

Go to page (83 pages in total): [First page] << < 1 2 3 4 5 6 7 8 9 10 11 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 501 |
PF04226 (Transgly_assoc) |
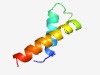 Estimated TM-score=0.76; hard target. |
>Transglycosylase associated protein (Length=47) ILLGIVGSFVATYLGSAMGWYQPGQTAGWIASVVGAVILLFIYGMVR |
| 502 |
PF04093 (MreD) |
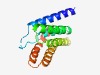 Estimated TM-score=0.76; easy target. |
>rod shape-determining protein MreD (Length=152) KRFILQWLILICTFIVALVMEIAPWPTDFQNFKPAWLVLVLLYWTLALPNKVSIGSAFVLGVIWDLVLGSILGVHALVLS LFTYLVALNHLVLRNLSLWMQSLLVIVFVFSIRFSIFLIELLLHSASFNWQESYGALASGLLWPWVFLLLRK |
| 503 |
PF03812 (KdgT) |
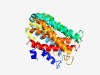 Estimated TM-score=0.76; easy target. |
>2-keto-3-deoxygluconate permease (Length=304) ILKAIERIPGGLMVVPLLLGAIINTFFPQALQIGGFTTALFKQGAMPLIGLFLLCNGAQITLREAGVSLSKGVALTAVKF FIGAALGWVVGKFMGPTGLLGLSSLAIIAAVTNSNGGLYTALASQYGDPTDVGAIAILSINDGPFLTMVAMGATGLANIP FIALVAVVIPILVGMILGNLDEEWKKFLAPGQKLLIPFFAFPLGAALDFRTILKAGFPGILLGVLTVVLTGLGGYLTIKY IFRENKGVGAAIGTTAGNAVATPAAVAAVDPTLEPLVAAATAQVAASVIITAILCPLLVSYLDK |
| 504 |
PF03761 (DUF316) |
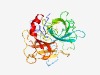 Estimated TM-score=0.76; easy target. |
>Chymotrypsin family Peptidase-S1 (Length=268) VIVIFFALTGCIGCDFYKLTADENKDRASTCGKEVLERKGNETTPANVSQTDWTLKMHNIDHNPDRYYLATMISTRHVLT SARLVRNESGWRDGSRDECDGKNLVVPPSLLEKVEIPQHAQDGSKSVSNSTRCKITAAYILDGCASSYPMLTAGPLIATI DYSSQSNISVPCLAQEENLMNVSEIGDLYFIDDSYKLKHSKTTINHTYSLFYKTSIPTVFKSRHDYGILTNVVDNKMTVM AFAASDDQNTFYTMSFLYKSLCEPTGIC |
| 505 |
PF03756 (AfsA) |
 Estimated TM-score=0.76; easy target. |
>A-factor biosynthesis hotdog domain (Length=136) LVHRSSIAEVFVTDGVRTGENAFSVGAQWPRDHALYHPDENGLNDPLLFAETLRQAHFYGAHTYFGVPVGSRFIGQDVSF EITDPTALRVGAAPLAVVLNGTWTEERDRRGRPAGARLDVTLTVDGRPCGRGHTRG |
| 506 |
PF03687 (UPF0164) |
 Estimated TM-score=0.76; trivial target. |
>Uncharacterised protein family (UPF0164) (Length=329) AARATTLRSRGGVVSSRASGGTLVVTAQKPKVMARNDVDYRPLSLQAGGRQGSLDLVATATADDASFFEANAAGSATIPR MTLAFFHTMRISDSHIDVLSFVGRAGRTGYGVSARAFYPDMSSKTTGFVGIFNVSHAFSSAYRFKGVSVGANLKVGYRHT RGGGSSQSKSSNGKENHHIVLTADVGVRGAWTVSKNFGAHEPNLWAGVAFRNIGASINATNLHGNNGAGGSGGGGGGNGD GKPAHVTDSRVILALAYQPVRYFLFGAGLEWLYNVGSIKAVNSLRYGAAFMLFPLRQLAFSSSVVMKGMGPQQVRASAGA EVQFSHVRC |
| 507 |
PF03665 (UPF0172) |
 Estimated TM-score=0.76; easy target. |
>Uncharacterised protein family (UPF0172) (Length=186) YSIAPLAYLKLVLHAAKFPSSTCVGLLIGSVAESSCTITDAIPLLHHWTDLSPMAEAGLGLAELYAKQQQLVFLGLYVAN ERLGDQAVPNGVQKMAEAIRRDRPEAIVLVVDNEKLASNEPALIPYLPPKSPSAAWQPTTLSSARISLADPSAPTKALEQ VRQGRHKLLGDFDEHLEDVRVDWLRN |
| 508 |
PF03659 (Glyco_hydro_71) |
 Estimated TM-score=0.76; easy target. |
>Glycosyl hydrolase family 71 (Length=383) VFAHVVVGNTAAHTQSTWQNDITLAHNAGIDAFALNIATPDSNTAPQVANAFAAAKALNNGFKLFFSFDYLGGGQPWPAT GSNSVVSYLTQYKNSGVYFYYNGLPFASTFEGTNNINDWAPGGPIRSGAGDVYFVPDWTSLGPSGVASHSANIQGAFSWD MWPAGANDMTTTPDQQWVSALAGKSYMMGVSPWFFHSASGGTDWVWRGDDLWAQRWQQVLQINPAFVEIVTWNDFAESHY IGPVYTTSEIAAGSSTYVNNMPHESWRDFLPYYIAMYKKSTFNISRDQMQYWYRTAPAAGGSTCGVVGNNPGWQTTMSPN SIVQDKVFFSALLMSPATVTVQIGNYAAVSYNGVQGINHWSQPFNGQTGSVKFSVVRNGATVK |
| 509 |
PF03400 (DDE_Tnp_IS1) |
 Estimated TM-score=0.76; easy target. |
>IS1 transposase (Length=131) VIVCAEMDEQWGYVGAKSRQRWLFYAYDRLRKTVVAHVFGERTLATLERLLSLLSAFEVVVWMTDGWPLYESRLKGKLHV ISKRYTQRIERHNLNLRQHLARLGRKSLSFSKSVELHDKVIGHYLNIKHYQ |
| 510 |
PF03382 (DUF285) |
 Estimated TM-score=0.76; easy target. |
>Mycoplasma protein of unknown function, DUF285 (Length=111) FAGKSTFNSDISRWITSQVTTMRYMFAWTAKFNADIGNWDTSRVKDMQSVFEGSSSFDRDISRWDTSSVTNMDAMFYQAV SFDSDVSGWDTANVMVFRLMFDGATAFQNNF |
| 511 |
PF02667 (SCFA_trans) |
 Estimated TM-score=0.76; trivial target. |
>Short chain fatty acid transporter (Length=447) MISRVSRFMTTFVSKYLPDPLIFAVLLTFVTFICAWGLTDSTPVDLVNMWGNGFWNLLGFGMQMALIVVTGNALATSPQI RKFLSTIASIAKTPAQGVVLVTFMGSIACIINWGFGLVVGAMFAKEVARRIKGSDYALLIACAYIAFMTWGGGFSGSMPL LAATPGNPVAHLMMSESNPQGIIPAVSTLFSGYNIFITLSLVICLPFITYMMMPKNGETKSIDPKLIAPDPTFDKKLDKD ATLAEKMEESRFLAYTIGALGYSYLGMYFYKNGFNLTINNVNLIFLITGIVLHGSPMAYMRAIINATRSTAGILVQFPFY AGVQLMMEHSGLGGLITEFFINVANKDSFPLLTFFSSAFINFAVPSGGGHWVIQGPFVIPAAQALGADLGKSTMAIAYGE QWMNMAQPFWALPALGIAGLGVRDIMGFCMTALIFTAPIFIIGLYFL |
| 512 |
PF02534 (T4SS-DNA_transf) |
 Estimated TM-score=0.76; trivial target. |
