

Go to page (83 pages in total): [First page] << < 1 2 3 4 5 6 7 8 9 10 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 401 |
PF13173 (AAA_14) |
 Estimated TM-score=0.77; easy target. |
>AAA domain (Length=133) GMIKVITGIRRCGKSYLVFHIFKNYLLESGVDQEHIIAIELDQRKNKELRDPDKILNYIDNQIVDDKQYYILLDEVQMLD DFEEVLNSLLHISNVDVYVTGSNSKFLSKDIITEFRGRGDEIHIYPLSFKEFM |
| 402 |
PF12976 (DUF3860) |
 Estimated TM-score=0.77; trivial target. |
>Domain of Unknown Function with PDB structure (DUF3860) (Length=92) MDSQNLRDLIRNYLSERPRNTIEISAWLASQMDPNSCPEDVTNILEADESIVRIGTVRKSGMRLTDLPISEWASSSWVRR HERARYNERKES |
| 403 |
PF11814 (DUF3335) |
 Estimated TM-score=0.77; easy target. |
>Peptidase_C39 like family (Length=204) PWYQQTTEFTCGPAALMMAMASLDSHFALEQTVELDLWREATTIFMTSGHGGCHPVGLALAAHRRGFEAAVYINTDQPLF VDGVRSAKKKAIMATVHDQFVYQAQRQSVALHYQDISQHQVEQWLQEGLGVVILISTYRLDGKKSPHWVTVTTIDEFCFY VHDPDLDDKQHQRPIDCQYLPLARDDFEKMSSFGSGRLRTAVTL |
| 404 |
PF11684 (DUF3280) |
 Estimated TM-score=0.77; easy target. |
>Protein of unknown function (DUF2380) (Length=129) MAVIDFDYVDTSGEAKDQTDEHEKRLDAFMSALRQDLAASGKYKIVAAACHPDACRVGRSSLADLQTAAREAGAKILLMG GIHKLSTLVQWAKVLAIDVDTNAVTFDRLLTFRGDTDSAWKRGEDFIVS |
| 405 |
PF11251 (DUF3050) |
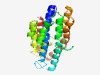 Estimated TM-score=0.77; easy target. |
>Protein of unknown function (DUF3050) (Length=231) ILNHSLYAEMETLADLRIFMQHHVFAVFDFMSLLKSLQRGLTCVDLPWIPVGNANTRFLINEIVCGEESDIDENGIRMSH FEMYLKAMRNADADSTAINAFLQQLQSGVAVQNVIEQADLAIGVTKFLDTTFNLIASGNVPLQAAVFTFGREDLIPNMFT SLLRDMAVNSPDALKSFVYYIERHIEVDGGHHSHLALEMTSELCGSDAALWEAATAATIEALKARIGLWDA |
| 406 |
PF11161 (DUF2944) |
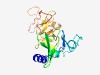 Estimated TM-score=0.77; easy target. |
>Protein of unknown function (DUF2946) (Length=184) MDDIVRQALAKWPNVPHCTGWLLLDRRGQWRLRDDAAQAAGEPGSPVRHAALNAFIARNYECDAHGQWFFQNGPQRVYVE LAYTPWIVRLAERDSRLALTDHTGAPFEPAQAWLDDAGGVLFRDTGTPPRIAALHDHDLGLFADHADFDAAPPVLRWRDG CALPLDSIARADVPARFGFVASPA |
| 407 |
PF10979 (DUF2786) |
 Estimated TM-score=0.77; easy target. |
>Protein of unknown function (DUF2786) (Length=40) RILDRIRALLAKAESTDFPEEAEAFTAKAQELMARHRIDH |
| 408 |
