

Go to page (83 pages in total): [First page] << < 3 4 5 6 7 8 9 10 11 12 13 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 701 |
PF14243 (R2K_3) |
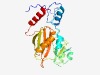 Estimated TM-score=0.73; easy target. |
>ATP-grasp domain, R2K clade family 3 (Length=207) QMGFEIKFFKNIKELEDNSKEDVIVSFVDDVRMALHRYNIITPEIDYPEELKNYLGRKVWKTKLSVIANNPDNWNVFIKP IEDKKFTGVVVRSTKDLIGCGTYNEDPDIFCSEIVNFVAEWRCFVWYGKILDIRRYRGDWKMHFDPSVVENAVAQFKSAP KGYAIDFGLTDKGETLLIEVNDGYSLGCYGLFDIDYAKLLSARWAEL |
| 702 |
PF14142 (YrzO) |
 Estimated TM-score=0.73; hard target. |
>YrzO-like protein (Length=46) MFEGLLFFISAGIVCELAAINRNGRKNIKQQAELIQILKENLYKDI |
| 703 |
PF14090 (HTH_39) |
 Estimated TM-score=0.73; easy target. |
>Helix-turn-helix domain (Length=69) TQRARMLAAMRELGSITTFEAMRFLDVFDPRPRIHELRHRHGHHITTAMRAEQTESGVLHRVGVYFLSS |
| 704 |
PF14013 (MT0933_antitox) |
 Estimated TM-score=0.73; hard target. |
>MT0933-like antitoxin protein (Length=49) FDKAKQFAEDNPDKVNQGIDKAGDLVDERTGGKHSEQVDKAQDAARKHL |
| 705 |
PF13875 (DUF4202) |
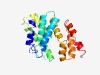 Estimated TM-score=0.73; easy target. |
>Domain of unknown function (DUF4202) (Length=183) EQALAGIDDAHRGDPNIVTVDGSEIPYELHYANKMTKYLEKHNPSASEALKLAVRAQHLRRWEIPRSSYPMTKPGYFSWR TALKNRQADIADEICRNCGYDEDAAGRVAALVRKENMKRDEECQVLEDVACLVFLDDQFEKFEKEHDEEKIVGILKKTWA KMSERGHELALQIDMSDRAKGLV |
| 706 |
PF13828 (DUF4190) |
 Estimated TM-score=0.73; hard target. |
>Domain of unknown function (DUF4190) (Length=65) LAIASLITSIAAIVTCYLGIILGIVGVILGIVALNQLKTKQQDGKGIAIAGIILGAVIPVLWILV |
| 707 |
PF13795 (HupE_UreJ_2) |
 Estimated TM-score=0.73; easy target. |
>HupE / UreJ protein (Length=155) YFQEGWQHIVDLSGYDHILFVMALCAVYLMKDWKKVLVLVTAFTIGHSITLALSVLNIVKVNTALIEFLIPVTIVITALS NVFRKTEETKKMQLNYFFALFFGLIHGMGFSNYLKSMLGSQTNIVQPLLAFNLGLEAGQLIIVLVILLLAQIIVN |
| 708 |
PF13788 (DUF4180) |
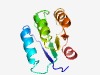 Estimated TM-score=0.73; hard target. |
>Domain of unknown function (DUF4180) (Length=110) NNIEIAVISSSEILIKDVQSALDFMATVQYETGCDRIILNKSAICEDFFHLSTKLAGEILQKFINYHVKIAVVGDFSVYS SKSLKDFIYESNKGKDIFFFPDEKKAIEKL |
| 709 |
PF13758 (Prefoldin_3) |
 Estimated TM-score=0.73; trivial target. |
>Prefoldin subunit (Length=96) QESLYHWRTWEAEYDGLKEEISNLDDNATTSDFLRIGRNFGGSLVNEDEVKVILGEKQGVTRSRQQVIDLISRRIDYVKE NVATMEKRLRKAESQL |
| 710 |
PF13730 (HTH_36) |
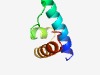 Estimated TM-score=0.73; easy target. |
>Helix-turn-helix domain (Length=53) ITDKAKILYSEISALCNKKGCCWATNDYFEKLYECSKITVIRAINALKENGYI |
| 711 |
PF13464 (DUF4115) |
 Estimated TM-score=0.73; hard target. |
>Domain of unknown function (DUF4115) (Length=72) VVLRARAESWVMLREQATGRVVFDRVLQPGESHAVPDRAGMLLTTGNAPGLEVLVDGEPLPALAGAGRVRRD |
| 712 |
PF13403 (Hint_2) |
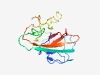 Estimated TM-score=0.73; easy target. |
>Hint domain (Length=146) PCFTPGTLIATPKGERPVEELREGDKVITRDNGIQEIRWIGTTNVSSERLAHDKHLMPVLIRAGALGHGLPERDMLVSPQ HRVLVANDRTALYFEEREVLVAAKHLVNNRSIQRVAPRALSYLHFMFDQHEVVLSDGAWTESFQPG |
| 713 |
PF13371 (TPR_9) |
 Estimated TM-score=0.73; trivial target. |
>Tetratricopeptide repeat (Length=67) ETYLKLEKYNEALTDFNNLLELEPNNVYILRLRGETYQKLESYNGALTDFNNLLKKDPYDTLVLRLR |
| 714 |
PF13366 (PDDEXK_3) |
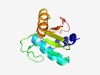 Estimated TM-score=0.73; easy target. |
>PD-(D/E)XK nuclease superfamily (Length=117) QLTGVVIGAAIAVHRELGPGVDEAACEQALSLKLTALGIPHQCQKPLPLLYKGASLDCGYRLDLLIDGRLPVELKCVDKI LPIHVAQTLTYLRLGGFPLGLLINFEVCALKQGIRRL |
| 715 |
PF13249 (SQHop_cyclase_N) |
 Estimated TM-score=0.73; trivial target. |
>Squalene-hopene cyclase N-terminal domain (Length=290) IQRAQDYLLSIQYPEGYWWGELETNVCMAAEYLLLTHFLGAADRRRWDKIVEYLRRQQLPDGTWSIYHGGPSDLNATVEA YFALKLAGVSPDEPSMAKARQFVLSRGGVPKVRIFTKIWLALFGQWDWRGVPVLPPELMLLPSWFPINIYEFASWARATV VPMLIILTRRPVCPIPGEAHIDELYPAPREQVDYSLPKSDRLLSWKTLFLTTDKLLRLYERWGWKPFRRRAARAAEEWIV EHQEADGSWGGIQPPWVYSLIALKVLGYPLDHPVMAKGLEGFEGFAIEDE |
