |
| 1005 |
PF13699 (DUF4157) |
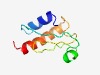 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF4157) (Length=78) PLPDPAREFMESRFENDFSGVRLHTDSEAVQLSHNLRAKAFTHENDIYFNAGQYNPNSVGGRRLLAHELTHTIQQTGA |
| 1006 |
PF13682 (CZB) |
 Estimated TM-score=0.70; easy target. |
>Chemoreceptor zinc-binding domain (Length=65) HLKWIRDLQMAFRGHGKAMRAQPSDECSLGIWIHGTAMKELGATTALKNLDTVHKQFHREVDQVI |
| 1007 |
PF13564 (DoxX_2) |
 Estimated TM-score=0.70; hard target. |
>DoxX-like family (Length=100) VTTGIIALETGVGSVWDIMKIPFVRTILEQLGYPSYMLTIMGVWKALGVVILLAPRFPRLKEWVYAGLVFVYSGAAASHL VIGHGADAVGPVIFVCLTIA |
| 1008 |
PF13562 (NTP_transf_4) |
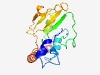 Estimated TM-score=0.70; hard target. |
>Sugar nucleotidyl transferase (Length=150) YILFDGPVRKALLPFTFTRPVADLRIGILTIREKWEKYLGYTTTTITEDYLSEKYPMVEMEENILINASFLPNEVLVEMI KSLEKNQAIFKGEEVVAFYTNDTQEEVDFDTYEIIDYNEEGLRIEHTWDIFSKNDAAIREDFELLTEGRT |
| 1009 |
PF13553 (FIIND) |
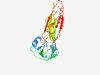 Estimated TM-score=0.70; easy target. |
>Function to find (Length=248) KVDTEGVYECTETGLIFEVNRKVDIKYCVLSWSKYADLVVKPWAVGGPMFDVKCDPACLTSIQFPHSLCLGHHDANMTFR VLHVKNSEASLECIVDHSATHVKWHVSSLSPVGPVIQSEETLYHHGAVILYKVIDQNPSLSFRVYVATNNESYIKDIAKA VKHSSKKFMKIDKPPVCLKLLEHGKKYRLVSEPEADITPEEIEFVDGSLLKLKSYIEVYLEQPMEFKLLLVEMDSDEIVW KAKLRECD |
| 1010 |
PF13547 (GTA_TIM) |
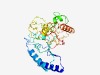 Estimated TM-score=0.70; easy target. |
>GTA TIM-barrel-like domain (Length=299) WGLRRMVLHYAHLCAAAGGVDAFLIGSEMRGLTTIRSGASTYPAVQAYRDLLADVRSILGSGTRIGYAADWSEYFGHQPS DGSGDVFFHLDPLWADPAIDFIGIDNYMPLSDWRDGFDHADAQEGWPAIYDRAYLQGNISGGEGFEWFYASAADRTAQVR TPITDGTAGKPWVFRNKDLRAWWSNPHYDRPGGVESGAPTAWVPQSKPIWFTELGCPAIDRGTNQPNVFFDPKSSESFTP HFSRGWRDDAIQRAYLEATYLFWSTQANNPVSSVYGGRMVHVSECAAWTWDARPYPYFP |
| 1011 |
PF13488 (Gly-zipper_Omp) |
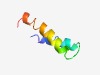 Estimated TM-score=0.70; easy target. |
>Glycine zipper (Length=42) GALGGAAAGAAVGTAVGGPVGTVIGGLAGAVAGGAGGQEVGE |
| 1012 |
PF13451 (zf-trcl) |
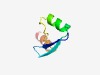 Estimated TM-score=0.70; hard target. |
>Probable zinc-ribbon domain (Length=48) EDKTLVCKDCGNEFIFTVGEQEFYKEKGFDNEPVRCPDCRRARKQQRN |
| 1013 |
PF13369 (Transglut_core2) |
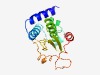 Estimated TM-score=0.70; easy target. |
>Transglutaminase-like superfamily (Length=150) LQALDAIAAAVAERLPTERYPLRVIQTLNRYLYEDLGFAGNAADYYDPRNSYLNDVIERRKGIPITLSLVYLEVAQRLDF PMVGVGMPGHFLIRPALAEMEVFVDPFHQGAVLFAEDCQERLSQVYGHPVALRPEFLQPVSKRQFLARML |
| 1014 |
PF13346 (ABC2_membrane_5) |
 Estimated TM-score=0.70; easy target. |
>ABC-2 family transporter protein (Length=209) GLIIKDILNLKNYMKQLILVLIFFIAYGIFLKNGTFVGTMITLMLSMQVITTMSYDEYAKWDKYALTMNINRKDIVLSKY VFFIMSIIIGMVVGVISSMTINQILQSDINMNEIIATSIVVPCIFAILFSIVIPVVFKNGVEKGRIVMMIVLFIPAVLVA AIVKISENNNIPMPNVSTLEMILNFGLIGVILLTIISIFMSYKISLSIY |
| 1015 |
PF13342 (Toprim_Crpt) |
 Estimated TM-score=0.70; hard target. |
>C-terminal repeat of topoisomerase (Length=63) GCNFQLGEIAGILLNEQQVKTLLEEGKTDTIRGFKSKAGKKFDACLKYEITEEGKGKITFDFE |
| 1016 |
PF13228 (DUF4037) |
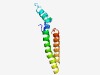 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF4037) (Length=101) WLHLPEDKLAAVTGGAVFEDGLGKFSAIREELRKYYPEDVRIKKIAARAAKMAQSGQYNYGRCMRRGDTVAAMLALNEFV RQAISMVYLLNRTYMPYYKWM |
| 1017 |
PF12928 (tRNA_int_end_N2) |
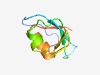 Estimated TM-score=0.70; easy target. |
>tRNA-splicing endonuclease subunit sen54 N-term (Length=67) QAISGTRIHNPKNHVLATYDPSSNMARVDQVKGQAFRTMGRHISGVMWLLPEEALYLLERGSLDIRW |
| 1018 |
PF12670 (DUF3792) |