PF10508 (Proteasom_PSMB) |
 Estimated TM-score=0.77; trivial target. |
>Proteasome non-ATPase 26S subunit (Length=504) MAAQVQSLLREVSRLEAPLEELRALQSVLQAVPLSDLREQVTELSLGPLFSLLNENHREQTTLCVSILERLLQAMEPVHV ARNLRVDLQRGLTHPDDSVKILTLSQVGRIVENSDAVVEILNNSELLKQIVYCIGGENLSVAKAAIKSLSRISLTQAGLE ALFESNLLDDLKSVMKTNDIVRYRVYELIVEISSVSPESLNYCTTSGLVTQLLRELTGEDVLVRATCIETVTSLAHTHHG RQYLAQEGVIDQISNIIVGADSDPFSSFYLPGFVKFFGNLAIMDSPQQICERYPVFVEKVFEMTESQDPTMIGVAVDTIG ILGSNVEGKQVLQKTGTRFERLLMRIGHQAKNASTELKIRCLDAVSSLLYLPPEQQTDDLLRMTESWFSSLSRDPLELFR GISNQPFPELHCAALKVFTAIANQPWAQKLMFNSPGFVEYVMDRSVEHDKASKDAKYELVKALANSKTIAEIFGNPNYLR LRTYLSEGPYYVKPVSTTAVEGAE |
| 409 |
PF10321 (7TM_GPCR_Srt) |
 Estimated TM-score=0.77; trivial target. |
>Serpentine type 7TM GPCR chemoreceptor Srt (Length=312) MTLFYMLTNSFELWPDAYECSENTKYPETRWPRFGIYLISSGSILIVLYSLCFIAILKVKKLTPAYQLMLLISVFDIKSL SIGSVMSGFFTYYGIFFCQYPRLFYISGCIVMLTWLTNCVACIILAIERCVEVNPRFFLYFLFGKRVFKLVILCLAVYGI NSLMFSKPVIFSPEYSSYVFDPMIGKDPTLYINSWLALNNLMIAVSTTTLYFYLCYYLIFQFGYSTSMWLYKTKRQRILR ISDCNARSDNVSVPFRGSLYLRVHDSPLYLIVTGQTVWQLACGCLSVVYLTLNRTIRNSVLKMVIPKAIRQK |
| 410 |
PF10237 (N6-adenineMlase) |
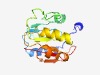 Estimated TM-score=0.77; easy target. |
>Probable N6-adenine methyltransferase (Length=162) EDWQLSQFWYSDETASCLANEAVAAAGKDGKIACISAPSVYQKLKEQDGKDFSVCILEYDRRFSVYGEEFVFYDYNNPLN LPENLLPHSFDIVVADPPYLSEECLQKTAETIKYLTKGKILLCTGAIMEEQAAKHLGVKICKFVPRHSRNLANEFRCYVN YA |
| 411 |
PF10125 (NADHdeh_related) |
 Estimated TM-score=0.77; trivial target. |
>NADH dehydrogenase I, subunit N related protein (Length=218) MLESIAGSLYGYIPFGDIVFYMTDFSMIAFVVALFFTIIVALTKPEKQIEAQIGELGDKFNTVSLKEMKARRLMAVICGI TTAFAMLTGDVFNFALFLAVIGMANIGIVSAVKKEWVLTAAFNYGIIAMISTLPLFGAASIILAKTGTLSIFELSRHPYD IFYQKILYAVGMVGETGIAPFYAAKAEMFRAPGAPYILMIHLSSLLVIVRTAEILLNI |
| 412 |
PF10096 (DUF2334) |
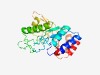 Estimated TM-score=0.77; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2334) (Length=232) AYIRIEDVHPLTDPKKLRAIADFLYSRHIPFMVAVIPVYTDAAGGNVVNFEDKPEFTAALHYMEDRGGSLIMHGYKHRYR DGETGEGFEFWDAQSDKPIPDEAAYMTDKLEKGIARMAKQGLYPLAFEPPHYAMSQTGYRIVNRYFSTLVGNLQLNDTTY LVSQQPPYRLQHSLAGLKVVPETIGYVADEAGAAAPMLEKAKQLMIVRDSVIGAFFHPYLKLSQLTELVDGL |