>Type IV secretory system Conjugative DNA transfer (Length=502) RWADKKDIQAAGLLPRPRTVVELVSGKHPPTSSGVYVGGWQDKDGKFHYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSW AHSAVITDLKGELWALTAGWRKKHARNKVVRFEPASAQGSACWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLESHWQ KTSQALLVGVILHALYKAKNEGTPATLPSVDGMLADPNRDVGELWMEMTTYGHVDGQNHPAVGSAARDMMDRPEEESGSV LSTAKSYLALYRDPVVARNVSKSDFRIKQLMHHDDPVSLFIVTQPNDKARLRPLVRVMVNMIVRLLADKMDFENGRPVAH YKHRLLMMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDINQLKSRETGYGHDESITSNCHVQNAYPPNRVETAEHLS KLTGTTTIVKEQITTSGRRTSALLGNVSRTFQEVQRPLLTPDECLRMPGPKKSADGSIEEAGDMVVYVAGYPAIYGKQPL YFKDPIFQARAAVPAPKVSDKL |
| 513 |
PF18869 (HEPN_RnaseLS) |
 Estimated TM-score=0.75; trivial target. |
>RnaseLS-like HEPN (Length=146) WWDYIHEDLRELLKEANLLEDKVGKWNEKFHDYSFIVFPAAKAYEGFLKTLFRDLGFISDEDYFGKRFRIGKALNPSLEH SIREEESVYDKLVNFCGGKDLADNLWETWKEGRNLLFHWFPNEKSAITFEEARLRIDKILATMDLA |
| 514 |
PF18846 (baeRF_family5) |
 Estimated TM-score=0.75; easy target. |
>Bacterial archaeo-eukaryotic release factor family 5 (Length=135) PKTGIILTQKEAIKILDTELGTLKETQMFELDLDTEDWRQHTGPHRAQPSMGSGGKSTKQEQFQERFEANRYRWYKSLAP KLDKLAKDNQWERIYIVGDKEEAADLTENMNKEVTDVVNKNMLDHEEMKVINEVV |
| 515 |
PF18712 (DUF5644) |
 Estimated TM-score=0.75; trivial target. |
>Family of unknown function (DUF5644) (Length=108) KAVLKNYEDFFKQVPYITKGEKEELEKFIQINFINPQTNPKYLGDGFFLYVKWLMKRYPTERNRLLEMISQPESGVMNFL SVAHYLYKNDDNIDHEIYELQEMLTNSK |
| 516 |
PF18154 (pPIWI_RE_REase) |
 Estimated TM-score=0.75; easy target. |
>REase associating with pPIWI_RE (Length=115) GLAEVELEDRLTDLGLDVTMWPGFDAYDLLVVFPDGHRWAVDVKDWAHPAFLGRAARPVRPEPPYDEAFWVVPVYRVRER PGYQQAFQRARPADAAVRLLTDDELTRAARQRLAE |
| 517 |
PF16760 (CBM53) |
 Estimated TM-score=0.75; easy target. |
>Starch/carbohydrate-binding module (family 53) (Length=87) VTVFYNPRDTPLNGRQQIYLMGGWNRWTHKRHFGPIAMHRPVDGGEHWQATLTVPKDAFKMDFVFADVPGGDGTYDNRGG FDYHLPV |
| 518 |
PF16529 (Ge1_WD40) |
 Estimated TM-score=0.75; easy target. |
>WD40 region of Ge1, enhancer of mRNA-decapping protein (Length=331) GSSKVKLKNIVDFTWEHHLYTGQLLAVHVSGKYLAYGIKVGTGGGLVRVVHKELEHRVLLRGMRGAIQDLAFAHVSNVVL ACIDHSGSLFIYTIEPTQSELLCSLVIQVDAEDVPSISHRVIWCPYIPEDDASDGDEVSKLLVLTRGSKVELWSIATVAS KFRSIDATNAAAKDCGGMIEIEQHSGTIVEATFSPDGTALATASTDGEVKFFQVFLHNNSHNGQPQPRCLHQWRPHAGRP ISCLFFLDDHKNYHPDVQFWRFAVTGCDNNSELKVWSCEVWSCLQTIKFAPTPTTGKLPVLKAGLDLSAGYLLLSDIYNK VLYILSLSKDR |
| 519 |
PF16244 (DUF4901) |
 Estimated TM-score=0.75; trivial target. |
>Domain of unknown function (DUF4901) (Length=223) REKASSIVSIPENYHLVVEDNIADERLFIWEDPQHPEANIEITLDLQTGELIKLSIDKNQDNACSGADCAIPSADQAKRI ADEFIKQHLPDLRGEYTWIHIEAGRRHMHISYRQETGGLPLPFSGCNCTVDSGGNITDFKHYGCKNKPAWPASITDVKTV KQQIVEDLRMKLCIMRLYPSMHEVEDLEYRLVYEPVPCLRSFNAITGRDLFEPDHYLMPRSHP |
| 520 |
PF16173 (DUF4874) |
 Estimated TM-score=0.75; easy target. |
>Domain of unknown function (DUF4874) (Length=148) NPERGFEFDFDPSLPQYEAAKSREAGYSIAFWNIDLEAFRSAPLDAAFLDGLEQGFVQVRQAGIKVQMNFAYGTTGQDAP KETVLGHIEQLKPLLTGNADVIVSAQAGFIGAFGAWTSSSNDLADPATARLIVQALLDALPDNRMIQI |
| 521 |
PF15801 (zf-C6H2) |
 Estimated TM-score=0.75; easy target. |
>zf-MYND-like zinc finger, mRNA-binding (Length=46) CMGDDCDNDAGSLQCPTCLKLGIKDSYFCSQDCFKRNWSTHKALHK |
| 522 |
PF15608 (PELOTA_1) |
 Estimated TM-score=0.75; easy target. |
>PELOTA RNA binding domain (Length=81) WQGLQDITSIQKHFGIENINLIKPGVGETTRVLLRRVPWKILIRDENNPDLKHILLLAKEKNVPVELYKDMTYSCCGLIK P |
| 523 |
PF15413 (PH_11) |
 Estimated TM-score=0.75; easy target. |
>Pleckstrin homology domain (Length=102) FLTKRGHVVTNWKTRYFVLRPHAILCYYADESMGKKLGQVHLVKVAPWEYVSSTSTNSILKSTRGSDDMKFGFMFFTTKR VAYYVYTNDKGERQKWLVALQD |
| 524 |
PF14885 (GHL15) |
 Estimated TM-score=0.75; easy target. |
>Hypothetical glycosyl hydrolase family 15 (Length=284) KALNPAITVLVYKDLSSTRSYETATLHGGLLPTGVDFAATEREHPDWFATDTGGRRVEWAPYPQHWQMAVWNADYQREWT TAVTTEAVRDGWDGVFADNDFASLGFYSDAVLAGTSSRAETDRLLRDGLDRMVAVAGSALAAQGKLFIPNLSEARLYPGR WTEHARFGGAMEENFAQYAGDELLTWQGTQWEEMVRTSSDGKHLNLLVTKTAGVPESSPGAAGRAGFAGAALLAGDRACW TGPDAGDYSRPAFSGYQSLALGEPRGPAVQQDSGVWTREFTGGW |
| 525 |
PF14667 (Polysacc_synt_C) |
 Estimated TM-score=0.75; easy target. |
>Polysaccharide biosynthesis C-terminal domain (Length=140) LSILAPAAVFLGLHQTTSGVLQGLGRTHLPVKNLLVGVVVKLGLNYYLTGLPVFGIKGPALGTLAGFCVSSVLNLADLHR VTDWRLSRQVLSKPAVAATIMAVGVYLTFGFLKLTYNMHLATVLTVLFGAGIYFVVLALI |