| 716 |
PF13244 (DUF4040) |
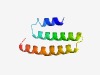 Estimated TM-score=0.73; trivial target. |
>Domain of unknown function (DUF4040) (Length=67) VLLILSAILAVRAKDLLAAVIIFSVYSLIMALVWQRLQAPDLALTEAAVGAGVTTVLFLVAIFKTHR |
| 717 |
PF12889 (DUF3829) |
 Estimated TM-score=0.73; easy target. |
>Protein of unknown function (DUF3829) (Length=274) VGKLNAYVGCINRLSERSYESRNRYFSWAKKTGPTGKERIIYGTYTIYDTTDCKKNVEMANASEPHDAELESAASAYAEA VGKLEPLLKEADEYYQQENYKDDKMAKGKEMHPRLVAAWDAFASADKKLRAGVEAIKDKQAEEQLAAIEAKEGRKTHYYA EALMIHAKRLLRAEDTSKPDIAAITKALTEYETTVKAAEDASGKDGDTKLGSSFLSSAKSYLTSAKQLMRRIRDKVPYSQ GDRMMLGSGGGWMVEGSPPRLLRDYNQLIESYNR |
| 718 |
PF12860 (PAS_7) |
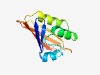 Estimated TM-score=0.73; easy target. |
>PAS fold (Length=109) LPLGVWVVGADGRLAFANAATRQVTGIDPAETPPGLPAAELVRRFAYRGIYGEGDPAALAAEHLAIDRSRPFRRQFRRTD GASFEQAGVPLPGGGYVAAVIELTGHLAR |
| 719 |
PF12742 (Gryzun-like) |
 Estimated TM-score=0.73; easy target. |
>Gryzun, putative Golgi trafficking (Length=60) LQNKTDLVQDVEISVEPSDAFMFSGLKQIRLRILPGTEQEMLYNFYPLMAGYQQLPSLNI |
| 720 |
PF12641 (Flavodoxin_3) |
 Estimated TM-score=0.73; easy target. |
>Flavodoxin domain (Length=159) SIVFSSSTGNTKKLADAVYEILPKDKCDYFGENDSKIPPSDLLYVGFWTDKGSADTKTLELLAKLKNKKIFLFGTAGFGG SDVYFEKILGQVKQSIDSSNAVIGEYMCQGKMPQSVRERYVKMKENPEHPANLDMLIANFDCALSHPDMDDLEKLKKII |
| 721 |
PF12525 (DUF3726) |
 Estimated TM-score=0.73; easy target. |
>Protein of unknown function (DUF3726) (Length=78) VSHNELVAAVNKAFLGMRRTCGEADVIANMVADLQMVGLDGVRHFNNASNFMGLEDDCPVDIQVRSDNTVEVDLHKAS |
| 722 |
PF12147 (Methyltransf_20) |
 Estimated TM-score=0.73; easy target. |
>Putative methyltransferase (Length=309) IRRFVGQCFQQPAERPSLLDADRQGASCAEAESLAAPLPKYSPRNLYWQATRASLRLGKQLSEGVKLGFDTGFDSGSTLD YVYRNQPNGKGKLGQLVDRNYLDAIGWRGIRQRKLHAEELLRNAIERLRQQGRPVHIVDIAAGHGRYILEALQGLEQRPD SILLRDYSELNVTQGNALISEKGLGDIARFVQGDAFDRASLAALDPKPTLAVVSGLYELFPDNDLVRTSLAGLAEAIEDG GYLVYTGQPWHPQLEMIARALTSHRGGEAWVMRRRSQAEMDQLVQAAGFRKLEQRIDEWGIFSVSLAQR |
| 723 |
PF12099 (DUF3575) |
 Estimated TM-score=0.73; easy target. |
>Protein of unknown function (DUF3575) (Length=182) LFVILLFIGISLPAQNVAVKTNLLYDATATLNLGAEIGLAPRWTLDLSANYNPFSFSDNRKWKHWMAQPEARYWFCERFN GHFMGLHLLGGQYNVGNVKFPLGILPSTEDYRYEGYYYGVGLSYGYQWVLGRRWSIEASLGLGYVRAHYDKYDCPKCGEW KEKGDKNYLGVTKAAISLIYVI |
| 724 |
PF11968 (Bmt2) |
 Estimated TM-score=0.73; easy target. |
>25S rRNA (adenine(2142)-N(1))-methyltransferase, Bmt2 (Length=225) AIEAEIEALGGIEKYQEASLQGQRHDRGGDSSNVLMEWVGPYLSSEEYKEGRLRMLEVGALSTKNACAQSNHFDITRIDL NSQEPGILEQDFMERPLPKDESEKFDIISLSLVLNFVPDHKDRGEMLVRTTQFLTSPKRFSGTTAAFFPSLFLVLPAPCV TNSRYLDQERLAAIMESIGYEMVESKTTQRLVYYLWRRTSDQVGNDRPFRKEELRSGSSRNNFAI |
| 725 |
PF11813 (DUF3334) |
 Estimated TM-score=0.73; easy target. |
>Protein of unknown function (DUF3334) (Length=229) PQIITTDDILIKLCNSVSHVLSSTTASQVTHAAMVQSITRTCLKPDLGCFSIFDGGFSGLVVINFSAPAAIEIYRSYMKQ MGMPESELAFSHTSDEVGDVLGELMNQILGNFISKVNKQLQTSINQSQPKMLTINKELTISIDTNLDEAVARRVSFRTQN NHIFYLEFAMDKTEFIQLNDFDAEEEFDPDDLLAEHGQNNKSKATPLTKVSDNKSMTDEADDLLNELGI |
| 726 |
PF11150 (DUF2927) |
 Estimated TM-score=0.73; easy target. |
>Protein of unknown function (DUF2927) (Length=214) DDLVRNFERIAFFDEYSRGGRGGGTLSRWEVPVRVGIEFGPSVPPAQRNKDDADIRAYVARLARITGHPISVTDRRTNFH VFVAGEDDRDFVQNRMRQLVSGVSDSELSLFRDLPRSFYCFVVAVSDRRNPNAYVHAAALIRAEHPDLVHLSCIHEELAQ GLGLPNDSPQVRPSIFNDDDEFALLTTHDEMLLRMLYDPRLSPGMTAEQARPVA |
| 727 |
PF10914 (DUF2781) |
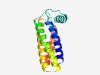 Estimated TM-score=0.73; easy target. |