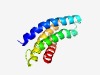 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF3792) (Length=113) QTILKGVLIAYVITGVFLLILALLLYKCNLSEQIINIGIIVAYVLSVFVSGFYMGKKMKNKRYLWGLLAGTVYFVLLFAV SLIVNREFTGVQGGFLTTMLLCLGGGMLGGMVS |
| 1019 |
PF12224 (Amidoligase_2) |
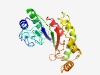 Estimated TM-score=0.70; easy target. |
>Putative amidoligase enzyme (Length=234) QDFGIEIEMTGLTRSRAAEVIAEYFGTTKEYEGSFYDAYFARDTTGRKWKVMSDGSLDCQRKEGRRKVSADRNYSVELVS PICQYKDIETVQELIRKLREAGAFVNSSCGIHIHINAAPFDAPHLRNLVNIMAAKEDMIYKALKVSRGRESHYCRKIDPA FLERLNRQKPATLDRLKSLWYNGRDGSREHYHDSRYHCLNLHSVFQKGTVEFRAFNGELHAGKIKAYIQFCLAM |
| 1020 |
PF11959 (DUF3473) |
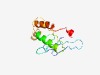 Estimated TM-score=0.70; hard target. |
>Domain of unknown function (DUF3473) (Length=127) SSVYPVKHDHYGLPDAPRFPYRLDSGLMEMPISTVRYMGRNLPAGGGGYFRLLPYGASRWLIRQVNERDGRPAMFYFHPW EIDPEQPRVQGTTAKTRFRHYINLSRTEARLRRLMDDFSWGRMDQVF |
| 1021 |
PF11911 (DUF3429) |
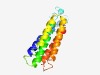 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF3429) (Length=153) VPRQAYYIGLAGVLPYAATSAATVYSAWEIHNGGYLMTEKTAELVLQILEPLQVGYGATIISFLGAIHWGLEWAKYGGEQ GYPRYAIGVISTALAWPTILLPVEYALISQFLIFNFLYYTDSRASKKGWAPGWYGVYRFVLTFIVGASIVVSL |
| 1022 |
PF11797 (DUF3324) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function C-terminal (DUF3324) (Length=136) KGLSIENKYAYVVGVTLQENDEKVKQDLKLHDVKAGQVNARNVINATLQNPTATYLNQFEVDAKITEKGKDKALYTSKKQ GMQMAPNSNFDYPISLNGEKLEPGTYTLHAKAKSTEETWEFAKDFTIKDTEAKQLN |
| 1023 |
PF11761 (CbiG_mid) |
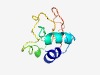 Estimated TM-score=0.70; easy target. |
>Cobalamin biosynthesis central region (Length=87) PGIDMLGVPFGWQRGEGDWTGVSAAVARGEAVEVIQDAGSTLWQDHLPQGHPFQFLATDVAKARIWISPTQRRFSSASDF PKIQWHP |
| 1024 |
PF11700 (ATG22) |
 Estimated TM-score=0.70; easy target. |
>Vacuole effluxer Atg22 like (Length=525) TSKKELIGWYSYGWAAEVFTVCAMGSFLPITLEQMARERGVLFSDKTTPCTATWKTPENLHANWQNTKAAVSGQCIVYIL GAEVNTASFAMYTFSVSVFIQAILIISMSGAADHGSYRKTLLVTFAAIGSISTMFFLSVASKLYLLGGLFAIVANTCLGA SFVLLNSFLPLLVRYHPSLLKHANPTESDVQATAMDGSRTPLLQADGVSPDEAPLSDASKELILSTQISSYGIGIGYISA VLLQGLCILVIVRTHQTTFSLRVVLFLIGLWWLVFTIPAAFWLRPRPGPPLLRMQDGKAQQSWLGYMAFAWKSLGRTAMR TRHLKDLLLFLASWFLLSDGIATVSGTAVLFAKTQLNMEPAALGMINVITMISGVLGAFSWSYVSRALDLRASQTIIACI LLFELVPLYGLLGFIPAVKNLGYLGLQRPWEMFPLGVVYGIVMGGLSSYCRSFFGQLIPPGYEASFYSLYAITDKGSSIF GPAIVGFITDRYGEIRPAFFFLAVLILLPLPLMLLVDADRGKRDA |
| 1025 |
PF11478 (Tachystatin_B) |
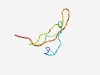 Estimated TM-score=0.70; trivial target. |
>Antimicrobial chitin binding protein tachystatin B (Length=42) YITCLFRGARCRVYSGRSCCFGYYCRRDFPGSIFGTCSRRNF |
| 1026 |
PF11459 (AbiEi_3) |
 Estimated TM-score=0.70; easy target. |
>Transcriptional regulator, AbiEi antitoxin, Type IV TA system (Length=149) RVMLFGYKDEKLPAWFLKHQWDVKTEYHSSSFLPAETGLVDLEFKTFSMQISGAARAMLECLYLVPKKQQLLECYQIMEG LNNLRPKIVQTLLEECRSIKVKRLFLYLANKAGHQWVQYLNLDNIDLGTGKRSIVKNGVYIHKYQITVP |
| 1027 |
PF11398 (DUF2813) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF2813) (Length=377) MHLESIEILGFRGINRLSLTLDENNVLIGENAWGKSSLLDALSLLLAPTLPLYHFDMQDFHFTPGDENSREKHLQIIFTF CETAPGHHLSPRYRSLSPVWVEGNESLYRVFYRLEGEVDESQSVFTWRSFLDADGHPIPLDNIDELAREIIRLHPVLRLR DARFMRRLRSGTLAATLDNSNEKLAQQFEQLIRELVQNPQKLTDKELRQGLVAMQQLLEHYFSEQNATGNDRRHHRHART HNGKSWRSLDNINRLIADPNSRSSRIILLELFSTLLQAKGSVVLDPHARPLLLIEDPETRLHPIMLSVAWGLLTQLPLQK VTTTNSGELLSLVPMEQVCRLVRESSRVATYRIGRQGMNAEDSRRIAFHIRLNRPAS |
| 1028 |
PF11360 (DUF3110) |
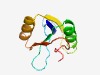 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF3110) (Length=84) VFVLLFNARTENEGIHTIQIGDRNKVLMFESEDDATRYALLLEAQDFPAPTVEAIDSEEIEEFCRSADYDWEIVKDGMLA IPPD |
| 1029 |
PF11287 (DUF3088) |