| 526 |
PF14634 (zf-RING_5) |
 Estimated TM-score=0.75; easy target. |
>zinc-RING finger domain (Length=43) CSVCLEDVRGGETVRRLPACGHLYHAACIDAWLRSRTTCPLCR |
| 527 |
PF14568 (SUKH_6) |
 Estimated TM-score=0.75; easy target. |
>SMI1-KNR4 cell-wall (Length=120) EEEIHKVEEELGISFPGDYKEYVKKCNGGFPNRDEFYFDDGFYSVSLRNLISLHHDMLATYEDVKSYIPEKVIPFMETRS GDFLCFDFRESLTSPPIVYMNHEEEGDESLSALCDTFTDL |
| 528 |
PF14196 (ATC_hydrolase) |
 Estimated TM-score=0.75; easy target. |
>L-2-amino-thiazoline-4-carboxylic acid hydrolase (Length=143) RRIEAEIIKPIYDAMSRHFGKEKAQAVLKEAIAADAIRTGKEMAEKQQDKGRATLSNFAAMQYLWEQDDALKVDILASDH QHLDYNVTRCRYAEMYREMGLGEIGSILSCTRDASFIEGYNPNIEMSRSQTIMKGADCCNFRY |
| 529 |
PF13835 (DUF4194) |
 Estimated TM-score=0.75; easy target. |
>Domain of unknown function (DUF4194) (Length=170) VSLLRQGVILSSQKAKLFELLCRYQSAVRKHLSEVYLRLVLDEKAGVAFIAGFQNEEDTESLGIDDEEVVSLISRRTLSL YDTLLLLVLRKHYQDRETSGEQRVIIDIERIESHLTPFLPLTNSTKSDRRKLKGALDKMVTKKILSSVRGSEDRFEITPV IRYVVSAEFL |
| 530 |
PF13750 (Big_3_3) |
 Estimated TM-score=0.75; easy target. |
>Bacterial Ig-like domain (group 3) (Length=157) GNYTYEFNMKEVPEGSYNVVINAQDTFNNTGNRPYQTVVVDNTAPSVSISYEGKPISKDVTVYGLENVRIQLTDALTKPS LSRMTLRGGPVSDAVELSWVNLGNNLYAPNYPKIFPSLNEGETYTLTVQAKDEMNNVKESSVEFNYLPNNLVRLENL |
| 531 |
PF13643 (DUF4145) |
 Estimated TM-score=0.75; easy target. |
>Domain of unknown function (DUF4145) (Length=88) EAKMCFSVNCYRACAVMARRSIQSACIDKGADKNSRLEKQIEELTQGGIITKDIKEWADVLRWVGNDAAHPESQEVTKED AEDILKLA |
| 532 |
PF13632 (Glyco_trans_2_3) |
 Estimated TM-score=0.75; easy target. |
>Glycosyl transferase family group 2 (Length=206) YILIIDSDTRVPVDCLLDAVSEMEQSPDVAIMQFASGVMQVVHTFFENGITFFTNLIYTAIKYTVSNGDVAPFVGHNAIL RWSAIQEIGYFDEDGYEKFWSESHVSEDFEMSLKLQCNGYIIRLAAWAGEGFKEGVSLTVYDELARWEKYAYGCNELLFE PIRTWLWRGPFTPLFRKFLFSNIRMTSKITIISYIGTYYAIGAAWI |
| 533 |
PF13444 (Acetyltransf_5) |
 Estimated TM-score=0.75; trivial target. |
>Acetyltransferase (GNAT) domain (Length=102) LRYKVFAEEMGAKMPGQEERIDQDRYDRYCDHLLVRDTDTNEVVGTYRILSPDQAKKAGGYYSEGEFDLSRLDNLRSSMV EVGRSCVHPDYRGGGVITLLWA |
| 534 |
PF13349 (DUF4097) |
 Estimated TM-score=0.75; easy target. |
>Putative adhesin (Length=243) IDSMVIESDSKDVRIIAEERSDISAGISGDSGKLFVTENNRKLELTVKEKEFQFLNGFNRSTLIVRLPYDYKGDLAVRTS SGDVSVAGNDHLALSRLNTVSASGNASVTDVRLQDLKVKASSGDVTISNTVSKTSGIDLASGDANLVHVSGSLDVKMTSG DFNAVLTKITGPVSVTLTSGDANLSLPKNGSFTVKAKSASGDVSSPYSFADKAHKEQHHITGTQGSGRHPIDIKTDSGDL AIR |
| 535 |
PF13303 (PTS_EIIC_2) |
 Estimated TM-score=0.75; easy target. |
>Phosphotransferase system, EIIC (Length=326) IKVLNGMALGLFSSLIIGLILKQIGIFLNIKLLVQFGQVAQFMMGPAIGAGVAYSLGASPLGIFASTVAGAIGGQTITVG EGGTFLANIGEPVGALLSSLLGVEASKIIQGKTKVDIVLVPAVTIIVGGVSGFYLAPFVAEFMTLLGKMINLATQQQPVI MGILVSVLMGIILTLPISSAAIGIALGLDGLAAGAAIVGCATNMIGFAVASFRENGVGGLIAQGLGTSMLQIPNIIKNPR IFIPPILTSAILGPISTVVFKMEGNKIGSGMGTSGLVGQFSTIEVMGSRGIFGIILLHFVLPATITFLISEYMRKKKWIK YGDMKL |
| 536 |
PF12965 (DUF3854) |
 Estimated TM-score=0.75; easy target. |
>Domain of unknown function (DUF3854) (Length=129) FWQWIINHPSIPICITEGAKKAGALLTAGYAAIALPGINNGYRTPQDEFGNRIGKSRLIPQLEKLAISGRKIYIAFDQDS KPNTIKNVNTAIRKLGYLFAQAGCQVKVITWSPKWGKGVDDFISNKGEK |
| 537 |
PF12834 (Phage_int_SAM_2) |
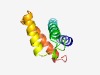 Estimated TM-score=0.75; easy target. |
>Phage integrase, N-terminal (Length=91) MSRLSREMKMLAKQAGGSHKTVHDRIRIMDRFSRHLLALNIQVRDVKHLKAKHIESYITDRFSQGIAIRSLHNEMAALRT VFRFAGRDKIV |
| 538 |
PF12700 (HlyD_2) |
 Estimated TM-score=0.75; easy target. |
>HlyD family secretion protein (Length=411) MGRKKRRAQRKIFNYLIFMGIFSYIVFNLVMSIINSSIQTHTVKKGQIVEYLQKDGYAVRQESVFYTSIGGIPKYLHNEG DRISINDYVCEIQTNEADQMLRQEADTIARRLEVVSSGGALAGVTPEKLDRGIEVKRKEIKNSITMGAFDSVERLKKELL FLMDEKSILYGSLSGQDSGMLQKKADAIQAKIQSSSRVIPTTISGVLSFQADGFEGSMTPAEIGKIDSDFLKTLSNNNVI KKDGALEKGDPAFKIANNHFMYLLFIVNEKENKELKVGQGIYVKSGKETVYCVIKDMYKDSQGKSIMVFKTASVPAEFLD KRILGCSIVTKNVSGIKIPKAAIATLDGKQGVYVISETGQATFKEIKCQIGENEEHIILDYKRIEAEKIDTAKLYDEIIM NPKRVKEGQKI |
| 539 |
PF12594 (DUF3764) |
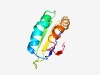 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF3764) (Length=84) ETSVFTFRISVPFEQWAAIFDSEDIRKMHDENGIQPLYRGVSEDDPMKVIVIHQAEAGVAKAFFEASREPIEAGGHIWDS TEIT |
| 540 |
PF12386 (Peptidase_C71) |
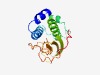 Estimated TM-score=0.75; easy target. |