>Protein of unknown function (DUF2781) (Length=137) DAAVVLFSLTVAVAAPLIDAQSVLPRHLFPAPLVSLKRWYAREFGDYLVARPPGFLRGLVWLELAFLWPLALATLYGILA RRRWAATTSLIAGVSTLTSMSAILGEIVGSKKATLKLLQMYVPFAVFAVIAILRGLC |
| 728 |
PF10824 (T7SS_ESX_EspC) |
 Estimated TM-score=0.73; trivial target. |
>Excreted virulence factor EspC, type VII ESX diderm (Length=98) KNLTVLTDHIRKLSTVHDKAVGEIDGANRSMVENGTNMWETHGVISALTNWAVADAVEARTAAGGALRRVSVELSEKLRA AATNYDNTDSTEAGNIDT |
| 729 |
PF10648 (Gmad2) |
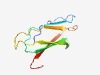 Estimated TM-score=0.73; easy target. |
>Immunoglobulin-like domain of bacterial spore germination (Length=89) VTISSPAPGAVLPNMNAITVTGTGTSLFENSLTVQALDSGGNILAQQPVTTNAPEVGGTGSYQATLSVSVAPGTAGVIRA FAVSANDGS |
| 730 |
PF10647 (Gmad1) |
 Estimated TM-score=0.73; easy target. |
>Lipoprotein LpqB beta-propeller domain (Length=262) GSMVSVTETGVTPVPGYFGSARNMRSLALSQDGKLVAAVADTGRPAPEPASSLMVGAYEDGAASVLEGGAITRPTWAPDN SAIWAAVNGNTVIRVLREPGTGRTSVVNVDAGAVTALGATITELRLSRDGVRAALIVDGKVYLAIVTQTPGGQYALTNPR AVAIGLGSPALSLDWSTSDTIVVARAASDIPVVQVAVDGSRMDALPSRNLTAPVVAVDASTTTEFVADSRAVFQLNNNDP AGDRYWREVPGLTGVKAIPVLP |
| 731 |
PF10626 (TraO) |
 Estimated TM-score=0.73; easy target. |
>Conjugative transposon protein TraO (Length=167) LPGMKGLQVTGGMADGVHWNSKSDFAYYFGAAMSTYTKNGNRWVVGGEYLEKHYPYKDLQIPVSQFTGEGGYYLNFLSDR KKTFFLSLGLSALAGYETSNWGDKLLPDGSTLTDKDGFVYGGALTLELESYITDRVVFLINARERCLFGSSVGKFHTQFG VGLKFIM |
| 732 |
PF10579 (Rapsyn_N) |
 Estimated TM-score=0.73; trivial target. |
>Rapsyn N-terminal myristoylation and linker region (Length=80) MGQDQTKQQIEKGLKLYQSNDTEKALYVWMKVLRKTSDPGGKFRVLGCLITAHSEMGKYKEMLQYSLAQINTAREMEDPD |
| 733 |
PF10404 (BHD_2) |
 Estimated TM-score=0.73; easy target. |
>Rad4 beta-hairpin domain 2 (Length=62) KSADKWYRLGREVKPNEIPVKWLPKRARPKNSRYDEEDAHVEDEVVGTPLYTEDQTELYEPP |
| 734 |
PF10317 (7TM_GPCR_Srd) |
 Estimated TM-score=0.73; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Srd (Length=292) LQIFYPVFFVASLLIQIILMYLIFYHSPKSLQLMKVFLGNTCFFQILLVFVTCAAQFRMISTSVPIELRSYGPCRYLEAW VGYSMYQMLQTSAFMSGMSILITFFFKYEVVRCIELPRCRIIVIVLAFHIPIFISMIMEVIMIITQSLPDDVRQRYKLLN ANVEEFSVIGALSLKTLPSIINFALISGSVVLAPFVCFFYRNKILLRINSQLNQHSKQRKTLIQVFVKGLTIQAFLPLIF YVPVFALYFYCILTHAEILFQQYFMTVVPSLPALFDPFVTLYFVTPYRRRLK |
| 735 |
PF10315 (Aim19) |
 Estimated TM-score=0.73; easy target. |
>Altered inheritance of mitochondria protein 19 (Length=122) SQSPLPAWAMSASLLYSGLNTHTAVDTKLGSGGSSRFSKALALSKPTKKSCFLFGGVNALGGYIIYDGDISNGAGFTFAW SVLYLLVNGKSALKSLTRGRLSPMALSILALGNTGLYGREFF |
| 736 |
PF10309 (NCBP3) |
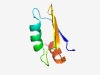 Estimated TM-score=0.73; easy target. |
>Nuclear cap-binding protein subunit 3 (Length=58) APNKVHIRGLDNLTTKDIRTFATEHYVENSPEHIEWIDDTSANLVYESEEVALQALQA |
| 737 |
PF10223 (DUF2181) |
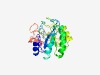 Estimated TM-score=0.73; easy target. |
>Uncharacterized conserved protein (DUF2181) (Length=236) DGLHIKWYHGANSLSEMHEAIAGDHMMLEGDIILRWWGLPNQTEEPVMAHPPAVNSDNTLDNWLTQILNREDKGIKLDFK GLQVVRPSLELLKEKENLVNHPIWANADVVAGPGSSADPIDPKQFLSIVNEVWPNMTLSLGWTTGCCEPFTRQMMEDMLE VCHGVTQPVTFPARAASFRASWEHFKWLLEQNPESYTITVWTPSGAEDWEGTDIYDLLYVRNDCDKTLIYYDLPGE |
| 738 |
PF10105 (DUF2344) |
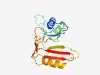 Estimated TM-score=0.73; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2344) (Length=182) KIRIKFKKEGSMKFIGHLDIMRYFQKAMRRADIDIAYSEGYSPHQIMSFASPLGVGITSDGEYIDIEIKNRISSKEAMHK LNEVMVEGIEILSFRQLPKDSKNAMSIVEAADYEIKFREGYEPKLWEERLKDFFTQEEITVTKKTKKSEKEVDIKPMIYS LNVSNGKVSMQIASGSANNLKP |
| 739 |
PF10087 (DUF2325) |
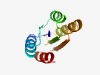 Estimated TM-score=0.73; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2325) (Length=89) SIVIIGGHDRMAGQYKTICKNYNCKAKVFTQMPSNFKNQIGNPDLIILFTNTVSHKMVKCAVNEAKNSNANIIRSHTSSA NALKQILEN |
| 740 |
PF10086 (YhfC) |
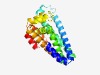 Estimated TM-score=0.73; easy target. |