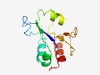 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF3088) (Length=110) RDRLFLLQPGFEDPAFPGQSFYCWHCALIEGVLASFPQLAEKLDVERIPWPRPRQAVIALVGEENQSLPLLVLATGATSP HQTGSHQGRAFIADKDAILAALSERHGFPD |
| 1030 |
PF11059 (DUF2860) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF2860) (Length=292) GFSGEISINTGVASSTSNFNTDADSNLSSIDQKASSESSFLVAPLGSVAYTFGEKLNHQVYTGTARDDVATGTVVLEVGY KYQLESGMVIDASLLPTIMSGETWADPYNTTSARTKTDETGNAFRLKLSNIAGSAFSLDMAYATKDVDKDDIQEQELKRD ANTLYLKGQYRLPLSRTMMLIPSVIYQTRDADGKAESYDQFGGEVSLFGGMGRHQYVLTAGYNQRSYDASNPIYDKKRSD DNINLFAAYEYDQFMDWENWSFISLAGYGTSDSNITFYDESQYIVSVGLNYK |
| 1031 |
PF10978 (DUF2785) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF2785) (Length=175) LFAHIDEPQNDAVYQRSFAVLLLSVLLYADHAGLKFMTPARLDLIVTQMTIYILLETDTRGFVGTTGWAHAYTHIGNILD ELADESKLARADKLLLLAALLSRYQKLTTPLIFGEPERLSTYLTLITNKDELYCDYLLAALKEWHRQLMMHTRPSSEAEW TQIFNRNRLLEALAL |
| 1032 |
PF10732 (DUF2524) |
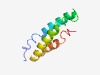 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF2524) (Length=84) MATRQSVDEFLQRCEDVIRYAKEQYTEAQKQEHYNDTEYTKAQQMLEEANNELAHLALSCNAQQREQLHRMRLQLQQLQN EMIL |
| 1033 |
PF10340 (Say1_Mug180) |
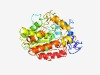 Estimated TM-score=0.70; easy target. |
>Steryl acetyl hydrolase (Length=375) LRFLSKFIGILPFKMTYYLIKVVMESERGVNIDIVSRIFMRESTKLIDENICQYVLNPIFEFIYKFEGNGISLINYSVPN DDLEQISGGIFDKPFVKSTRTQSKFFWKRQINPHTFNPEIDPILIYYHGGGYALQLTPTTLSFLTNTGKYFPDMPILISD YATTASKLDSNKYPRQLLDILAVYDYVTQSLGCKNVIVMGDSAGGNAVLVLLLYLHKHNRPLLPRKAIPISPWLNTTYIG EQERTYMKQSEKWDGICTKGSEMFGDLYIPKILRESGYNPDNDEYINIEHNFDAETWNLITEKCDLLVTYGDDEILSYQI KLFVDKLIECNPKRYNNVENVLAHFQGCHTGPILAWDGNVESWSKQTMIKPILDF |
| 1034 |
PF10327 (7TM_GPCR_Sri) |
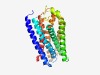 Estimated TM-score=0.70; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Sri (Length=302) DVDFETPQFLVHHYYISGSISIIINTFVFYLLLFHKGKMDSFRYYMLAFQLTCSLCDIHLTFLMQPVTLFPMASGYCTGV LIKLIDVTPHFLMTILGFFVGYQVNVLNLCFLRKHQAIAKISNKYVLSEKVYNAIVLFFMTYTFNYVIPFYLAHLTKEEE YQIIDRNYPKLRHKFETLSNFDIFEFNIMIQLSVAMIMAGCMQSTITVSVLAFQMYQVLMQCKLNLSKSTLEKHKSALKS LVGQFMTTPIAILPAMLIVSTLFFPFKGTQVFTWIMLMVMTTHSTINCFVVIFTVPEFRAFV |
| 1035 |
PF10031 (DUF2273) |
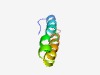 Estimated TM-score=0.70; easy target. |
>Small integral membrane protein (DUF2273) (Length=48) FFEHHRGKIIGVALGLAFGWFAIVYGFWKAVFVAVCVGIGYYVGKRVD |
| 1036 |
PF10027 (DUF2269) |
 Estimated TM-score=0.70; hard target. |
>Predicted integral membrane protein (DUF2269) (Length=149) VVKWLHILSSTFLFGTGIGSAWYMLFANISRDVRAIAVVSRNVVIADWVFTATTAIIQPATGFYMIHLAGYPWHSKWIMW SIALYVLAGACWLPVVWIQIRLRDLAAEAVRNNTELPPQYWRLFKTWVALGVPAFIAFVVIFWLMVAKP |
| 1037 |
PF09945 (DUF2177) |
 Estimated TM-score=0.70; hard target. |
>Predicted membrane protein (DUF2177) (Length=122) LKSYLLTAIVFFAIDLVWLGVVAKNLYNKHLGGLLADQVNWTAAIIFYLLFIVGIYIFAIVPAVNKESLKYAIMMGALFG FFTYATYDLTNLATLKDWPIKIVFIDIIWGSVLTAAVSASGY |
| 1038 |
PF09939 (DUF2171) |
 Estimated TM-score=0.70; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2171) (Length=65) IKEHAEVVGSDGQHVGTVDHMEGADRIKLTKRDQDADGKHHYIPLSWVQSVEGSQVHLSKTRDEA |
| 1039 |
PF09892 (DUF2119) |
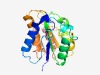 Estimated TM-score=0.70; easy target. |
>Uncharacterized protein conserved in archaea (DUF2119) (Length=187) KGPMRLFVGGVHGKEGLTTIKALSQIHEDDIPNGKLLIYNCERSPYISTLDKRYYQSKIGKEILSIINYYKPVVYVEPHC YKPESYKKLIDLDREKRVGVPPLIELENGVLIGSVSPFIRTTCFKREDICITLEMPCYPSEESLNAYVNVLKAVAGSRDR AELENKLKVKYPQQIKTARRYAREFFG |
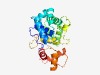
| 1040 |
PF09674 (DUF2400) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF2400) (Length=228) DPICIPHSFTKKQDIEIMGFWAAVLAWGNRTTIIKKCKELVKLMDGAPHDFVCNHQESDLKAFLSFKHRTFNATDTLYFI HFFQDFYQKHSSLEEAFTQDFSAEEPHIEKALINFHELFFSLPEYPERTRKHVATPARKSACKRVNMFLRWMVRNDEKGV DFGIWEKIKPSQLICPCDVHVERVARKLHLLDRKQMDWKTALELTDNLRKLDPQDPVKYDFALFGLGV |