>Pseudomurein endo-isopeptidase Pei (Length=141) LKGHWTTKVIEKIGTFHDATSLYERVKKTCKYKYYYNDQVPNHVAVMRMTTSGINCTDACQLFSKVLEEMGYEVKIEHVR VKCNDGKWYGHYLLRVGGFELKDGTIWDYVSATKTGRPLGVPCCTAGFQHLGWGIVGPVYD |
| 541 |
PF12085 (DUF3562) |
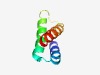 Estimated TM-score=0.75; trivial target. |
>Protein of unknown function (DUF3562) (Length=59) EDSQKIVHEIAESTDAPEEVVSQMYTDAVQAYERDARILDYVPLFAAKRVRETLRSRTA |
| 542 |
PF11325 (DUF3127) |
 Estimated TM-score=0.75; easy target. |
>Domain of unknown function (DUF3127) (Length=84) EISGKIIAILPLQSGTGRNGSEWKKQDYVVETHDQYPKKMCFNLWGDRIDQFAVQMGEEVTVSFDIDCREWNGKWFNDIR AWKI |
| 543 |
PF11253 (DUF3052) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF3052) (Length=125) AARLGLQPGQVVQELGYDEDCDEELREAIEELTGNELVDEDYDDVVDVVLLWWRDGDGDLVDALVDALTPLADGGVIWLL TPKAGRDGHVEPSDIGEAAPTAGLSPTKSINAAKDWSGTRLVAPK |
| 544 |
PF11017 (DUF2855) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF2855) (Length=316) FALTSNNITYAAFGDAMSYWQFFPTGEEGWGSIPVWGFGSVVQSLHPGVAVGERLYGYWPMSSTAVLSPDRLTSARFTDA AAHRAALPAVYNQYFRCNADPLYTADTEDLQALLRPLFITSWLIDDFMADNDFFGASTMLLSSASSKTAYGTAFQLHQRK GIEVIGLTSPANVAFCESLGCYDRVVTYDALDTIAADTPAVYVDFAGNGELRNAIHARFANLKYSCSIGGTHVEQLAAQG AGKDLPGPRATLFFAPAQIKKRTAEWGADEFGRRMVAAWHDFLATVTDAKTPWLRAEHHHGGEAVQAAYARVLAGK |
| 545 |
PF11008 (DUF2846) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF2846) (Length=85) AGKAGVYMYRNSFMGQALKKDIYMDGKRLGESANKTYFYNQVDPGEHTVSTESEFSDNDFKFTVQSGMNYFIRQYIKMGV FVGGA |
| 546 |
PF10963 (Phage_TAC_10) |
 Estimated TM-score=0.75; trivial target. |
>Phage tail assembly chaperone (Length=82) ITLEIAGKDIRFEPNMTAYNGFINEMAMDNKVAPAHTYLRRIVVKDDKEALDELLQMPGAALQIATSINDQYAPKLEISV KN |
| 547 |
PF10962 (DUF2764) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF2764) (Length=276) MGKYYCLIAGLPNIALDDSKLTYSISQFRQELDGILTKADKKLIDLFFLKFDNKNLIAHAKQPDRDPDPRGRITYDEFDA LYKALKEEEKPPKNDRIPPYFKEFFKLYLEAQGKENKQEIPWEDRLAALYYAYAMKNANKFVSDWFCLNLNINNMLTAIT CRKYGFDKAGYIVGDNEVAQALRTSNARDFGLGDTVDYLPDLWRIAEETDLMVREKKVDLLKWEWLEEHTFFKPFDIESV FAYLLKLEMIERWVMLDKATGEKTFREIVGAMKKGS |
| 548 |
PF10665 (Minor_capsid_1) |
 Estimated TM-score=0.75; easy target. |
>Minor capsid protein (Length=106) PIEFLVDSFEYKDYLGEGDWNKPVYAEPILIENCRIDKTSQYTATTSGKQLLFKAVIFCYPGLTSPIGPFKEQGLVIYDG QEHVITSAIPIKEAYSDDLYSYELEV |
| 549 |
PF10474 (DUF2451) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function C-terminus (DUF2451) (Length=236) MFGLSERIIATESLVFLAGQFESLHAHLEAMIPANKRAFLSQFYSQTVSTAQELRRPVYRSVASRVIDYDQTLQLMTGVR WDIRDIMSQHNAYVDILVRELQVFSMRLTQLANKQIPVPQETRAVLWDHCIRLINRTFVEGFASAKKCSNEGRALMQLDF QQYLMKLEKLTDIRPIPDREFVETYVKAFYLTENEMERWIREHKEYTAKQLTSLVMCGVGSHVTKKGKQKLLAIIE |
| 550 |
PF10335 (DUF294_C) |
 Estimated TM-score=0.75; easy target. |
>Putative nucleotidyltransferase substrate binding domain (Length=144) IFLACLAANALEHTPPLGFFRQFVLATHGEHEDTLDLKHDGLVPIIDLARLHALAHGVPAVNTRRRLRQLAEQGALNDGD AADLQDAFAFIAQIRLRHQVRQIADGEAPDNYVSPDALSAFDRRHLKDAFRIVKTAQAAVEQRY |
| 551 |
PF10328 (7TM_GPCR_Srx) |
 Estimated TM-score=0.75; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Srx (Length=260) LTSIFGSTVNFYLFYKFLKRDGKANGFQKICLVKTLPNFMICFAFLLWVVPVTALDLTYSQLPYRLNSIVGSLAGSWAYL FTPFLQVSLSCNRFYVLYFPFGIKILKRVPVTNISIGSSILLVTCVCSVGLQETCGYVYDPNYFTWRPEGLPCADKLSEV ILFMIFSITFTSNAFNVGTGARLLVNKMVGMNREEASRRRKRWMIMFTQSVIQDCLHLVDIINATYIWKLSEELWFQFLF LTLSFIIIYTLDGVVMFIFN |
| 552 |
PF10324 (7TM_GPCR_Srw) |
 Estimated TM-score=0.75; trivial target. |
>Serpentine type 7TM GPCR chemoreceptor Srw (Length=320) KYILAIFGVVINSFHLFILTRKSMRSNCINLLMIAVSICDIYNMMYIVHELNFVFDSKPPESYFQVFLNHIMNILNGLLR RLSSWLGILMAAIRYFAIKYPMSPTIDVISRPLFAGKSTFLTLLISALVSFLYWVHFSVQPVFLWKPVEELVHLICLEII LNLCRCGFIDNYTVAIYGAELSEFIQEYPSILGICQSADGLSQVIPAVTLPVLTFLLIKQLDITEKNRSNFLKSQKNNEN SKTDTTKMIMMMTIASTLAEGPIGVLPIVEIFACPFLMVISANLMVIFGAVVALNTSTHCFVCCAISSQYRKTVTQVFLW |
| 553 |
PF10118 (Metal_hydrol) |
 Estimated TM-score=0.75; easy target. |
>Predicted metal-dependent hydrolase (Length=241) NLDFGIDESLPKYWWGGNPYRTRLFDAVQCTFPDGERYFIKSVVAFRQKVSDPVLLADVRAFSRQEGQHGIAHQQYNALV EGRGVPLTKIIQPAIDRFNRFSARLSPEFNVAITAAAEHFTAMMAECFFAKKATMAEVEPHMKAMLAWHAIEEMEHRAVA FDVMKKIAKVGYFKRVWALIYLQGGMYEVMVRMPHAMLKADGFTRLQRWRMQIRNIPWLLRLFGPLNRKALSYFKPGFHP L |
| 554 |
PF10116 (Host_attach) |
 Estimated TM-score=0.75; easy target. |