>YhfC intramembrane metalloprotease (Length=223) AFLLPIGLIVWFKRKYGASLKVFFIGALTFFVFAQLLEGGVHLYVLQVNEVTKEAMQHPLWYGIYGCLMAGIFEECGRYL MMRFFMKRHHTWADGLAFGAGHGGLEAILITGLSSISLIVYAFAINSGTFEQLLVNDDVKQALLPIQEQLLHTPSYEWLL GGIERISAIAVQIGLSLLVLYAVKSRRPLFLLYSILLHALFNVPAVLYQRGIVEHAAAVEIIV |
| 741 |
PF09200 (Monellin) |
 Estimated TM-score=0.73; trivial target. |
>Monellin (Length=40) GEWEIIDIGPFTQNLGKFAVDEENKIGQYGRLTFNKVIRP |
| 742 |
PF08823 (PG_binding_2) |
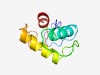 Estimated TM-score=0.73; easy target. |
>Putative peptidoglycan binding domain (Length=74) TLLQREDPLDVVLKKDVAGEVQEALRRLGFYKREPTGVWDEETERAFRNWAGYENFENKIRNDDKIWGSLYRYL |
| 743 |
PF08670 (MEKHLA) |
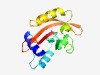 Estimated TM-score=0.73; easy target. |
>MEKHLA domain (Length=139) TLAHWICNSYRCYLGVELLKSNNEGNESLLKSLWHHSDAILCCTLKALPVFTFSNQAGLDMLETTLVALQDTPLEKIFDD HGRKILFSEFPQIIQQGFVCLQGGICLSSMGRPVSYERVVAWKVLNEEENAHCQKLRFF |
| 744 |
PF08631 (SPO22) |
 Estimated TM-score=0.73; easy target. |
>Meiosis protein SPO22/ZIP4 like (Length=257) SAVAQGDFQKAVICVQRCKDMLMRLPKETCYLSLLCYNFGVETYEQKRYEQSSFWLSQSYEIGKMDKRYSTGQEMQAKVL RLLATVYLEWDCKVYQDKALQAISLANKENLHPAGFFLKIQILLKSGAADEDLSTAIAEIMHHEVSLDFCLNTAKMLLEH NRELVGFDFLKSVAGRFESSPDLGKVILLHIEFLLQRMREPLAKQKIEDIITGHYTGKQLSAETLNQLHIILWDRAAKNY EAKNYPEALQWYNYSLS |
| 745 |
PF08461 (HTH_12) |
 Estimated TM-score=0.73; easy target. |
>Ribonuclease R winged-helix domain (Length=66) IEIMRIISESDKPVGARNIADELNNRGYNIGERAVRYHLRILDERGFTKKHGYAGRTITERGKEEL |
| 746 |
PF08277 (PAN_3) |
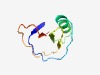 Estimated TM-score=0.73; easy target. |
>PAN-like domain (Length=69) MVVTCGKPLQASAPIILNTTWDECLQTCWADSNCAVIYDTTPTCSYYSIELITSVQKLPKSSGERVAFR |
| 747 |
PF08269 (dCache_2) |
 Estimated TM-score=0.73; easy target. |
>Cache domain (Length=297) VIEKFKADAYQSKQEELENYVSLAMKSVEAYHARTSKEKVKAEVQSYLKEQTGFMLSIMEGTYEKYKGTVSDDELKELIR NTVKSTRYGKTGYFWINDTAAKIVMHPIKPQLDGKDLANYKDKGGKKIFSEFARVASSSGEGFVDYVWPKPGFEAPQDKV SFVKLFKPFNWVVGTGEYVDNVSDKLKKEALANVKSMRYGAKGNGYFWVNDSNHVVIMHSIKSSIDGKSMYDLQDPNGKY LYREIVKAANQNAKGGVVDYMWSKPGSETPQPKISYVKKFEPWDWIIGTGVYVDDIE |
| 748 |
PF08086 (Toxin_17) |
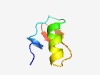 Estimated TM-score=0.73; trivial target. |
>Ergtoxin family (Length=41) DRDSCVDKSRCAKYGYYQECQDCCKKAGHNGGTCMFFKCKC |
| 749 |
PF08011 (PDDEXK_9) |
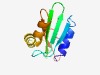 Estimated TM-score=0.73; easy target. |
>PD-(D/E)XK nuclease superfamily (Length=96) FEGYYASVVYAYLASLGYEIIPEDTTNKGRIDLTVKTRPYIWIFEFKVKGIDHSGNKDPLKQIKERGYAEKYAADSRPVI EIGIVFDPKTRNIEAW |
| 750 |
PF07878 (RHH_5) |
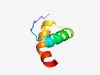 Estimated TM-score=0.73; easy target. |
>CopG-like RHH_1 or ribbon-helix-helix domain, RHH_5 (Length=43) GKRISITLSDEILEDLEIWAQQRGQTVAGLAALLVEKAVTEAQ |
| 751 |
PF07875 (Coat_F) |
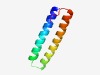 Estimated TM-score=0.73; trivial target. |
>Coat F domain (Length=61) DATILGDMLMTEKFVSGAYDTAIFESANSDVRQTLQHIQKEEQQHGEGIFNYMSQHGMYNP |
| 752 |
PF07862 (Nif11) |
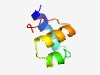 Estimated TM-score=0.73; hard target. |
>Nif11 domain (Length=48) MSKESVLALIKAAEADQSLKAQLESAENPEQVLVIAASQGYNFTEEEL |
| 753 |
PF07672 (MFS_Mycoplasma) |
 Estimated TM-score=0.73; easy target. |
>Mycoplasma MFS transporter (Length=275) QRRKSVLSNANLWGFNSGIVVAFVPFLFQSVQQAGTKYWVFILTALILIGFGILCVFAWFEKQMDPFMPQKQTKEQMQLG NQPSAGDILKRKATWKMIGMYGICLVVLVNPLTGGWWNILQAVSPASSFNVKDGVKTLKPLEGAGGYFAGLPTLAILWVL GYGMGYMVFSPFNKTVYDRKRWLSFMFFMNALMVIVIVLFAATLGVGTAVGFAFVAIATFIGGSFAWSMQSTILILPHEF KEYKRSEVSVLFGYIWGFGYVIYTAFDITNSMFLE |
| 754 |
PF07576 (BRAP2) |
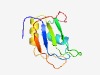 Estimated TM-score=0.73; easy target. |