| 1041 |
PF09650 (PHA_gran_rgn) |
 Estimated TM-score=0.70; hard target. |
>Putative polyhydroxyalkanoic acid system protein (PHA_gran_rgn) (Length=87) IHIERPHSLGLDAARAKAEKLAERLAKEHGVQCEWQGDELTFKRSGVDGRIAVGADKVTVDVKLGLMLSAMSGLLKGEIE KALDKAL |
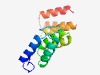
| 1042 |
PF09613 (HrpB1_HrpK) |
 Estimated TM-score=0.70; easy target. |
>Bacterial type III secretion protein (HrpB1_HrpK) (Length=131) PDYMQCRPEIVGGLIETVSTALLRTFPKVTADPYDIELILDALRVMRPNVVEIQMLDGVLYMACGRWDDAIRILSEVCAT APHFGYGKALLAFCLSTKGDDAWKQHAAEALESSPNDDTLVLVKALQARED |
| 1043 |
PF09553 (RE_Eco47II) |
 Estimated TM-score=0.70; easy target. |
>Eco47II restriction endonuclease (Length=213) LDFISEEDFKKHVRATIMKYGEKLESYDLKRFNSNLIDPIKLIFDKSVYRTSWEEIVNNEIFRQRDKSNNNDIGYFHQNI FSYFKGCEVPQTGWDVIYRNPDGIQMPDGDIVHTIYVEMKNKHNTMNSASSAKTYIKMQGQILEDDDCACLLVEAIAKKS QNIKWSTKVDGKNVQHRLIRRVSMDQFYAILTGEEDAFYKMCMALPEVINSVV |
| 1044 |
PF09539 (DUF2385) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF2385) (Length=96) PFEPDLMRLAEILGALQYLRTLCGANEGQKWREQMQALLDAEATTADRKNRMVTSFNRGYRSFQQSYRTCTPAANVAVRR YLEEGSKISREITARY |
| 1045 |
PF09520 (RE_TdeIII) |
 Estimated TM-score=0.70; easy target. |
>Type II restriction endonuclease, TdeIII (Length=239) KQEIKQVEEVLKNSLRNKFNNYKPEPSVMPFHTRLLGKDRMALFSFIHSLNTNFGTSIFEPVAITLAESQFKIAKSQAIA GTQISKKAQRVIQDIMDNLTAANLKPNKQEEIEAIRKVCQEGEMQIVKPTKVDVWLESKNEELFLIDIKTAKPNKGGFKE FKRTLLEWAACVLAEDPKAKINTLIAIPYNPYSPKPYSRWTMAGMLDLDNELKVAEEFWDFLGGAGAYEQLLDCFERVG |
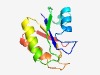
| 1046 |
PF08821 (CGGC) |
 Estimated TM-score=0.70; hard target. |
>CGGC domain (Length=104) KIAVVRCETVSEVCPGTGCFKAFNEKKVHFDRYDKKDEIIGFFTCGGCPGRRVYRLIKALKKHGASAVHLSSCMKTKKYP PCPHIDSIIKIIEDAGMEVVEGTH |
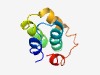
| 1047 |
PF08707 (PriCT_2) |
 Estimated TM-score=0.70; hard target. |
>Primase C terminal 2 (PriCT-2) (Length=71) KLVDILDAARGESYNDWVRVGWCLRNVDHRLLDKWIDFSRRSPKYVEGECARMWTHMRMGGLGMGTLHMWA |
| 1048 |
PF08497 (Radical_SAM_N) |
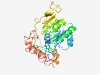 Estimated TM-score=0.70; easy target. |
>Radical SAM N-terminal (Length=316) FLPMSRAEMDRLGWDACDIVLVTGDAYVDHPSFGMAIIGRLLESQGYRVGIIAQPDWHSAEPFKALGKPRLFFGVTGGNL DSMVNRYTSDRRLRSDDAYTPGGEGGKRPDRCTIVYTQRCREAYKDVPVILGGIEASLRRIAHYDYWSDKVRRSILADAK ADLLIYGNAERAVIEVANRIAAGEAPRDLTSVRGVALFRKVPEHFTELHADDLDSADEAATRHPGDTVIRLPSFEQVEND REAYARASRVLHREANPGNARPLVQRHGDRDLWLNPPPIPLTTEEMDQVYDLPYARAPHPAYGDAKIPAWDMIKFS |
| 1049 |
PF08472 (S6PP_C) |
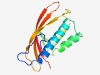 Estimated TM-score=0.70; easy target. |
>Sucrose-6-phosphate phosphohydrolase C-terminal (Length=133) GPNTSPRDVPHFSECKPDNVNPGHEIVKFYLFYERWRCAEVEDTDPCMENLKVDYHPAGVFVHPSGVERSLHDCINAMRS CYGDKQGRKFQVWVDRVSPVQMSSDTWIVKFDKWELSGDERHCCITTVVLSSK |
| 1050 |
PF08161 (NUC173) |
 Estimated TM-score=0.70; easy target. |
>NUC173 domain (Length=193) MSYQYNKAWKQILHILALMFKAAGATCHEFMVKCLQSLAELRDSYNFPYVNELDYAVGAAVRTMGVDVVLRHVPYQISFN EKGNEIKRSWMLPVLKANIQNASIGFFIEHFLPLASQCRKKSEEFRQSDSTAAAVSYDLLQTQIWSLLPSFCNKPKDIES FPKLAKVLGNVLINFKDLRLTVMLALRKLMEDN |
| 1051 |
PF07914 (DUF1679) |
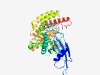 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF1679) (Length=406) LFKNGDGLFNTHVYLEDLQEVIRDQMKTNAQLGPGTKYTIVGDGNGFMSRVVLVDPDWTVSDENLPKRFVLKITSCMHVL NVLDQMNLPDKRESALWSIFEYEAQGLHNREVNLYEIFGKWDIDNLLLSPKVYFSKKFDSENQTKGFFGMEFVENAITRH LYINLKPYELHAVLKALAIFQAEGLKLNKEEQESVTGYDLEKIVGKMFSVSGLKGIFDQAREINPEELSETANRVEAFGL ELVNFDMVKNLNTYLGIKKDVLVHGDLWSANIMWTEKEDGELKVNKIIDYQSIQLGNPSQDLVRLFTSTLSGSDRRAYWE KLVEQFYEYFLEAVKDKNVPYTLEQLKESYRFYFVTGSLLMLPMLGPIAKVKLAEMTDPKEIEECRNILTEKAEIILEDM EHWHLM |