| 413 |
PF10012 (DUF2255) |
 Estimated TM-score=0.77; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2255) (Length=115) ELDTIGAAEELEIAPRRRDGTLRTPVTIWVVHVGDSLYVRSYRGRGGAWFRAAQVRHEGRIRAGGVEKDVTFVEETDPGI NDQIDAAYRTKYRRHAARYVDPMVASEARATTIKL |
| 414 |
PF09956 (DUF2190) |
 Estimated TM-score=0.77; easy target. |
>Uncharacterized conserved protein (DUF2190) (Length=101) IQPGDTITVPAPADVVSGAVVLVGKLFGVANFDAKSGEDVEISTKGVFTLPKTSAQAWTVGAAVYWDATNSEVTTTSTSN TLIGHAVAAAANPSATGTVRL |
| 415 |
PF09865 (DUF2092) |
 Estimated TM-score=0.77; trivial target. |
>Predicted periplasmic protein (DUF2092) (Length=211) EELLKKACSFLRDQKSFTVEVDITYDDVIDSGEKVQYSAYQKVWVKKPNQLRSDYVGDERNTNFYYDGKSFTLFTPDLNL YGTKMAPLNIDESIEDLEQNYGITIPLSNLFVSDPCVALDSDIQKSLFVGTNLVNRVETYHILLTREDRDVQLWISKDEQ PVLLKAIITYKNLPSSPQYTAVFSQWNFNPSIEKDTFTFTVPDDAAEIEFL |
| 416 |
PF09693 (Phage_XkdX) |
 Estimated TM-score=0.77; easy target. |
>Phage uncharacterised protein (Phage_XkdX) (Length=40) FEIIKKNYERGLWSKQMVATAVRKGVITAEQYREITGEDY |
| 417 |
PF09570 (RE_SinI) |
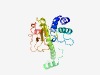 Estimated TM-score=0.77; trivial target. |
>SinI restriction endonuclease (Length=217) AQTIAENTAITEDERSLIVSFTEVCCFLSDNPIFLSWRGKKKPSVFNDEGLNILAQKYFDGYRKSDFPAIPQTVPDEIVS VVMQVAYGYSEEQCNQIKLEHQHSMCAENCVGALLERYLDSVLRSNGWHWCCGNFVKAIDFIRRNDDGTWLPLQIKNRDN SENSSSSAIRNGTPIQKWFRSFSKTGKTNWDNLPPSMQGYSLNEQGFINFVRQYLDE |
| 418 |
PF09318 (Glyco_trans_A_1) |
 Estimated TM-score=0.77; trivial target. |
>Glycosyl transferase 1 domain A (Length=199) YFISGGLPSNYGGLTKSLLLRSKLFGEECNQNTFFLTFRFDLELSSKIDELYSNGKIDKKFTSVINLFDDFLSVRTNGKR SYEERIGLDQIKKQVGMGKFAKTLLRLFGKKNNEMSVVYYGDGETIRYVDYWNDKNQLIKREEYTKNGNLVLVTHYDVQL NKMYLQEYINDQNQVYLDKLYVWNNEEKDVQLSHIIWYS |
| 419 |
PF08449 (UAA) |
 Estimated TM-score=0.77; trivial target. |
>UAA transporter family (Length=317) ALLAMLLVFVGCCSNVVFLELIIKIDPGAGNLITFSQFLFIAVEGLIFTSKFFTVKPKIALKDYVKLVLLFFGANVCNNY AFNFNIPMPLHMIFRSGSLMANMIMGIILLKKRYNLRQYSSVAMITVGIILCTLVSSGDVKDNTHHSLKADSSYSVFFWW SVGIALLTVALLVTAYMGIYQEVIYKRYGKHPDEALFFTHMLPLPGFLIMASNIAQHWQIAVASEAVSYTLPVVNWSVGM PLMLFYLVCNMVTQYVCISSVYVLTTECASLTVTLVVTLRKFVSLLFSIMYFRNPFTLSHWLGTILVFMGTVLFANV |