>Protein required for attachment to host cells (Length=136) WVIVADGEKALFLRNEGDALYPNLQVFRQMNDDNPPTREQGTDRPGRMPDAGPNHRSAVQETDWHRIEKERFAAEIAERL YKLAHRHEFKKLIVVAPPTVLGDMRKEFHKEVSDRIIAEVPKTLTNHPVDEIEKHL |
| 555 |
PF10108 (DNA_pol_B_exo2) |
 Estimated TM-score=0.75; easy target. |
>Predicted 3'-5' exonuclease related to the exonuclease domain of PolB (Length=210) SDFLPHHLQRVAAISCVLREGDNFKVWTLGEPDEDEGSIIKRFFDGIEKYTPQIVSWNGSGFDLPVLHYRGLIHGIQAPR YWDQGEDDKDFKWNNYISRYHSRHLDLMDLLAMYTGRANAPLDELAKLIGFPGKLGMDGSKVWDAYQQGQLAEIRNYCET DVVNTWLVFARFQLMRGQFSKARYEQELELVRSTLKKSDEKHWQEFLAQW |
| 556 |
PF09983 (DUF2220) |
 Estimated TM-score=0.75; easy target. |
>Uncharacterized protein conserved in bacteria C-term(DUF2220) (Length=179) GFRDKPLRVRFRILDPKLALLPTNTDQDITLTQATFARLEIPVTKIFITENEINFLAFPEVPEAMVIFGAGYGFENMASV EWMRDRVIHYWGDIDTHGMAILNQLRRFFPQAASLLMDHETLMEHQPLWGAEPSPETGTLTRLTAQEGALYDQLRRNELG SRIRLEQEKIGFEWLVEAL |
| 557 |
PF09664 (DUF2399) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function C-terminus (DUF2399) (Length=154) VLTLRQIRDSMAAFDTADTGRYIVSVCENAAVVAAAADRLGPDCPPLVCTAGQPSAAVIGLLDLLTAAGGRLRYHGDFDW PGLTIAGGLYRRYRWQPWRFTAADYASCEPPVATAPLTGIPVQAEWDTELTVAMSRRGIQIEEELTLPQLLADL |
| 558 |
PF08928 (DUF1910) |
 Estimated TM-score=0.75; easy target. |
>Domain of unknown function (DUF1910) (Length=123) RDPLCSESYLLETIEFDKEEICERKKKIIVLKDDMEKGIQRYPKDNQSIIYATYRGMFMYNTEILIAKYSLGSHPDEMIE DYLNGIEYLENVGEEKVWYIDLLWMLSLGILLEVDKQDLKRLA |
| 559 |
PF08643 (DUF1776) |
 Estimated TM-score=0.75; easy target. |
>Fungal family of unknown function (DUF1776) (Length=314) GRMDVVVIAGSPALPLTRSLSLDLERKGFVVFVVANSHEDEIMIQHLARPDIRPLSIDITDPPSTGTSIDTFARYLQSTH AAIPNGKRHHLLLKAVILIPSLNYQTSPIATIPPSSFADLFNTHLLQPILTIQAFLPLLTARLSPPSDKERPSPKVLVFT PSIISSINPPFHAPEATICSALSAFTEVLTGELRPLGIPVTHMQLGTFDFAGFTPAANSRLLQPHGLLTAPEDAEALPWP DSARHAYGKNFVSQSGSAIGAGRIRGMRGSSLKELHNAVFDVIDGSLTNGTVQVGLGANIYGFVGRWVPKGLVS |
| 560 |
PF07961 (MBA1) |
 Estimated TM-score=0.75; trivial target. |
>MBA1-like protein (Length=237) KQDTKKIQDFNPRHLGVASELYVPASLSRLPSVITSPKLWFDNVIRRIYMLGLNTVQIGIFRFQTGIKPAFLLWKNKAIE TYVQVNTNFASKSVEKLKPEVSVWVEEALNARSKQIPDNVKLDWQLLKFNDVPKLISIQPMMIPGRPLEHIQLLYRFNTK QRLVKLNKTNNKVEKLDRDVVDYMVFLCDATTNDLILMGSVFESKPDAKLPKNYDDNTEKAVRRMKECGDIYRVPPS |
| 561 |
PF07617 (DUF1579) |
 Estimated TM-score=0.75; trivial target. |
>Protein of unknown function (DUF1579) (Length=155) QPQKEHQWLQKLVGEWTYEIEAMMAPDQPPVKSTGTETVRSLGGLWILAEGQGEMGGCGSATTLMTLGYDPQKQRYVGTW IGSMMTYLWIYDGELDASETALTLNSDGPVMTGEEKLGKYKDVIEFKSDDHRVLTSHMLSDDGQWQQFMTAHYRR |
| 562 |
PF07606 (DUF1569) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF1569) (Length=147) TNKFDLKISSALNKLELLSDKNLVQTGKWELYKIFTHCAQSVEYSMSKFPEHKSSFFKNTIGKLAFSLFSSKGKMTHSLS EPIPGSPLIDSTLDTTEALNHLKKSLTDFDNYKGELAPHFAYGELTKSDYEIAHVMHLYNHLQEVKS |
| 563 |
PF07505 (DUF5131) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF5131) (Length=238) SKIEWTEQTWNPTTGCTKVSPGCTHCYAETMAYRLQAMGAKGYENAFKLRVIPERLNDPLKRKKPTVYFVNSMSDVFHEG VPFEYIDQIFETIAKTPHHTYQILTKRADRMAEYFANRDVPQNAWLGVSVEDKKYGVPRINYLRTVNAHIRFLSVEPLLE DVGELNLKDIHWVIVGGESGRKARPMKPEWAESIRDQCEEAGVAFFFKQWGGWGADGVKRAKKHNGRELHGRTWDQMP |
| 564 |
PF07281 (INSIG) |
 Estimated TM-score=0.75; trivial target. |
>Insulin-induced protein (INSIG) (Length=216) AVLFLLGMGYGALVTRLHGKHQLAALPAEGTSGPGFNWKYLTLWGICGVLLGALLPWFDGVWEEAFGGDTAVVDGGRDET LDNANRSSTDWALGVRSIGAFVGIVFAIRRLPWASTLQLSLTLALVNPFLWYLIDRSKPGFLLSAAVGLIGSAVLMGVDP EMVPTPSASSFRNETLSDSLTFGGLASQKTMETGIWMLSVLFCSCVCFGNIGRRLA |
| 565 |
PF07250 (Glyoxal_oxid_N) |
 Estimated TM-score=0.75; easy target. |
>Glyoxal oxidase N-terminus (Length=236) MHMQLLPNDRVVMYDRTDFGISNISLPDGKCRPNSTDCSAHSVEYDIASNSIRPLMVLTNVWCSSGTLMPDGRLIQTGGW DDGYRVVRIYKSCDSCDWQEIPNGLNQQRWYATNHILPDGKQIIIGGRQAFNYEFYPKMSTTENSPSLPFLVQTNDPNVE NNLYPFVFLNPDGNLFVFANNRAILFDYSKNQVLKTYPTIPGGQPRNYPSTGSAVLLPLMIKNGTVNTVEVLVCGG |
| 566 |
PF07014 (Hs1pro-1_C) |
 Estimated TM-score=0.75; trivial target. |
>Hs1pro-1 protein C-terminus (Length=259) MRRCPYTLGLGEPNLNGKPNLEYDAVCKPSELHSLKRNPYDHIHNHENQTVYTAHQILESWIHVSQQLLKRITERIESQK FEKAASDCYLIERIWKLLSEIEDLHLLMDPEDFLRLKNQLNVKSFNETKPFCFRSKALVEIAKQCKDLKHKVPYILGVEV DPMGGPRIQEAAMKLYSEKREFEKIHLLQALQAIESAMKRFFFAYKQLLVVVMGSLEANANRVVVSSDSCDSLSQIFLEP TYFPSLDAAKTFLGDFWNH |
| 567 |
PF06687 (SUR7) |
 Estimated TM-score=0.75; easy target. |