| 1052 |
PF07799 (DUF1643) |
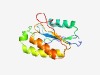 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF1643) (Length=133) RYLLWREWDLDKSKLVFVMLNPSKADADIDDPTLRRCINFTKSWGYGSLRVVNLFAYRSASPLDLKKVDDPIGKQNDRYL KKAIKSADQVVVAWGNNGKLMQRDRFVLELLSKLNIQPHCLGITKAGYPRHPL |
| 1053 |
PF07561 (DUF1540) |
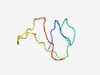 Estimated TM-score=0.70; hard target. |
>Domain of Unknown Function (DUF1540) (Length=44) VLCEVNTCTHWLLGNLCTAANIDILYEEEGKMAQKDAHTECKTF |
| 1054 |
PF07448 (Spp-24) |
 Estimated TM-score=0.70; easy target. |
>Secreted phosphoprotein 24 (Spp-24) cystatin-like domain (Length=64) VLDQDSLNMDLEFGIRETTCRRDSGEDPATCDFQRGYYMPSAICRSTVRVSAEQVQDVWVRCHW |
| 1055 |
PF07386 (DUF1499) |
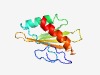 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF1499) (Length=111) LAPCPNTPNCVSSQSLDAQHRIEPLTYKSTPKEAMANLKKVIQNMERTKIITETDNYLYAEFTSKLMGFVDDVEFFLDES AKVIQVRSASRLGQSDLGVNRKRIEDIRAQM |
| 1056 |
PF07205 (DUF1413) |
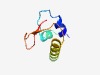 Estimated TM-score=0.70; trivial target. |
>Domain of unknown function (DUF1413) (Length=68) KTALSEAKNLQEGESFLLKDLFKGYLWNRISRGDRLLIGSLFLRYVSSHDCHIAVIHKGASGQQRYQK |
| 1057 |
PF07162 (B9-C2) |
 Estimated TM-score=0.70; easy target. |
>Ciliary basal body-associated, B9 protein (Length=160) VHVIGQLIGASGFPDNRLFCKWGIHYNGGWKVLEGLTEGQTQIDNPQNEDTAYWCHPIDIHFATRGLQGWPKFHFQVWHQ DSYGRVELWGYGFCHVPTSPGFHQLECVTWRPTGSIHEEISHFFLGGGLELKHSDIIYNGLDRYHLQTVAMGKVHLEINI |
| 1058 |
PF07129 (DUF1381) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF1381) (Length=44) TQYLVTTFKDSSGLPHEHFTAARDNQTFTVVEAESKEEAKEKYE |
| 1059 |
PF07042 (TrfA) |
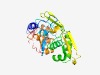 Estimated TM-score=0.70; easy target. |
>TrfA protein (Length=282) KKRTAGGELAEKVSEAKQTALLKHTKQQIKDMQLSLFDLAPWTGSHAALPNDFGRSAIFTVRNKKVPRPALQGQSIYHVN KDVEITYTGIELRADDDELVFAQVLEYANGTALGEPVSFTFYELCQDLDWSINGRYYTRAEECLTRLQASAMQFSSQRIG RLESVSLIRRFRVLDRGKRTSRCQVEIDAEIVVLFAGDHYTKFVWEKYRKLSPNRDGRMFDYFATHKEPYPLKLETFRLM CGSDSTRPKKWREQVGEACDELRENGLVESAWVNDDLVHCKR |
| 1060 |
PF07002 (Copine) |
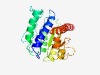 Estimated TM-score=0.70; easy target. |
>Copine (Length=219) SLHHVSADGRPNQYELAIISVGEIVQDYDSDKQFPGLGFGALLPPMFSGVQDAFSLTLQPENPYCAGMPGLLAAYRSCIR AVRLYGPTNFSPVINHVASMAAAHQTGAQYFVLLIITDGIITDFEKTKQAIIKASSLPMSIIIVGVGDEDFSAMEELDSD DKRLRCGSMIAERDIVQFVPLRRFIQGGSIDRSRYALAKEVLAELPNQVCEWMRRRGIR |
| 1061 |
PF06906 (DUF1272) |
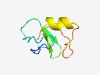 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF1272) (Length=57) MLELRPNCECCDKDLPPDSTQAMICSFECTFCQDCADQRLHGTCPNCGGELVRRPIR |
| 1062 |
PF06904 (Extensin-like_C) |
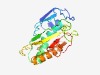 Estimated TM-score=0.70; easy target. |
>Extensin-like protein C-terminus (Length=172) RCAETLATSGLGYTVQADSPASATCPLSNVVRVQDAPVRLSSSFLATCPVAVAFALFERHGLQPAAQRVFGQPVVQVDHL GSYACRNMYNRAEGRRSQHASANALDIAGFRLKDGQRIELQKDWNGEGPKAQFLRQVHAGACESFNTVLGPDYNAAHHNH FHLDMGGWQICR |
| 1063 |
PF06250 (YhcG_C) |
 Estimated TM-score=0.70; easy target. |
>YhcG PDDEXK nuclease domain (Length=155) QETLKDPYIFDFLTIEEPFHERELETGLVGHVEKFLLELGAGFAFVGRQYHLEVSEQDFYIDLLFYHLKLRCYVVIELKK GKFQPEYAGKMNFYCSVVDDKLRHEQDNPTIGLILCQTKDKIVAEYALRDVNKPIGISEYELTRALPDNLKSSLP |
| 1064 |
PF06211 (BAMBI) |
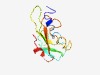 Estimated TM-score=0.70; easy target. |
>BMP and activin membrane-bound inhibitor (BAMBI) N-terminal domain (Length=107) MDRLVSLWFQLELCAMAVLLTKGEIRCYCDAPHCVATGYMCKSELNACFTKVLDPLNTNSPLTHGCVDSLLNSADVCSSK NVDISSGSSSPVECCHDDMCNYRGLHD |
| 1065 |
PF06021 (Gly_acyl_tr_N) |
 Estimated TM-score=0.70; trivial target. |