| 420 |
PF08444 (Gly_acyl_tr_C) |
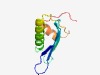 Estimated TM-score=0.77; easy target. |
>Aralkyl acyl-CoA:amino acid N-acyltransferase, C-terminal region (Length=81) FCLLGPKGTPVSWCLMDHTGELRMGATVPEYRGQHLIYHSSLYQIQSLVKLGFPLYAHTDKANHIIKKTSVRLRYVPMPS E |
| 421 |
PF08284 (RVP_2) |
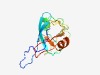 Estimated TM-score=0.77; easy target. |
>Retroviral aspartyl protease (Length=128) NRLYAITSRHEQENSPNVVTGMIKVFVFNVYALLDPGASLSFVTPYVANQFEVLPERLCEPFCVSTPVGESILAERVYRD CPVSISHKSTKADLVELDMVNFDVILGMDWLHACYASLDCRTRAVKFQ |
| 422 |
PF07870 (DUF1657) |
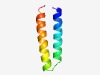 Estimated TM-score=0.77; easy target. |
>Protein of unknown function (DUF1657) (Length=49) VKQTLAGLKSAQASLETFALETENEQAKQLFQQAAQQTQGVVDGLEPRL |
| 423 |
PF07788 (PDDEXK_10) |
 Estimated TM-score=0.77; easy target. |
>PD-(D/E)XK nuclease superfamily (Length=74) SDVEYDIVIKDGSVILVEITSSLKRGDLPTIKRKKEFYEKVKNIKVNLVYVVTPFIHDRYPERVKAMAKDMGIE |
| 424 |
PF07694 (5TM-5TMR_LYT) |
 Estimated TM-score=0.77; easy target. |
>5TMR of 5TMR-LYT (Length=159) ERGFSMADRALIVIVFGTLSIYGTTQAILFDGAFINFRDLGPMIAGLIGGPVLGGLAGLIGGLYRYSLGGWTALPCAVAT ILAGVLAGYAAICWKGRLDYLRLTGLGVFVEAIHLLVIFPLLVYPAPLEDIFNLIRSFLLPMILTNVFGLAIFLYILKE |
| 425 |
PF07603 (DUF1566) |
 Estimated TM-score=0.77; easy target. |
>Protein of unknown function (DUF1566) (Length=118) GTVTDKNTGLMWGNDSVVTGTWESALASCEASGANGYDDWRLPSAAELLSIVDYGRKDPAVDDTVFTNTVSEFYWAATLI FDGSDRAWIVNFLNGWASASTYPRNLVESDTHYMRLVR |
| 426 |
PF07433 (DUF1513) |
 Estimated TM-score=0.77; easy target. |
>Protein of unknown function (DUF1513) (Length=307) ALPGRGHGVARHPSGEEFVLVGRRPARFAAIVRRQADGEPSYEELSSAEDRHFFGHACYSRDGRYLYTTENDYASGRGVI GVRDAQDGYRQVGEWSSGGIGPHELHLSADGKALVVANGGILTHPDNPREKMNPDAMEPSLAYIDIASGALLERWELEDK KLSIRHLDVADDGAVCFGCQYEGPPTDTPPLVYMHKSGSPIQAASAEYSLWRSLKNYTGSVVINSRLRVAGVTSPRGDSI TLWSLDAQKCVGKISLPDVCGLSLTPDGKEFLASSGEGKVIRVDPLTMKVEQHWSVAGTRWDNHMTL |
| 427 |
PF07062 (Clc-like) |