>SUR7/PalI family (Length=211) FPCALTIGSLICLVIVGLGCTRAHKAENLYFLRFIQALDEVNNTTGLNDFYSIGLWSYCTGNITDNGTYKTTYCSKPRGG FWFDPIDTWGLNGTNTNDFLPGKLKKSLNMYRNVSLWMAIIYLIAVIATVLEIMTGLMAVGGNRLVSCFAWLLAGVSLLF TTAMSITSTTIFATLAATVKTVLKPHGITGHMGRHIYAAAWLAVALSFLAS |
| 568 |
PF06525 (SoxE) |
 Estimated TM-score=0.75; easy target. |
>Sulfocyanin (SoxE) domain (Length=149) SGAYALPYNSNNKTVFIYLVVSSSSSNLFNFNGTSDGSLKIYVPAGWNVMVILKNTESLPHNANIVQNNTPIPNSINISS DGKIILYVGDGPSNYYSSGVSSGNEASGMLENIPAGYYWIACGLPGHAKNGMWVDLIVSSTISTPYAII |
| 569 |
PF06356 (DUF1064) |
 Estimated TM-score=0.75; easy target. |
>Protein of unknown function (DUF1064) (Length=116) RSKYGAKKIVIDGITFDSKDEGKYYEYLKKLKSQEKILNFELQPKYELQPGFKKNGKTYRAITYAPDFLIYHLDGTEELI DVKGMSTQQGEMRRKMFDYKYPDLKLTWVARSLKYS |
| 570 |
PF06347 (SH3_4) |
 Estimated TM-score=0.75; trivial target. |
>Bacterial SH3 domain (Length=52) SSLNVRSNASTSGSIIAKLSSGSKVSVQSESGGWAKVTVNGKTGYVSSAYLS |
| 571 |
PF05992 (SbmA_BacA) |
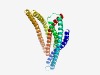 Estimated TM-score=0.75; easy target. |
>SbmA/BacA-like family (Length=314) PHPWQRWSILGSALILFATYFNVQVSVAINTWYGPFYDMVQKGLTTPGAVSAAEFYWGLADFAGIAFLAITIGVLNLFFV SHYIFRWRTAMNDYYMSHWPKLRHIEGAAQRVQEDTMRFSTTLEQLGVSLVKSVMTLIAFLPVLFTFSQKVSTLPIIGYV PHALVWAAVIWALFGTGFLALVGIKLPGLEFNNQRVEAAYRKELVYGEDHADRAEPITMRELFKNVRHNYFRLYFHYVYF NVARIFYLQVDNIFPLIILIPSIVAGKLTLGLMSQITNVFDQVRGSFQYLINSWTTIIELLSIYKRLRAFEAAI |
| 572 |
PF05977 (MFS_3) |
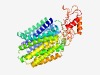 Estimated TM-score=0.75; easy target. |
>Transmembrane secretion effector (Length=521) SPLAPFRHDIFRTIWIASLASNFGGLIQAVGAAWLMTSISQSVNMVALVQASTSLPIMLFSLVSGALADNFDRRRIMLVA QSFMLAVSALLTVCAYYGIVTPWLLLIFTFLLGCGTALNNPSWQASVGDMVPRDDLPAAVALNSMGFNLTRSVGPAIGGA IVAAAGAAAAFAANTLSYFAILFALARWKPVAPENRLPRETLGRAVSAGLRYVAMSPNIRKVLVRGFAFGLSASAILALL PLVARDLVGGGPLTYGVMLGAFGVGAIGGALLSARLREFLTSEAIVRYAFAGFAFSALVTAISPQAWLTCLVLAVSGACW VLALSLFNTTVQLSTPRWVVGRALSLYQTMTFGGIAGGSWLWGVTAEQYGAANALIGSCLLMLAGAAIGLRFALPEFKSL NLDPLNRFNEPLLELDLKPRSGPIVVMIDYEIADEDIPEFLKTMAERRRIRIRDGAGHWALMRDLENPTTWTETYHVPTW VEYVRHNQRRTQADAAVGDKLTAFHRGPNPPRVHRMIERQT |
| 573 |
PF05912 (DUF870) |
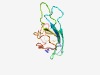 Estimated TM-score=0.75; easy target. |
>Caenorhabditis elegans protein of unknown function (DUF870) (Length=110) EMLIRCDLSIAHYCGKLVYYEMDYQGQHDVFKTHQFCSDYEYQQFQYEKFWPGGDPGKEYEFSFTLEHNCTLTGQRLCIR PHQYVRKYIDGEQYVEFYVKAYNRGENFDC |
| 574 |
PF05631 (MFS_5) |
 Estimated TM-score=0.75; trivial target. |
>Sugar-tranasporters, 12 TM (Length=354) FYYLVFGGLAAVVAGLELGKSGKDRVATTPAFNAFKNNYILVYSLMMSGDWLQGPYVYYLYSQYGFDKGDIGRLFIAGFG SSMLFGTIVGSLADKQGRKRACITYCISYILSCITKHSPEYKILMIGRVLGGIATSLLFSAFESWLVAEHNKRGFDPQWL SITFSKAIFLGNGLVAIIAGLFANLLADNLGFGPVAPFDAAACFLAIGMAIILSSWSENYGDTSDNKDLIAQFKVAAKAI ASDEKIALLGAIQSLFEGSMYTFVFLWTPALSPNDEEIPHGFIFATFMLSSMLGSSIASRLLARKLKVEGYMQIVFSISA FTLFLPVVTNFLVPPSSVKGGGISFGGCLQLLGF |
| 575 |
PF05576 (Peptidase_S37) |
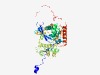 Estimated TM-score=0.75; trivial target. |
>PS-10 peptidase S37 (Length=448) AEPKAVDIKDRLLSIPGMSLIEEKPYTGYRFFVLNYTQPVDHRHPSKGTFQQRITVLHKDVNRPTVFYTGGYNVSTNPSR REPTQIVDGNQVSMEYRYFTPSRPAPADWSKLDIWQAASDQHRIFKALKPLYSKNWISTGGSKGGMTATYYERFYPRDMD GVVAYVAPNDVVNKEDSAYDRFFARVGTDECRDKLNGVQREALVRRAPLEKKYAAYAAENGYTFDTIGSLDRAYEAVVLD YVWGFWQYSTLADCADIPADAKNATDDAIWGSVDAISGFSAYTDQGLETYTPYYYQAGTQLGAPTIHFPHIEKKYIRYGY QPPRNFVPRSIPMKFEPWAMRDVDTWVRHNARHMLFVYGENDPWGAERFRLGHGARDSYVLTAPGMNHGANVAGLVPDQK ARATARILDWAGVAPAKVQENPSAARPLATFDARLDQRDVEREPALRP |
| 576 |
PF04586 (Peptidase_S78) |
 Estimated TM-score=0.75; easy target. |
>Caudovirus prohead serine protease (Length=159) NLKTRSEEDSQTQIVTGYAAVFNSPTELWEGLNEVIKPGAFSRALSNSDVRCLFDHDWGKVLGRTRSGTLKLEEDDKGLR FEVELPNTTVANDLIQSMSRGDINQCSFGFYPTEETWDYRSDPVLRTIHEVELYEVSIVSLPAYEDTEAALSRNKQEMK |
| 577 |
PF04182 (B-block_TFIIIC) |
 Estimated TM-score=0.75; easy target. |
>B-block binding subunit of TFIIIC (Length=70) NPFQLLCEIARYGASGIQAPDLCKATGQDPRSLTPRLKKLEELGYIRKKNVYNDKTRQHTSLCVHSKFAE |
| 578 |
PF03982 (DAGAT) |
 Estimated TM-score=0.75; easy target. |
>Diacylglycerol acyltransferase (Length=293) LIFTFQWHILLAYSLWYLYDRKSPQNGGYRVNFPQRWRIQKWFANYFPITLHRTAELPADRNYIVGCHPHGIIGMAVTSN FASEGTDKSKVFPGIRFSVCTLASNFQVMFTREFLLLCGLIDCSKESIANALAEQKTGRAIVLAVGGAEEALEARPGAHK LKLLTRKGFVKQAIRNGASLVPVYSFGENDLYNQLDNPEGSLVRKIQTLSKRWLGISLPLFYGRGVLQLNFGFLPHRRPI HTVVGAPIAVSRIPEPTDEEVDNVHRQYCKALEELFEQHKTRFGVSKEAKLIL |