>Aralkyl acyl-CoA:amino acid N-acyltransferase (Length=205) MIHLQSAQKLQMLEKSLRMVLPESLKVYGTVFHMNQGNPFKLKALVDSWPDFNTVVIRPQEQEMTDDLDHYTNTYLIYSK DPEQSQEFLGSPAVINWKQHLQIQSSQSNLNRVIQNLVDINLSKVKHTQCNLYMVYDTAKKLAPSLVDVKNLSPKGDKPK PLDQEMFKLSSLDVTHAALVNQFWHFGGTERSQRFIERCIRTFPS |
| 1066 |
PF05868 (Rotavirus_VP7) |
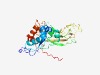 Estimated TM-score=0.70; trivial target. |
>Rotavirus major outer capsid protein VP7 (Length=249) MASLLLLVLAAAVTAQLNIVPSTHPEVCVLYADDHQADANKFNGNFTQIFHSYNSITLSFMSYSSSSYDVIDIISKYDLS SCNILAIDVFNASMDFNVFLQSTNNCSKYNANKVHHVKLPRGEEWFSYSKNLKFCPLSDSLIGMYCDTQLSDTYFEISTG GTYEVTDIPEFTQMGYTFHSSEEFYLCHRISSEAWLNYHLFYRDYDVSGVISKQVNWGNVWSGFKTFAQVLYKILDLFFN SKRNVEPRA |
| 1067 |
PF05136 (Phage_portal_2) |
 Estimated TM-score=0.70; easy target. |
>Phage portal protein, lambda family (Length=340) GRRLGNWQPEEAGINALLFRDATLMRARSRDLVRRNAWASNAVDSLVGNLVGTGIKPQSTVKDPQIKAQIQSLWLAWTKE ADAHGMLGFYGLQALIARAMIEGGEVLVRLRNRDARDGLLVPLQLQVLEPEHLPASDSRDLPNGHRVRAGIEFDKIGRRV AYHLFREHPGEQPMLFKAGDVVRVPGSEVCHIFKPLRPGQLRGEPWMAQALVRLHELDQYDDAELVRKKTAALIAGFITK PDPELGVGGEDGQDPDEQGAVPVTWAPGTMQVLLPGEDVKFSDPADVGGQYGEFMRTQLRAVAVGLGLTYEQLTGDLTGV NYSSIRAGMVEFRRRMEQIQ |
| 1068 |
PF04606 (Ogr_Delta) |
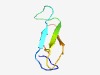 Estimated TM-score=0.70; easy target. |
>Ogr/Delta-like zinc finger (Length=47) HCPLCQHAAHARTSRYITDTTKERYHQCQNVNCSATFITYESVQRYI |
| 1069 |
PF04601 (DUF569) |
 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF569) (Length=142) MEFFNQAKAVRLQSHLDKFLTANPDEETVRQSRNGSSHKACWTVELVEGKSHIIRLKSCYGWYLTASDEPFLLGMTGKKV VQSDTREASEEWEPIREGNTHVKLRSKDGGMYLRANGGTPPWRNSVTHDFPYTSATQGWVLW |
| 1070 |
PF04298 (Zn_peptidase_2) |
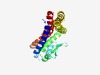 Estimated TM-score=0.70; hard target. |
>Putative neutral zinc metallopeptidase (Length=217) TYMLIIISALISLFAQFLVNSRFSKYSRVRSRSGMTGAQAAERILQSQGIYDVAIQRVSGKLTDHYDPRNKTLNLSDAVY ASTSVAAVGVAAHECGHAIQHARGYAPLSFRSALVPVANIGSQLSWLFIILGIFFGGSHTLIMIGILMFSAAVLFQLVTL PVEFNASGRALKLLSETGILQKDEVSDTRKVLSAAALTYVAAAATAVLQLLRLLRLF |
| 1071 |
PF04282 (DUF438) |
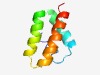 Estimated TM-score=0.70; hard target. |
>Family of unknown function (DUF438) (Length=68) LRDILLELHNGASPESVQDRFDATFTGVSAIEISLMEHELMNSDSGVTFEDVMELCDVHANLFKNAIK |
| 1072 |
PF03790 (KNOX1) |
 Estimated TM-score=0.70; hard target. |
>KNOX1 domain (Length=42) ALKAKIVSHPHYSNLLQAYMDCQKVGAPPEIAGRLTAVRQEY |
| 1073 |
PF03779 (SPW) |
 Estimated TM-score=0.70; easy target. |
>SPW repeat (Length=48) DGPVFLLGLYCAVSPWILHYTTSQPALVTHNLIVGIAIGLLALGFTAA |
| 1074 |
PF03616 (Glt_symporter) |
 Estimated TM-score=0.70; trivial target. |
>Sodium/glutamate symporter (Length=366) TFSTYETLALASLVLLLGYFLVKRINVLKTFNIPEPVVGGFIVAIGLLIWHKIDGTSFNFDKNLQTTMMLVFFTSIGLSA NFSRLIKGGKPLVVFLFIAALLIFGQNVIGIASSMALGIHPAYGLLAGSVTLTGGHGTGAAWADTFAHQFNLQGATEIAI ACATFGLVFGGIIGGPVARFLLNRQKQGENPENDEVDDIQEAFEHPTYKRKITARSLIETIAMISVCLLIGQYLDVQTKG TALQLPTFVWCLFTGVIVRNILSNIFRFQVAESAIDVLGSVGLSIFLAIALMSLRLWELAGLAIDVLIVLAIQVAFMAAF AIFITYRAMGKDYDAVVLSAGHCGFGLGATPTAIANMQAVTSRFGP |
| 1075 |
PF03522 (SLC12) |
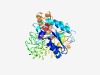 Estimated TM-score=0.70; easy target. |
>Solute carrier family 12 (Length=394) RPNLLNFANLITKNQSLMIVGNVVEQKLNYKERKAHVQAGKKVLNDLKIKAFYNILDGLPFDESVRAMIQSTGFGRLTPN ILMVGYKQGWRTCGTEELHTYYNVLHNAFDNRLALTILRLPNGLDCSHLVPPKPELDLADSALNIMSSMAYINTPQGLRI QKGLMPVNSNLDLHGMAASSEQISVNVPNEDQDHRMNPNSTTPGKELNMFREKQPAGYIDVWWLYDDGGLTILLPYIIST RAKWSECQIRVFALASQQTNVEEERENMTILLDKLRINYVSLIMVTLSDKPQEATIQAHRALLSTLVEGQETDVFVSASE QAQLEEKTYRQLRLRELLQQYSKHASLIVLSMPIPRKGIVSAPLYMSWLEMLTKDMPPFLLVRGNQTSVLTFYS |
| 1076 |
PF02521 (HP_OMP_2) |
 Estimated TM-score=0.70; easy target. |