 Estimated TM-score=0.77; easy target. |
>Clc-like (Length=208) SITASIILGLIGMGLTIAALLTPSWQVVNLREYNSIHEHGLWLDCTRHSRDGERPLLQRYATITEPLHCVYKFDYDKYSG TFDLEDDNSPVGEVNRHKFYGWHTSTLILLFLAILCTFLATCLGICSCCYGSISVVYTCITLGTTIMSSLAVGIFFFYSH RADNRFIKGIVGTYEQRVGTAFFLQLAACFFYFFSFIVSMLGVYFSFQ |
| 428 |
PF06940 (DUF1287) |
 Estimated TM-score=0.77; easy target. |
>Domain of unknown function (DUF1287) (Length=162) VNDARKQINVTVSYDSTYRKMAFPMGDVPQHTGVCTDVVIRAYRQQQIDLQKLVHQDMKKAWVSYPKIWGLKTTDTNIDH RRVPNLETFFKRHGKSLSLTDLSSFKAGDIVTWRLPRNLPHIGIVSDKKNKEGIPLIIHNIGQGTQEEDILFKYKMIGHY RY |
| 429 |
PF06813 (Nodulin-like) |
 Estimated TM-score=0.77; easy target. |
>Nodulin-like (Length=247) HWFMLFASLLIMAASGAGYMFGMYSNEIKTSLGYDQTTLNLLSFFKEVGATVGIISGLVNEITPPWVVLSIGIIMNFLGY FMIWLAVTARIAKPQVWQMCLYICIGSNSQTFSNTGALVTSVKNFPGSRGSLLGILKGYVGLSGAIIAQLYHAFYGDHNP QALILLIAWLPAAVSFLFLPTIRILNTVHQTNDNKVFYHLLYISLGLAGFLMVLIIVQNKLSFTRSEYIADGVVVFFFLL LPLVVVV |
| 430 |
PF06808 (DctM) |
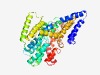 Estimated TM-score=0.77; trivial target. |
>Tripartite ATP-independent periplasmic transporter, DctM component (Length=403) IVVFALTMTLGVPIAFCLGLAGLSFIVLSGTVPIAVLPTLMFGGMDSFPLLAIPFFIIAGDLMQRCGVLPKLIEFANSLV GHLRGGLAYVVVLTSMLFAGVTGVALAECAAIGSMLAPAMVRQGYPGGFVSSVIAGSAVMGPIIPPSVAMLIYAYVYGGG ISVGKLFLMGVVPGVLVAVALMALVWVTARQRHFPKSDTKFSLGRVVRSLRGALLGLVVPVIILGGILGGVFTATEAGAV AALYAVLIGVFVLRTLGPRELGQSLLVGAKTSAIIYLLLGTSNVVSWILVRHQVPKIVAQWSLELTDSATVFIIVTIAVL FLLGLALEAVPIMIMLIPVLAPAAAAYRIDPHHLGLIIVMTVQTALLSPPVALSLYVVATVIRCPMREANREVWKFVAAV LAI |
| 431 |
PF06379 (RhaT) |
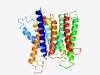 Estimated TM-score=0.77; easy target. |
>L-rhamnose-proton symport protein (RhaT) (Length=344) MSNAITMGIFWHLIGAASAACFYAPFKQVKQWSWETMWSVGGIVSWLILPWTISALLLPDFWAYYGQFNLSTLLPVFLFG AMWGIGNINYGLTMRYLGMSMGIGIAIGITLIVGTLMTPIINGNFDVLIHTEGGRMTLLGVFVALIGVGIVTRAGQLKER KMGIKAEEFNLKKGLLLAVMCGIFSAGMSFAMNAAKPMHEAAAALGVDPLYVALPSYVVIMGGGALVNLSFCFIRLAKVQ NLSIKADFSLARPLIISNILLSALGGLMWYLQFFFYAWGHARIPAQYDYMSWMLHMSFYVLCGGLVGLVLKEWKNAGRRP VAVLSLGCVVIIIAANIVGLGMAS |
| 432 |
PF06241 (Castor_Poll_mid) |