| 579 |
PF03942 (DTW) |
 Estimated TM-score=0.75; easy target. |
>DTW domain (Length=182) RPLCPRCQRPETQCYCSALKEETACMDIIILQHPRESRHPLNTARILELGIGNCQVWVGEDFSDHTHLQTTLAEKDCYLL FPGANAHASQDILRTAMPEALIILDGTWRKAKKIYYRNPQLQQLPSIELTELAISDYRIRKTPGAGALSTVEAAVTLLRQ ASQDSKAHQQLLDTFSLMINQQ |
| 580 |
PF03901 (Glyco_transf_22) |
 Estimated TM-score=0.75; easy target. |
>Alg9-like mannosyltransferase family (Length=414) AFKCLLSARLCAALLSNISDCDETFNYWEPTHYLIYGKGFQTWEYSPVYAIRSYAYLLLHAWPAAFHARILQTNKILVFY FLRCLLAFVSCICELYFYKAVCKKFGLHVSRMMLAFLVLSTGMFCSSSAFLPSSFCMYTTLIAMTGWYMDKTPIAVLGVA AGTIVGWPFSAALGLPIAFDLLVMKHRWKSFLRWSLVALILFLVPVVVIDSYYYGKLVIAPLNIVLYNVFTSHGPDLYGT EPWYFYLINGFLNFNVAFALALLVLPLTSLMEYLLQRFHVQNLGHPYWLTLAPMYIWFIIFFIQPHKEERFLFPVYPLIC LCGAVALSALQQYRLEHYTVTSSWLALGTVFLFGLLSFSRSVALFRGYHGPLDLYPEFYRIATDPTIHTVPEGRPVNVCV GKEWYRFPSSFLLP |
| 581 |
PF03354 (Terminase_1) |
 Estimated TM-score=0.75; easy target. |
>Phage Terminase (Length=478) EFQKFIVGSLYGWRRGQYRMFTKAYISMARKQGKSLIVSGMSVNELLFGQYPKFNRQIYVASSTYKQAQTIFKMASQQVN LMRSKSKFIREKTDVRKTDIEDVLSSSVFAPLSNNPDAVDGKDPTVVILDELASMPDDEMYSRFKTGMTLQKNPLTLLVS TAGDNLNSQMYQEYKYIKRILNEEVRADNYFVYCAEMDSQEEVQDETKWIKAMPLLESKEHRKTILQNVKADIQDELEKG TSYHKILIKNFNLWQAQREDSLLDISDWEQVITPMPNINGKDVYIGVDLSRLDDLTSVGFIFPNDDKKVFLHSHSFIGLR TNLEQKSKRDKINYELAIERGEAETTQSDSGMIDYKQVIDFIVKFITTHDLNVQAVCYDPWNAQSFITTIESMALDWPLI EVGQSFKALSQSIKEFRMWVADERIQHNDNMLLTTSVNNAVLIRDGEDNVKINKKMNRQKIDPIISIITAFTEARMHE |
| 582 |
PF03215 (Rad17) |
 Estimated TM-score=0.75; easy target. |
>Rad17 P-loop domain (Length=186) YLSENEPWVDKYKPETQHELAVHKKKIEEVETWLKAQVLERQPKQGGSILLITGPPGCGKTTTIKILSKEHGIQVQEWIN PVLPDFQKDDFKGMFNTESSFHMFPYQSQIAVFKEFLLRATKYNKLQMLGDDLRTDKKIILVEDLPNQFYRDSHTLHEVL RKYVRIGRCPLIFIISDSLSGDNNQR |
| 583 |
PF03090 (Replicase) |
 Estimated TM-score=0.75; easy target. |
>Replicase family (Length=128) RIRPREYAIRYPYMQVNRPGMVSWLVFDLDHSNPLIWEDEGLPAPNIIVSNRKNGHAHLFYAIPPVCTTESARSKPIAYM KAVYEAMAARLGADPSYSGPVAKTPDHPWWNTWEIHNAVYDLGELADY |
| 584 |
PF02628 (COX15-CtaA) |
 Estimated TM-score=0.75; trivial target. |
>Cytochrome oxidase assembly protein (Length=324) VWLFICCFLVFAMVVVGGVTRLTDSGLSIVEWQPIVGTIPPISEQDWEELLEKYRATPQYQQVNKGMSLDEFKGIFWWEY FHRLLGRAIGLVFFIPFVYFLIRKQIDRPLGWKLAGIFVLGGLQGLMGWYMVMSGLVDDIRVSQYRLTAHLGLAFVIFAA LFWVATGLLSPENKHTGQPNNAPKTLQRFSVVLTFLIFIMVLSGGLVAGIRAGLAYNTFPLMNGHLIPPEILLLEPWYRN FFENMATVQFDHRLLALILAILIPIFWFNARKYHLSDSARLACHLLLVMLAIQIGLGIATLLHAVPIPLAAAHQGGAVLL FTAA |
| 585 |
PF02544 (Steroid_dh) |
 Estimated TM-score=0.75; easy target. |
>3-oxo-5-alpha-steroid 4-dehydrogenase (Length=149) PMPLLACTMAIMFCTCNGYLQSRYLSHCAVYADDWVTDPRFLIGFGLWLTGMLINIHSDHILRNLRKPGDTGYKIPRGGL FEYVTAANYFGEIMEWCGYALASWSVQGAAFAFFTFCFLSGRAKEHHEWYLRKFEEYPKFRKIIIPFLF |
| 586 |
PF02474 (NodA) |
 Estimated TM-score=0.75; easy target. |
>Nodulation protein A (NodA) (Length=194) SKVQWRLCWENELELTDHIELARFFRTTYGPTGAFNAKPFEGGRSWAGARPELRAIAYDSRGVAAHMGSLRRFIKVGSVD VLVAELGLYGVRPDLEGFGIGHSIRAIYPVLQELRVPFAFGTVRHALKNHFARLFRNGMGTILDGVRVRSTLPNVHLDLP PTRIEDVLAVILPIGSPLNEWPDGAAIERNGPEL |
| 587 |
PF02447 (GntP_permease) |
 Estimated TM-score=0.75; trivial target. |
>GntP family permease (Length=437) MPLVIVAIGVILLLLLMIRFKMNGFIALVLVALAVGLMQGMPLDKVIGSIKAGVGGTLGSLALIMGFGAMLGKMLADCGG AQRIATTLIAKFGKKHIQWAVVLTGFTVGFALFYEVGFVLMLPLVFTIAASANIPLLYVGVPMAAALSVTHGFLPPHPGP TAIATIFNADMGKTLLYGTILAIPTVILAGPVYARVLKDIDKPIPEGLYSAKTFSEEEMPSFGVSVWTSLVPVVLMAMRA IAEMILPKGHAFLPVAEFLGDPVMATLIAVLIAMFTFGLNRGRSMDQINDTLVSSIKIIAMMLLIIGGGGAFKQVLVDSG VDKYIASMMHETNISPLLMAWSIAAVLRIALGSATVAAITAGGIAAPLIATTGVSPELMVIAVGSGSVIFSHVNDPGFWL FKEYFNLTIGETIKSWSMLETIISVCGLVGCLLLNMV |
| 588 |
PF02175 (7TM_GPCR_Srb) |
 Estimated TM-score=0.75; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Srb (Length=235) YHPVYRFAQFWTFFVSSLAIPALLYFLFKRIFSLPFHGNIKFMLICYFFSSFLFAVVMLFDFGYHFVVPFFVFSKCSLII DSTLFKIGHLSMTLFMTIPMIMPIGFSIERFVALGMAKNYENVRTLLGPVLVVILITTDVNLIYNVFQNEKFNDPFISFI LIPSTSAVQFNNFFWFLLYVEITNFISNCVLLLVHSRLKTRFLRQKSTLSVRYELEEISQSSKFTLIVSFTHLLF |
| 589 |
PF01914 (MarC) |
 Estimated TM-score=0.75; easy target. |
>MarC family integral membrane protein (Length=201) QYFILAFSSIFSILNPFGAVPVFITLTESYPKKERDLVAKKTVIYALAILLAFALFGEWILKFFGISLDAFKIAGGILLL LISLDMVRGQQEAKIHRKEIEAAYEIDEIALMPLATPLLAGPGSITACMVAMAEASDIGDKFLVILAILLSLGITYLTLL SAESVLDRIGRLGIRILTRMMGLILTAIAVQMIVNGIRGAL |