>Putative outer membrane protein (Length=441) GVSSLYAFDYTVSGKAESFSKIGFNNTPINSTEGKYPTDSFVTVVGALQVDMNLLPKSTTSHRLTGGIGGMIGGLAYDST RNLIDTATGQRFGSVIFNYIGYWAQYNGTAPWQSPDYNVGMTQNTRTYVIYNAYLDYDYEGVFGIKGGRYESGADFMSGF TEGFELYAKFSKFKLWWFSSYGRAFAYDEWLYDFYAPKTYTTPEGKTINLGIHAFKGTYRHNGFSVSPFIYFSPKTYTAP SIQFDYDTNPNFEGIGFRSQTTVIGLFAIFDPRVYDVNRYGNPTGKNGQTLLIKQRFDIDNYNIGGGIYKNFGNANAQVG TYGNPVGIDFWTASAYDIGASISDMMGEDALTGFIYGGGVHGDFRWGILGRLTTSKRSNEQSIAANIAYDFTRYVSAGLK LEYFNDLTKAGYKVGDTGRVPGQPQNISDRSHMMTFIRTKF |
| 1077 |
PF02440 (Adeno_E3_CR1) |
 Estimated TM-score=0.70; easy target. |
>Adenovirus E3 region protein CR1 (Length=89) FSQAGFHTINATWWANITLVGPPDTPVTWYDTQGLWFCNGSRVKNPQIRHTCNDQNLTLIHVNKTYERTYMGYNRQGTKK EDYKVVVIP |
| 1078 |
PF02101 (Ocular_alb) |
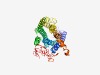 Estimated TM-score=0.70; easy target. |
>Ocular albinism type 1 protein (Length=398) MASPRLGTFCCPTRDAATQLVLSFQPRAFHALCLGSGALRLALGLLQLLPRRRPAGPGNPATSPPASVRILRAVTACDLL GCLGIVIRSTVWLGFPNFVDSISDVNSTDIWPTAFCVGSAMWIQLLYSACFWWLFCYAVDAYLVIRRSAGLSTILLYHIM AWGLATLLCMEGAAMLYYPSVSRCERGLDHAIPHYVTMYLPLLLVLVANPILFQKTVTAVASLLKGRQGIYTENERRMGA MIKIRFFKIMLVLMICWLSNIINESLLFYLEMQTDINGGSLKPVRTAAKTTWFIMGILNPVQGFLLSLAFYGWTGCSLGF QSPRKEIQWESLTTSAAEGAYPSSVDSRVPHENPASRKVSHVGGQTSDEALSMLSEGSDASTIEIHTASESCNKNDGD |
| 1079 |
PF01856 (HP_OMP) |
 Estimated TM-score=0.70; easy target. |
>Helicobacter outer membrane protein (Length=159) GVQLGYKQFFGKNKFFGIRYYGFFDYNHAYIKSNFFNSASNVFTYGAGSDLLLNFINGGSDKNRKVSFGIFGGIALAGTT WLNSQLVNLKTTTSIYNAKINNTNFQFLFNTGLRLQGIHHGVELGVKIPTINTNYYSFMGAKLAYRRLYSVYLNYVLAY |
| 1080 |
PF01617 (Surface_Ag_2) |
 Estimated TM-score=0.70; easy target. |
>Surface antigen (Length=248) VGGNFYISGKYVPSVSHFGVFSAKQERNTTTGVFGLKQDWDGSTISKNSPENTFNVPNYSFKYENNPFLGFAGAVGYLMN GPRIELEMSYETFDVKNQGNNYKNDAHKYYALTHNSGGKLSNAGDKFVFLKNEGLLDISLMLNACYDVISEGIPFSPYIC AGVGTDLISMFEAINPKISYQGKLGLSYSISPEASVFVGGHFHKVIGNEFRDIPAMIPSTSTLTGNHFTIVTLSVCHFGV ELGGRFNF |
| 1081 |
PF01385 (OrfB_IS605) |
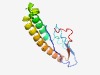 Estimated TM-score=0.70; easy target. |
>Probable transposase (Length=114) SIQTESEVSTPVHPSASMIGLDAGVAKLATLSDGTVFGPVNSFQKNQKTLARLQRQLSRKVKFSNNWQKQKRKIQRLHSC TANIRRDYLHKVTTTVSKKHAMIVIEDLKVSNMS |
| 1082 |
PF18818 (MPTase-PolyVal) |
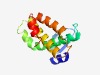 Estimated TM-score=0.69; easy target. |
>Metallopeptidase superfamily domain (Length=126) HADAFYANLGIATVYGGNEAYYLPSADRVHVPSFSSFRDAASHYSVLFHEGLHATAAKHRLDRDLSGRFGSEAYAMEEIV AQVGSSMILADLRIAAGPRPDDASYISSWLKVLRNDSSAIFTAASK |
| 1083 |
PF18743 (AHJR-like) |
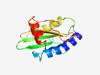 Estimated TM-score=0.69; easy target. |
>REase_AHJR-like (Length=124) MATPTANLERERLLQLAEEYRDKGYEVSFHPNPEDLPDFLRNYRPDMIVRRGDEAVIIEVKSRRSLNSSSRQYLQNLAQS VEQHPGWRFELVMTNPEDAVYSPKAESSLQQPEIETRLQVARQL |
| 1084 |
PF18731 (HEPN_Swt1) |
 Estimated TM-score=0.69; hard target. |
>Swt1-like HEPN (Length=115) RALDLLKDGLYPFVEREMRSAYSDKWLVAATPFVSEDRTLRRSVQQILKQDISELLKLMSNQWREVFKKTLGNAERSLVG ELISTRNSWAHNDPFSTDDAYRALDSVSRLLTAIS |
| 1085 |
PF18676 (MBG_2) |
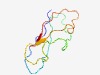 Estimated TM-score=0.69; easy target. |
>MBG domain (YGX type) (Length=78) LTVTAVSQTIPFGGTIAPYTDTITGFVNGDAPSVVTGAASLTTTPPTPTAPNTYTITAANGTLAASNYSFTFVNGTLT |
| 1086 |
PF18647 (Fungal_lectin_2) |
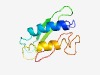 Estimated TM-score=0.69; easy target. |
>Alpha-galactosyl-binding fungal lectin (Length=99) ENRKAFASWERLNKKYRDFCEDVEPPQDTINWKWEKTYDKGTPDETQFVVHLSNEASAFDRSQCFESFDLILNSCDGNDP ENPMNWKFGGKYVRGSYRY |
| 1087 |
PF18478 (PIN_10) |
 Estimated TM-score=0.69; easy target. |
>PIN like domain (Length=84) MNFLIDNNLPPAIARALHQLCKAEGHAVVPLRDKFQASASDIHWITALSTEGGWAVVSQDKFSKGNAEREAFRECGLPIF CLAK |