>BRCA1-associated protein 2 (Length=98) LPVGRKPLVCVLAVPNHMTYADFCQFCGSFIQHILEMRIVRNDGDEDQYSVLIRFDGQESADNFYKHFNGKRFSSMEVEV CHVLFTVDVQFTASIEHA |
| 755 |
PF07381 (DUF1495) |
 Estimated TM-score=0.73; easy target. |
>Winged helix DNA-binding domain (DUF1495) (Length=89) ILRSINKSVLRKKVLFYLYSIYPNYDYLANISRRVRSDPTNVLGCLKGMGNRYNGSSSLIELGLVEVVEKDGYKYYRITD LGRKVVEYV |
| 756 |
PF07310 (PAS_5) |
 Estimated TM-score=0.73; easy target. |
>PAS domain (Length=136) MKHPSNRELFAYWNDRRGHAKAPERSEFEPSAVRELLGDIFVLSCDAELGFPFRMAGTRLCALLGGDVKDKSFPAQFTPA SRREIEEIVTVVSEETLPAIAGVKAQAPDRTTASFELLLLPFATRAHEPISLTGLL |
| 757 |
PF07030 (DUF1320) |
 Estimated TM-score=0.73; trivial target. |
>Protein of unknown function (DUF1320) (Length=121) YCTKQDLIDRFGEDELIDLTDRDNMSVIDETVLDQAIADGSAEMDGYLGGRYQLPLVTVPPVLKALCCNIARYKLYDEQA SEQVTKRYDSAIKFLFSVSKGEISLGVDGSGAKATSTDLAD |
| 758 |
PF06316 (Ail_Lom) |
 Estimated TM-score=0.73; trivial target. |
>Enterobacterial Ail/Lom protein (Length=199) MRKLCAVILSAVVWLVAAGTPASAAEHQSTLSAGYLQTHTDMPGSDDLNGINVKYRYEFTDTLGLITSFSYANAEGEQKT HYNDTRWHEDSVRNRWFSVMAGPSVRVNEWFSAYALAGVSYARVSSFAGDYVTLTSDEGKKQEHLTRSDSGRRSHTALTY SAGVQINPTENIVVDLAYEASGRGDWRTDAFIVGTGYRF |
| 759 |
PF06301 (Lambda_Kil) |
 Estimated TM-score=0.73; easy target. |
>Bacteriophage lambda Kil protein (Length=42) NHQALMAAQSKAVIARFLGDAGMWLQANQQMKQAVSMPWYRR |
| 760 |
PF06259 (Abhydrolase_8) |
 Estimated TM-score=0.73; easy target. |
>Alpha/beta hydrolase (Length=190) DPNQQVLAAVAVGNPDTAANVSVTVPGVGSTTRGALPGMVTEARDLRSEVIRQLNAAGKPASVATIAWMGYHPPPNPLDT GSAGDLWQTMTDGQAHAGAADLSRYLQQVRANNPSGHLTVLGHSYGSLTASLALQDLDAQSAHPVNDVVFYGSPGLELYS PAQLGLDHGHAYVMQAPHDLITNLVAPLAP |
| 761 |
PF06162 (PgaPase_1) |
 Estimated TM-score=0.73; trivial target. |
>Putative pyroglutamyl peptidase PgaPase_1 (Length=164) MLSMCNKYPDFDDEERVRRVDDDFRDVITVFDAPFEGKSPSPAVIVFDELVDSDGSSKYLNFKMEQSYEKVDEIPEKIDA ERIRYAIHLGSHSQKNVLQIFQSAYSDGYTEEDKNGKVPVGGKVKCAETETGFRTKINCENVVTAVNEYMDSNREKFGEL RIET |
| 762 |
PF05677 (DUF818) |
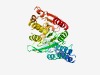 Estimated TM-score=0.73; easy target. |
>Chlamydia CHLPS protein (DUF818) (Length=361) AILDMHPKPSIAMFSSEQARTSWEKRQAHPYLYRLLEIIWGVVKFLLGLIFFIPLGLFWVLQKICQNFILPGAGGWIFRP ICRDSNLLRQAYAARLFSASFQDHVSSVRRVCLQYDEVFIDGLELRLPNAKPDRWMLISNGNSDCLEYRTVLQGEKDWIF RIAEESQSNILIFNYPGVMKSQGNITRNNVVKSYQACVRYLRDEPAGPQARQIVAYGYSLGASVQAEALSKEITDGSDSV RWFVVKDRGARSTGAVAKQFIGSLGVWLANLTHWNINSEKRSKDLHCPELFIYGKDSQGNLIGDGLFKKETCFAAPFLDP KNLEECSGKKIPVAQTGLRHDHILSDDVIKEVAGHIQRHFD |
| 763 |
PF05651 (Diacid_rec) |
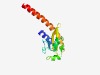 Estimated TM-score=0.73; easy target. |
>Putative sugar diacid recognition (Length=134) LNATLARQIVERAMKIIRYSVNVMDEYGRIIGSGDPTRLNQRHEGAILAINENRVVEIDPGTAQQLKGVKPGINLPIVFQ HRVIGVVGISGEPDKVRNYGELVKMTAELIIEQEALMAQVQWNQRHREELVLQL |
| 764 |
PF05272 (VirE) |
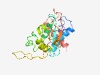 Estimated TM-score=0.73; easy target. |
>Virulence-associated protein E (Length=203) YLNTLVWDGTERIRFCLRHFLGADADDYTYEALKLFLLGAISRAFQPGCKFEIMLCLVGGQGAGKSTFFRLLAVRDEWFS DDLRKLDDDNVYRKLQGHWIIEMSEMMATANAKSIEEIKSFLSRQKEVYKIPYETHPADRPRQCVFGGTSNALDFLPLDR SGNRRFIPVMVYPEQAEVHILEDEAASRAYIEQMWAEAMEIYR |
| 765 |
PF05241 (EBP) |
 Estimated TM-score=0.73; hard target. |
>Emopamil binding protein (Length=181) LTSISAVGVAAIVATRRLLPKTASVKDKFTFIWLAFDVGIHAAFDSAFLYFSLFGRQVNTSAGPLAQLWKDYARADARWG VADPTIVSMELITVIGTGATILYIMQKLVQDDPARHYWIIVLCTAELYGGWMTFAPEWLSGSQNLNTGDAFYLWVYLFFL NTIWVFVPIWLMFDSYRYIAR |
| 766 |
PF05018 (DUF667) |
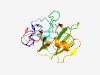 Estimated TM-score=0.73; easy target. |
>Protein of unknown function (DUF667) (Length=183) MYRQSYQRGVFSIFYSVGSKPLALWDTVTRDGYITKFLDEDIKSTVLEIGGTNVSTTYMICPKGNAVLGICMPFLVMIVK NLRKYFSFEVTILDETGTRRRFRVSNFQSQTQILPLCTVMPIGLSEGWNQIQFNLAEFTRRAYKKQFIEVQKVKINANIR LRRIYFAERLIPEDQLPAEYKLF |