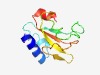 Estimated TM-score=0.77; easy target. |
>Castor and Pollux, part of voltage-gated ion channel (Length=96) HDIIGRLILLAARSPGLSKVYNSLLGFEGDEFYCKAWPSAVGASFGSLAERFPLATPIGVKTASGKIVIKPKMDYVIADK DEIVVLADDNDSYTCT |
| 433 |
PF06039 (Mqo) |
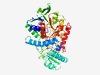 Estimated TM-score=0.77; easy target. |
>Malate:quinone oxidoreductase (Mqo) (Length=486) DVVLIGAGIMSATLGTLLKELQPELTIEIFERLDMAAAESSDAWNNAGTGHSAFCELNYTPQKQDGSVDIKKAVNIAEQF EVSRQFWAYLVENNIISNPQAFIQSIPHMSFVWGEENVDYLKKRYKALQQCHLFKKMEYSEDQEQLKAWMPLVMEGRDPN QKVAATHMEIGTDVNFGALSRALFNHLEQLDGVNIHFNHDVRDLKKADDGGWIIKVKDRETREKQTIKASFVFIGAGGGS LPLLEKSDIPEGKGFGGFPVSGQWLRCTNPDIIAQHQAKVYGKASVGAPPMSVPHLDTRMIDGERALLFGPYAGFSTKFL KNGSIFDLPKSIKVDNIHPMVSAGLDNIPLTKYLIAQVRQKPEDRLEALREYYPEARMEDWKLEIAGQRVQVIKKDPEHG GILEFGTEVVSAADGSIAALLGASPGASTAVSIMLNLLKRCFKDYVETEAWQRKLKQMIPSYGQSLLNNPQHCDELRDWT TSVLEL |
| 434 |
PF05150 (Legionella_OMP) |
 Estimated TM-score=0.77; easy target. |
>Legionella pneumophila major outer membrane protein precursor (Length=304) TVPCERTAWDFGVQALYLQPTLSAGNGYDFYNTDFNGVRRYHDFDPDWGWGFKLEGSYHFHTGNDLTVSWYHMNKSTDNH FGDFVLPNLNIVTANGIQLGNSLRTSFEPKWDAVNVELGQHVDFSEWKVIRFHGGVQYARIEHDYDANLTTPVSFPRYGV VAGVPFRLASAETKFNGFGPRVGADMSYNLGNGFAVYANGATALLIGQGKFRSSFGYVPSLTTPLFGIPGHGSKTTMVPE MEAKLGAKYTYAMAQGDLTLDAGYMWVNYWNANHILVGHSDGTYSSRESNFALSGPYVGLKWVG |
| 435 |
PF04339 (FemAB_like) |
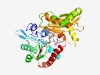 Estimated TM-score=0.77; easy target. |
>Peptidogalycan biosysnthesis/recognition (Length=384) VGAIAAADWDACARAALTPPAACASEAAAAAYNPFLSHAFFSALEQSKSASPRTGWGPRHLVAKQGDRVIGIVPCYLKSH SQGEYVFDRGWADAYERAGGNYYPKLQVSVPFTPATGPRLLIREGEDSATVAAALTTGLTALCQMSEASSVHVTFARESE WRFLAEQGFLQRTDQQFHWHNAGYASFDDFLGSLNSRHRKAIKRERRDAVANGITIHHLTGADITEDAWDAFFEFYMETG SRKWGRPYLTREFYSLIGQSMSQDVLLVMAKRDGRWIAGAINFIGGDTLFGRHWGAVEQHPFLHFEVCYYQAIDFAISRG LKVVEAGAQGEHKIARGYLPQTTYSAHYIADPALRRAIADYLKRERMYVDEMGRELTEAGPFRK |
| 436 |
PF04179 (Init_tRNA_PT) |
 Estimated TM-score=0.77; easy target. |
>Rit1 DUSP-like domain (Length=118) EMEVGVGKHKVASRNLRAALPDICGFVTRFLESQKKDDGSLAEGTTVLVACETGKDLSVAVALAICLWCFDQQDNYRAPD TKTIFNKDMVRHRLGSIMTACPEANPGRASLQSVNSFL |