| 1088 |
PF18143 (HAD_SAK_2) |
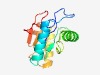 Estimated TM-score=0.69; easy target. |
>HAD domain in Swiss Army Knife RNA repair proteins (Length=132) YIFLDIDGVLHPATAGTDRQFSPNCLRALRTIVGATGAALILSSSWQSSQAAAEVVDEELARWGLPRCSGRTSAGPTGVG AAARAGEILAWLAAKTEVEAWVALDDLPLLAHRSYGRFVQTDPAVGLTEADA |
| 1089 |
PF18017 (SAM_4) |
 Estimated TM-score=0.69; easy target. |
>SAM domain (Sterile alpha motif) (Length=82) MVSVTMATSEWIQFFKEAGIPPGPAVNYAVMFVDNRIQKNMLMDLNKEIMNELGITVVGDIIAILKHAKIVYRQEMCKAA TE |
| 1090 |
PF17736 (Ig_C17orf99) |
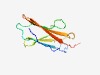 Estimated TM-score=0.69; easy target. |
>C17orf99 Ig domain (Length=95) PVVSIAYKVLEVFPKGRRVLITCHAPQASPPITYSLCGTKNIEVAKKVVKTHEPASFNLNITLKSSPDLLTYFCRAATTS GAHVDSARLQMYWEL |
| 1091 |
PF17619 (SCVP) |
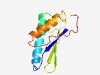 Estimated TM-score=0.69; hard target. |
>Secreted clade V proteins (Length=98) VHVIVTSSEPFDLSKAEENMVTVEKKLKEFADIEGIAFKQLQELPRTAENVGGKFGVHFTVEGAFERCARVLQFIQAACN EIKEVVSGTVKCGAFEPV |
| 1092 |
PF17263 (DUF5329) |
 Estimated TM-score=0.69; hard target. |
>Family of unknown function (DUF5329) (Length=93) KATAEIDALLNFVEHSDCQFIRNGSEYSGSKARAHLQQKLDYLENKDLVNSAEDFIERAASKSSMSGKPYQVSCPGGKQE ASTWLTTELKRLR |
| 1093 |
PF17181 (EPF) |
 Estimated TM-score=0.69; easy target. |
>Epidermal patterning factor proteins (Length=52) RLGSRPPRCEHKCKGCAPCSPIQVPTAHFGPQYTNYEPEGWKCRCGSILFNP |
| 1094 |
PF17120 (Zn_ribbon_17) |
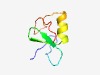 Estimated TM-score=0.69; easy target. |
>Zinc-ribbon, C4HC2 type (Length=53) TSHSCAYCGLQAKKRIFLCGNCQHVLHASCAAEWWENDDECPSGCGCNCPYMF |
| 1095 |
PF16980 (CitMHS_2) |
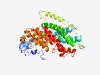 Estimated TM-score=0.69; trivial target. |
>Putative citrate transport (Length=441) LSVWWGIPFAGLLLSIALCPLLTPHFWHHHFGKVAIAWALAFLLPCAAYFGIGTAAGGAVHALVSEYIPFIILITSLFVV AGGICVRGNLQGTPALNTGLLALGTALASLMGTTGAAMLMIRPLIRANDNRKHAAHIVVFFIFLVANAGGSLTPLGDPPL FLGFLKGVDFFWTLKNIFPETLFLCVTLLLIFYALDTYYYRNREGELLPTPDPTPDSAIRFEGKLNFLLLAAIIGLVLMS GFWKSGIAFSIFGVEVELQNLIRDALLLVVIFVSLRLTPETARSGNDFSWGPIQEVAKLFAGIFITIIPVIAMLRAGQSG AFATIVSAVTDASGQPINAMYFWAAGILSSFLDNAPTYLVFFNTAGGDAQVLMTTLASTLAAISCGAVFMGANSYIGNAP NLMVKAIAEERGIKMPSFFGYMAWSCAILIPLFVLMTFFFF |
| 1096 |
PF16927 (HisKA_7TM) |
 Estimated TM-score=0.69; easy target. |
>N-terminal 7TM region of histidine kinase (Length=224) LAIAAITSASLALLGWRRRDVSGAQAFLGLMGAVTVWSLGYALELGSTKEAVKLLWAKTEYLGIVIVPAAWLIFALQYTG RGRVVTRRLLALLAIEPLVTLALVWTNEGHGLIWREVRLVTWGAFSLLDPTHGPAFWVHTAYSYLLLLAGTALLFRAFLR APTLYRRQVGAILIGALVPWAQNALYLSGLNPFPYLDLTPFTFTITGLAVAWALFRFRLFDIVP |
| 1097 |
PF16409 (DUF5017) |
 Estimated TM-score=0.69; easy target. |
>Domain of unknown function (DUF5017) (Length=179) SCMEDEAPSVELNVALDKQVYQVGEPVTFKLNGNPDNIVFYSGEVGHNYAYKERYHADGDLLVKFNSWVRYGDIYHNLKF LVSSDFSGIYDKENVEAATWIDLSDKFRFSVGDDQTPSGEVNLKEYVGAEEDAKLFVAFRYEDEQKARQNNWIIRSITLD CVSAEGVRSNLATMSTMGW |
| 1098 |
PF16408 (DUF5016) |
 Estimated TM-score=0.69; easy target. |
>Domain of unknown function (DUF5016) (Length=121) MKTLKYIALSLLVAASTTACKDDPELLTTDVGPEMTVVSADASGVYGGKVDFEVTMTDRYALSTLKAQVFFDDEMVAEEV IRTKSDGTYTGAVTLPFYKNIPDGEATLRFVGQNVRFGTTT |
| 1099 |
PF16400 (DUF5008) |
 Estimated TM-score=0.69; easy target. |
>Domain of unknown function (DUF5008) (Length=109) LVTSCNSDLNSNIKDDPYSGGKAPLGIGLLAESPSPESAYPNDTVVFKAKGMLNWCDPQSGRYDFKFYISDEETKIVTAT DTTITVKVPGNLSSGTAYIVLKEQVFYGP |
| 1100 |
PF16267 (DUF4920) |
 Estimated TM-score=0.69; easy target. |
>Domain of unknown function (DUF4920) (Length=86) YKSMPVGDSINSKMTAKVTDVCQAKGCWMTLDLEDGSEVMVKFKDYGFFVPKDIKGKEVIINGKAFVNEVPVDEQRHYAE DAGKSK |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218