| 767 |
PF04155 (Ground-like) |
 Estimated TM-score=0.73; easy target. |
>Ground-like domain (Length=73) VCCDSMIPLFIDNQLSTTDGIGAITRAVQQRVQSQFNRSFEVIISESDFVVNTYYSGPRQCKFETDRYFVALY |
| 768 |
PF03606 (DcuC) |
 Estimated TM-score=0.73; trivial target. |
>C4-dicarboxylate anaerobic carrier (Length=487) MPSSYTVLLIIIVIMAVLTWFIPAGAFIEGIYETQPQNPQGIWDVLMAPIRAMLGTHPEEGSLIKETSAAIDVAFFILMV GGFLGIVNKTGALDVGIASIVKKYKGREKMLILVLMPLFALGGTTYGMGEETMAFYPLLVPVMMAVGFDSLTGVAIILLG SQIGCLASTLNPFATGIASATAGVGTGDGIVLRLIFWVTLTALSTWFVYRYADKIQKDPTKSLVYSTRKEDLKHFNVEES SSVESTLSSKQKSVLFLFVLTFILMVLSFIPWTDLGVTIFDDFNAWLTGLPVIGNIVGSSTSALGTWYFPEGAMLFAFMG ILIGVIYGLKEDKIISSFMNGAADLLSVALIVAIARGIQVIMNDGMITDTILNWGKEGLSGLSSQVFIVVTYIFYLPMSF LIPSSSGLASATMGIMAPLGEFVNVRPSLIITAYQSASGVLNLIAPTSGIVMGALALGRINIGTWWKFMGKLVVAIIVVT IALLLLG |
| 769 |
PF02677 (QueH) |
 Estimated TM-score=0.73; easy target. |
>Epoxyqueuosine reductase QueH (Length=177) LLLHSCCAPCSSYVLEYLSEYFNITVYYYNPNIHPEKEYRKRVNEQENFIKKFNTKNKVSFIEGEYKPQHFFNMAKGLEK VREGGERCFKCYEMRLRKAAELAKQRGFDYFTTTLSISPHKNAQKINEIGQELEKEYGIKFLYSDFKKKNGYKRSIELSK EYNLYRQDYCGCVFSRR |
| 770 |
PF02666 (PS_Dcarbxylase) |
 Estimated TM-score=0.73; easy target. |
>Phosphatidylserine decarboxylase (Length=216) LTEFFSRRLKYDARPIDEHPLSIVSPVDGVVSQVGKIRHGTIIQAKGIDYSVSSLLGESPGETGFEDGYYMTIYLSPKDY HRIHMPLEGKVTRSTYIPGRLFPVNEIGVTQVKGLFTKNERVITHIQTEAGEMAMVKVGAFIVGSVQVAYKESFKRKKRT VHKNTMHTKPYFKKGSEIGHFEFGSTVILLFQKERVMINQTIQPGAAVKMGEKIGE |
| 771 |
PF01531 (Glyco_transf_11) |
 Estimated TM-score=0.73; easy target. |
>Glycosyl transferase family 11 (Length=311) LSLLCPDRSRVTSPVAILCLNPNTTFSCPKHPASVSGTWTIDPKGRFGNQMGQYATLLALAQLNGRQAFIQPSMHAVLAP VFRITLPVLAPEVDRHAPWQELELHDWMSEEYTHLKEPWLKLTGFPCSWTFFHHLRGQIRSEFTLHQHLRQEAQRSLSGL RLPRTGDRPSTFVGVHVRRGDYLEVMPLHWKGVVGDRAYLQQAMDWFRARHEAPVFVVTSNGMEWCRENIDTSRGDVIFA GDGQEDAPVKDFALLTQCNHTIMTIGTFGFWAAYLAGGDTVYLANFTLPDSSFLKIFKPEAAFLPEWVGIN |
| 772 |
PF01313 (Bac_export_3) |
 Estimated TM-score=0.73; trivial target. |
>Bacterial export proteins, family 3 (Length=72) VMTLGRQAMEVTLMVSAPMLLVALVVGLVVSIFQAATQINEMTLSFIPKLVGIFVTLVVAGPWMLSVMLDYM |
| 773 |
PF00978 (RdRP_2) |
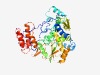 Estimated TM-score=0.73; trivial target. |
>RNA dependent RNA polymerase (Length=438) RDISLNVKDCRIDFSKSVQVPKERPVFMKPKLRTAAEMPRTAGLLENLVAMIKRNMNAPDLTGTIDIEDTASLVVEKFWD AYVVKELSGTDGMAMTRESFSRWLSKQESSTVGQLADFNFVDLPAVDEYKHMIKSQPKQKLDLSIQDEYPALQTIVYHSK KINAIFGPMFSELTRMLLERIDTSKFLFYTRKTPTQIEEFFSDLDSSQAMEILELDISKYDKSQNEFHCAVEYKIWEKLG IDEWLAEVWRQGHRKTTLKDYTAGIKTCLWYQRKSGDVTTFIGNTIIIAACLSSMIPMDKVIKAAFCGDDSLIYIPKGLD LPDIQAGANLTWNFEAKLFRKKYGYFCGRYIIHHDRGAIVYYDPLKLISKLGCKHIRDEVHLEELRRSLCDVTSNLNNCA YFSQLDEAVAEVHKTAVGGAFVYCSIIKYLSDKRLFKD |
| 774 |
PF18851 (baeRF_family8) |
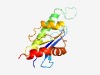 Estimated TM-score=0.72; easy target. |
>Bacterial archaeo-eukaryotic release factor family 8 (Length=140) PQAAFVLALAQNSVRLVEVSPDADPFEVSVSDLPEDAASAVGLASISGRSPSGRIQGSEGRKVRLHQYARAVDRAVRPVL SGSGVPLILAATEPLAGIYRSVNSYVHLAAPGIDGNPENVSDAELAGEARTVLDELYRIE |
| 775 |
PF18760 (ART-PolyVal) |
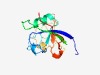 Estimated TM-score=0.72; easy target. |
>ADP-Ribosyltransferase in polyvalent proteins (Length=142) DKEGKPLVVYHGTGAEFESFDNKKTGANDGGLWGRGHYFSAVVGNANSYALRNDIGARVIPAYVSIKKPLILRTGNDLVT RLPDGSDYRSLVGSNLNGAKIKEIALEGGHDGIVQIKPDGLIGDLVAFSPAQIKSVFNRGTF |
| 776 |
PF18751 (Ploopntkinase3) |
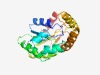 Estimated TM-score=0.72; easy target. |
>P-loop Nucleotide Kinase3 (Length=192) MKPRMIYLVGQPGAGKSTLMAALTKGLVRSPWHEHTVPHDQLVDRVTGETVAAELGVQRGAFSGTDALASSIIDKAVPWI QSRPYDLVLAEGARLANRRFIDAALDAGYAVTIGLLDHDQADTWRKRRSKALGREQSPSWVKGRLTASRNLAAHYPGALV VAGSVIPAEPGKAIVLAGHPDELEPILAAVMA |