| 590 |
PF00780 (CNH) |
 Estimated TM-score=0.75; trivial target. |
>CNH domain (Length=259) AAIIDHERIALGNEEGLFVVHVTKDEIIRVGDNKKVHQIELIPSEQLIAVISGRNRHVRLFPMAALDGRETEFYKLAETK GCQSXVSGHVRHGALTCLCVAMKRQVLCYELNQSKTRHKKIKEIQVQGNVQWMSIFSDRLCVGYQSGFLKYPLHGEGSPY SLLHPDDHTLSFISQQPTDAICAVEISNKEYLLCFSSVGVYVDCQGRRSRQQELMWPATPSSCCYNAPYLSVYSENAIDI FDVNSMEWIQTIPLKKVTA |
| 591 |
PF18845 (baeRF_family3) |
 Estimated TM-score=0.74; easy target. |
>Bacterial archaeo-eukaryotic release factor family 3 (Length=167) PFWVLSLSADRVTLWSGSADHVTEDRTGGFPLTRDLAENFDAERQERIGDAPSTFRDERTRQFLREADAAMSAVLREHPR PLYVTGEPAALSAFEEAGTVAGNAVQVQHGGLAHGTPEAVWRALGPVREEEARKETEAVLRELEAARGRRTYVSGVDEVW RTAREGR |
| 592 |
PF18726 (HEPN_SAV_6107) |
 Estimated TM-score=0.74; easy target. |
>SAV_6107-like HEPN (Length=101) ATALETPNERYATAHLAALRTAAAVLAARGRPEPTPRRRARIRSAWEVLPEIAPELTEWSALFASGADRRARAEAGIRGA VGTRDADDLIRDVAMFLRLVE |
| 593 |
PF18180 (LD_cluster3) |
 Estimated TM-score=0.74; easy target. |
>SLOG cluster3 family (Length=170) EYAATADVVAITAAVSALVHVTLGRRQLVWGGHPAISPMIWVVAQEMGIEYGRWVRLYQSRFFQDEFPEDNDRFQNVIYT PNVPEDREKSLLLMREQMFSENQFKAGVFIGGMAGIVDEFELFSRLQPNAAVVPVVSAGGAALQVAERIGVIAPDLRDDL DYVALFHRYL |
| 594 |
PF17660 (BTRD1) |
 Estimated TM-score=0.74; hard target. |
>Bacterial tandem repeat domain 1 (Length=50) GSAWEARHGLTSAQYQATFDKLVGEGYRLVDVSGYDVNGQGRYAAIWVKS |
| 595 |
PF17236 (DUF5309) |
 Estimated TM-score=0.74; easy target. |
>Family of unknown function (DUF5309) (Length=288) DLTDVIYNIAPTETPFLSMCGTTEATSTKHEWQTDDLAPAANNKQVEGDDAAAAAASPTVRVDNQTQISSKTVSVSGTQD AVSSAGRKKELAYQVSKRGKEIKRDMEFALTQNTTAIAGDSATARQLRGLEGWIATNNSLGATGVAPNPINNTAPTDGTQ RAFTESLLKDVLQKCWNNGGDPDVIMVGGTQKQVFSTFDGGATKQTDSKDKKVVATVDIYVSDFGTLKVVPNRFQRARTA FALQSDMWEVAYLRRFKTEPLAKTGDSEKRMLLAEYTLKAKNEKSSGA |
| 596 |
PF17201 (Cache_3-Cache_2) |
 Estimated TM-score=0.74; easy target. |
>Cache 3/Cache 2 fusion domain (Length=293) TSRSATKEAERTLEQQAQALVESMASYHGALADSAGKLAAVFRTHFPNSFTIDTSKTVTVGDKQTPLLASGSMAINLNTD IVDRFTSVTKAVATVFVRTGDDFVRVSTSLKKEDGSRAIGTALDRSHPAYGGLLKGDEFVGKATLFGKDYMTKYLPVKDA QGKVIAVLFIGLDFTDGLKALKEKVKAIKVGKTGYIYVFDAKEGKDQGKLVIHPAKEGTSVIDSKDADGREFIREMIKNK NGIIRYPWMNKEVGDRFQRDKVAAYRHLKEWNWIIGVGSYQDEFNGLARLVRN |
| 597 |
PF16933 (PelG) |
 Estimated TM-score=0.74; trivial target. |
>Putative exopolysaccharide Exporter (EPS-E) (Length=453) MAGIGFELRKMLRRDNLSGMLSAYAYAGIISSGPWILSIVGILLIGMLSLPFVVPGTLITQFQVSVTYLIAGSLILTGPM QLSFTRFTSDRLFEKRDHLILSNYHAVALLATVLSGGIGVACAAFSFGEQSVLYRLLMVAGFVVLSNIWIAAIFLSSMKQ YKAIVWTFLLGYAVSVAAALGLRRYGMEGLLGGFVAGQLCLLTGMVTLIYRNYTSRQFISFEVFRRRYAYPSLVAIGLLY NLGIWIDKFMFWYAPSTGHQVIGPLRASIIYDIPVFLAYLAIIPGMAVFLVRIETDFVEYYDAFYDAVRGGSSLEHIEDM RNTMVQTIRLGLYEIVKVQAIAALLLFAVGTWVLRLLGISELYLPLLYVDVIGASLQVVLLGVLNIYFYLDRRREVLLLT ALFVVLNCALTLVTLQLGPAWYGYGFAVSLLVVVALALAMLDHKLERLEYETY |
| 598 |
PF16212 (PhoLip_ATPase_C) |
 Estimated TM-score=0.74; easy target. |
>Phospholipid-translocating P-type ATPase C-terminal (Length=251) VMASDFAIAQFRFLTDLLLVHGRWSYLRICKVVMYFFYKNLTFTLTQFWFTFRTGFSGQRFYDDWFQSLYNVFFTALPVI VLGLFEKDVSASLSKRYPELYREGIRNSFFKWRVVAVWATSAVYQSLVCYLFVTTSSFGAINSSGKIFGLWDVSTLVFTC LVIAVNVRILLMSNSITRWHYITVGGSILAWLVFAFVYCGITTPRDRNENVYFVIYVLMSTFYFYFALLLVPIVSLLGDF IFQGVERWFFP |
| 599 |
PF15072 (DUF4539) |
 Estimated TM-score=0.74; easy target. |
>Domain of unknown function (DUF4539) (Length=84) KLSLLRCIVKACEHNGLGDMKITIRDPTGTVKASVHHKAIDHPEIGVHLKVGSVIILQDVSIFSPIRETYYLNITLRNIV KVFG |
| 600 |
PF15024 (Glyco_transf_18) |
 Estimated TM-score=0.74; trivial target. |
>Glycosyltransferase family 18 (Length=594) CYAFYGVDGSDCSFLIYLSEVEWFCPPLAWRNHSSPPVLQKNPPKRQAAFRTDLSALLDQVGTGKESLSFMKRRIRRLAS QWATAAHRLDIKLRRSWREQKKILVHVGFLTEESGDVFSPKVLKGGPLGEMVQWADILTALYVLGHNLKISISVKELQGF LGVPPGRGSCPLTGPLPFDLIYTDYHGLQQMKQHMGLSLKKHKCHIRVIDTFGTEPAYNHEEYATLHGYRTNWGYWNLNS RQYMTMFPHTPDNSFMGFVSEELNETEKRNIQENKVKNMAVVYGKEASMWKGKEKFLQILHRYMEVHGTVYYETQRPPEV PAFVKNHGLLPQQELQQLLRKAKLFIGFGFPYEGPAPLEAIANGCIYLQPKFHPPHSSLNHEFFRGKPTSREVSSQHPYA EQYIGQPHVMTVDYNNSEEFETAVREIMRTKVEPYLPYEYTCEGMLERVHAYIQHQDFCSIEAPFPPANASRVWPGGLFT LLPNSTLLGWAANSTVPPCWPPLSALRLTVSKEGQSCVEACQTSGLVCEPAFFRFINNKEALIKLEIQCEVVELEVNHIF PAFSVARHECGLQKESLLFSCAGFSAKYRRICPC |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218