| 777 |
PF18748 (Ploopntkinase1) |
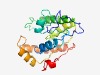 Estimated TM-score=0.72; easy target. |
>P-loop Nucleotide Kinase1 (Length=196) HLFHKPPAVVGEYPARGELYYVKGSNGSGKSTVPSWMAENDPQAYVVTHNGKVMLTVCPSFNIICVGKYDKSKSKGVDSL KDTEQMLFAVSITEQPEYIGYDVIFEGIIPATLLNTWIERLNRPSRRLVVLFLDTPKEVCLARVSSRNGGEDFKHDLVLE KWKRVNSHRERHKELFPNIPAGMMKSNGLTVDQAVS |
| 778 |
PF18405 (Serine_protease) |
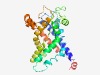 Estimated TM-score=0.72; easy target. |
>Gammaproteobacterial serine protease (Length=276) YFLANLGVKTAAISFSVSTLTQPFQAMLTRLQLNAPSGFNGGLFRSMYRGFVPYAIAGQKRGAVSVTAKQANKEVIEEEE IEIPGRQRWLGTFVFSQADLLISNALGGKSKLENAGIITKTNFKWSFANYWKLTGVNWGSRSFAGFVNFSALGFVGDYIS SFYKFDNDFYNKLAGGATAGVIATIFTTIPNSYADRKVLATKVENGRLLTVSPFTMFSQAKSHVNSVGMKQALSHFVKVN FLKEVVVRSPMSALTFGIIFGLDDLMGPEPLKRVWP |
| 779 |
PF17924 (TetR_C_19) |
 Estimated TM-score=0.72; easy target. |
>Tetracyclin repressor-like, C-terminal domain (Length=117) DLFCAFEEMFKHLLMCFKDKGNADFFKNTFLNMNHRIEKTFTPNICKEKAMEQRREFLSLVDLSKLNINSKEEVPYVLQI MMAITMHNLIQIFAKNLSFEEAIKIYKIEIDMIKKGI |
| 780 |
PF17741 (DUF5578) |
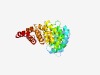 Estimated TM-score=0.72; easy target. |
>Family of unknown function (DUF5578) (Length=268) MTSAKEQAAIGRLLAFLQEWDNAGRAARSRILDNFIEANQGKTGPELELEFSQGASLFLARLTAWLRLSYMNGTCLDQLL KAIGIFLSAASGHRYLIEFLEVGGVLTLLEILGLSQLKEEDKKESIKLLQLVANAGRKYKELICESYGVRSIAEFMASSK SEKTQRQVQILLDSLGHGNPKYQNQVYKGLIALLPCTSPHAQQLSLQTLRVVQAIVGKSHPSIVEAVLGVLRSMELEVQY EAIQLIKDLTSYDVRPALLRGLVALLKP |
| 781 |
PF17175 (MOLO1) |
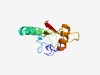 Estimated TM-score=0.72; easy target. |
>Modulator of levamisole receptor-1 (Length=115) DENSFPDPFTHPEECVLSLGISHTWLCDPSRFLSIEQQINIEAELLKIRDTNFHKCSNNSVYYYQVSVAIVPEIFVSKNE TYENAAQQFSEKLLRKWGIGNSPCHDGILLVYIKN |
| 782 |
PF17171 (GST_C_6) |
 Estimated TM-score=0.72; easy target. |
>Glutathione S-transferase, C-terminal domain (Length=63) SKNLRAVSALLGDKKFILGERPCQDDCAIFGQLGQAVWGLPNSNYEKLVHNELTNLRDYCFRI |
| 783 |
PF17159 (MASE3) |
 Estimated TM-score=0.72; hard target. |
>Membrane-associated sensor domain (Length=224) SFYSYLLFHSLIELFSIVVAFAIFILAWNSRQFLENNYLLFIGIAYLFIGGIDLIHTLAYKGMGVFPGYDSNLPTQLWIA GRYVESISLLIAALVLGRKLRPYLVFLGYAVGISLLLMSIFYWGVFPQCFIEGVGLTPFKKVSEYIISLILAGSMILLFR SRRQFDKGVLRLLVASIAITIGSELAFTFFVDLYGLSNLIGHLFKVISFYLIYKALIETGFRKP |
| 784 |
PF16946 (Porin_OmpG_1_2) |
 Estimated TM-score=0.72; trivial target. |
>OMPG-porin 1 family (Length=304) DDVSFLRLRQEDPPVHWNSIGSNMDAGGLHGNIGSQIEIDDVRWGNKNRNSGKFKLATIQAFLRHDELPNWYFGFWNARE DDYKGQFSNQDYSGSNTINEFFVGHIFENWRGNIGTEVMIGSETATERWKTRFKVWQDLRLSDRWSVAGYAYGEYQPQGN QPGNGDLEQYIFETEPAIQYRVNPDLGLYFRPYYYYNRQVRENWGDIVEQEWKVTTGLWKNWYPLLTSMYIGFGQDKIVN ASNSKELFYDGRYKFIGSTISYPVFGEVRLYGEFKASFTKEAGQWTYNGHSWNPFTIVGVTYNF |
| 785 |
PF16371 (MetallophosN) |
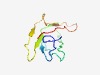 Estimated TM-score=0.72; easy target. |
>N terminal of Calcineurin-like phosphoesterase (Length=77) GYVHDSEKPLSGVLVTNGYSFATTDENGAYYLPSDADAEFVYLVTPAGYNADCRDGLTRFYQTIDKNGGIYRADFQL |
| 786 |
PF15569 (Imm40) |
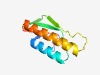 Estimated TM-score=0.72; easy target. |
>Immunity protein 40 (Length=93) LLSELGTQEYALDQQDAMNAIVIIEDLAIPILGGDVYLQSLEGIEISYENWSINRQPGESLDNYVRRSCAYTRNYIARFP VQAEKLPLFVLVL |
| 787 |
PF14771 (DUF4476) |
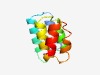 Estimated TM-score=0.72; hard target. |
>Domain of unknown function (DUF4476) (Length=91) ITDANFNTLVTNVKNKWSQSLKGEAIREALVNSNYYYTSTQVRQLMGLVSSETDRMDLIKLAYPVVSDTNNFPGLYDVFA STANRTEFNEF |
| 788 |
PF14440 (XOO_2897-deam) |
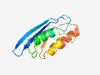 Estimated TM-score=0.72; easy target. |
>Xanthomonas XOO_2897-like deaminase (Length=100) PGYLAVLRYRAQDGSEQQVMRRSAPGTPHPEWQILHELRGMGVPPQQVLELHTELEPCDLPGGYCSRMIRETWPQVRLSH TASYGRDHASRQQGMQQLIE |
| 789 |
PF14361 (RsbRD_N) |
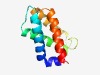 Estimated TM-score=0.72; easy target. |
>RsbT co-antagonist protein rsbRD N-terminal domain (Length=104) LAEIIAKHEQDLLAQWIKEQTSAVTQRPDLMSEAELREQSRALLNGIRNAVQRGRLDDITGSEWESVRELLDEVSRGRAR TGFSPTETAMFVFSLKQPLFSRLR |
| 790 |
PF14272 (Gly_rich_SFCGS) |
 Estimated TM-score=0.72; easy target. |
>Glycine-rich SFCGS (Length=114) VTVVIADRLGKGQNVAKGVEAAGGKAVVVPGMGADMRLGDVMQQENADLGISFCGSGGAGALTAATKYGYPERHGMRSVE EGITAINDGKTVLGFGFMDQEELGKRIVEAYVKK |
| 791 |
PF14252 (DUF4347) |
 Estimated TM-score=0.72; easy target. |
>Domain of unknown function (DUF4347) (Length=160) VVFIEDNVADYQTLIDGIGAGIEVVLLDSTQDGLAQMALWAQSNSDYDAIHIISHGAEGQVNLGGFSLDNFTVNTRSADL AQLGAALNEDGDLLLYGCEVASGEGQDFIIALAQATQADVAASDDLTGSAVLGGDWQLEFQQGQISETQFLDDSVEDIYH |
| 792 |
PF14016 (DUF4232) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF4232) (Length=135) CKSSDLKLTLGHGDAGAGTVYRPLIFTNVSDRPCVIQGFPGVSYVAGEDGHQVGPAAVRQGEKGGAFTLNNGDTAYAEIG FVNVQNYDTVTCQPQPVRGLRVYPPQETASMFVELPTTGCASEKIPGDQLTVRTI |
| 793 |
PF13924 (Lipocalin_5) |
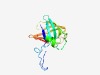 Estimated TM-score=0.72; easy target. |
>Lipocalin-like domain (Length=129) VGTWKLVSTERVSANGERLPPTSTNQIGFLIYDSAGNMGVTIMQGGRQKYAGAQPTPEEARAALTSYTSYFGTFIVNAAE GVVTHRLQGSLNPGMATDQRRGFELKGNLLTLKPPRGANGVQSQLTWER |
| 794 |
PF13752 (DUF4165) |
 Estimated TM-score=0.72; easy target. |
>Domain of unknown function (DUF4165) (Length=119) AHAELLEYTFKAPDGTQRSLTPNANYANPTGNISFALSAGIDRKVKISVIRSDGTVVSTATSHLLGATDRITVGGKSYYG AELQLPAPSAGSYTVKAEILASDGSTVQTDEYPLTVDVT |
| 795 |
PF13612 (DDE_Tnp_1_3) |
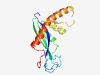 Estimated TM-score=0.72; easy target. |
>Transposase DDE domain (Length=155) KGKPTGLAFIDSTHLRVCHNIRIPRHKVFDGVAKRGKSTMGWFYGFKLHLIVNHQGEIMAAKLTPGNVDDREPVHELAKG LTGSLYGDKGYISQSLVDTLAETGVTFITKKRHNMKAKALSAWDNMMLSKRFIIETINDQLKNISQIEHSRHRSL |
| 796 |
PF13421 (Band_7_1) |
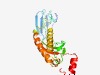 Estimated TM-score=0.72; easy target. |
>SPFH domain-Band 7 family (Length=211) VWRFERYDNEIKNGAKLIVRQSQVAIFVNEGKIADCFESGTYELETQNLPILSTLKGWKHGFSSPFKAEVYFVNIRNFTD YKWGTKNPVMLRDPEFGPLRIRAFGNYAIRVADPKKFLIDIVGTDGHFTTDEISDQLRNLIVTRFTDCIAESKIPVLDMA ANYDELSEYVTNKISPEFEELGIGITKFLIENISLPEAVEKALDKRSSMGI |
| 797 |
PF13386 (DsbD_2) |
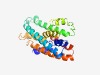 Estimated TM-score=0.72; easy target. |
>Cytochrome C biogenesis protein transmembrane region (Length=209) AAFLIGLMGAGHCVGMCGGVAAAITIGMPNNTHQKRRWIYLLNYNIGRLVSYTIAGGLIGAALAGISTLNGTSNPLVYMR LFAALMMIVLGLYIGQWWAGLSQIERVGQKIWRHISPIATSLLPLKSPVKALPFGFLWGWLPCGLVYSTLTWAAVSGSAL NGAAVMFAFGLGTLPAMLAVGGFADKLKNWLKNRIFRRISALLLIIYGV |
| 798 |
PF13367 (PrsW-protease) |
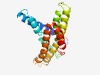 Estimated TM-score=0.72; easy target. |
>PrsW family intramembrane metalloprotease (Length=192) EPPWMLATAFFWGALVAIFIAFVMNTISGGIIALLAGDKAGEFFATVISAPFVEETSKALVLFGLFWWKRDEFDGVIDGI VYAAMVGLGFAMTENIQYYGGAALEGGVAGSLGTFILRGLMAPFSHPLFTSMTGIGLGLASQTNKRGIRILAPLVGLFVA MSLHGLWNLSASLNPVLYFVVYFFIMVPVFIA |
| 799 |
PF13157 (DUF3992) |
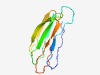 Estimated TM-score=0.72; hard target. |
>Protein of unknown function (DUF3992) (Length=95) VCSPWSATVVATAIDNVLYTNNINQNVVGTGFVKYDVGPGPITVEALDSAGTVIDTQTLNPGTSIGFTYRRFDIIQVVLP ATPAGTYQGEFCITT |
| 800 |
PF13151 (DUF3990) |
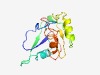 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF3990) (Length=147) MILYHGSYLEIKSPDLEHSRKNVDFGCGFYLTPIYEQAVKWCEKFKRRGKNGIVSRYEFDEKAYQELKILKFDSYSEKWL DFILTCRSGKDATDYDIVIGGVANDKVFNTIELFFDHLIDKEEAIRRLRFEKPNLQLCFRTQKALES |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218