| 437 |
PF04082 (Fungal_trans) |
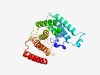 Estimated TM-score=0.77; easy target. |
>Fungal specific transcription factor domain (Length=247) RFLVHIYPCIPIINMSRFLAYQWDPKASPPDPLLLNAIYLVTARQLSPNDEVFSTLNISRESFIALCENTTRSLMDQEYA QPKMSTVQAMLLMTNLSNSKDSNVRWLTSGLAIRMAQDLGLHRSASRFKMSKEEEETRRRVWWACYCLDRWTCASLGRPL AIHDSDCDVEMPSTADLGFQDISRFAGANNIVKSMDQTYGAFTHLVRLSTILGHILKKLYTPKANAMGYGKNMHSTLKDL SDQLKSW |
| 438 |
PF03923 (Lipoprotein_16) |
 Estimated TM-score=0.77; easy target. |
>Uncharacterized lipoprotein (Length=160) PRIQLPSQDPSLMGITVSINGADQRTDQALAKVNRDGQLVTLTPSRDLRFLLQEVLEKQMTARGYMVGPNGAVDLQIVVN NLYADVTEGNVRYSITTKADISIIATAQNGNKMVKNYRQTYSQEGAFSATNDKITKAVNSTLGDVIADMAQDTTIHNFIK |
| 439 |
PF03444 (HrcA_DNA-bdg) |
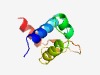 Estimated TM-score=0.77; trivial target. |
>Winged helix-turn-helix transcription repressor, HrcA DNA-binding (Length=78) ELTPSQKNILQELVNLYRESESAVKGEDIADKVDRNPGTIRNQMQSLKALQLVEGVPGPKGGYKPTATAYDALQIQEM |
| 440 |
PF03269 (DUF268) |
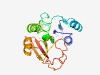 Estimated TM-score=0.77; easy target. |
>Caenorhabditis protein of unknown function, DUF268 (Length=176) GMSGVVIGSMQPWVEVMALRNGAEKILTVEYNRLTIQEEFRDRMSSILPVDFVQNWHTYADKFDFAASFSSIEHSGLGRY GDPMDPIGDLREMLKIKCILKKGGLLFLGFPLGTDAIQYNVHRIYGPIRLAMMFYGFEWLSTFSGGLENAFDLNSQSLHS NVKFGMHQYTMVLKKL |
| 441 |
PF02988 (PLA2_inh) |
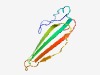 Estimated TM-score=0.77; easy target. |
>Phospholipase A2 inhibitor (Length=82) DCEHCVVWGQNCTGWKETCGENEDTCVTYQTEVIRPPLSITFTAKTCGTSDTCHLDYVEANPHNELTLRAKRACCTGDEC QT |
| 442 |
PF02530 (Porin_2) |
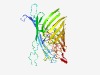 Estimated TM-score=0.77; trivial target. |
>Porin subfamily (Length=359) DAVVVAEPEPAEYVKICDVYGAGYFYIPGTETCLRIGGYVRYDIGLGDVGSFDGARSNDVQDGKNQSTWYKNTRFTLKTW TGQETELGTLKTYTETRFNFGNSNADPDFGSNSAHNKDVSLNFAWIQLGGLRVGKDESAFDTFIGYAGNVIQDTLVPYGD FDTNVVQYYFDAGNGFSAVVSLEEGSGAGVDGQYGVGTIDSYVPHVVGGVKWTQGWGAISGVVAYDSNYEEVAGKVRLDV NVSNELSLFIMGGYGTDDNYNDATYDQPAGGRGFYKQWGGNWAVWGGGTYKFNEKTSFNAQVSADDWKNVGVAANVAYDI VPGFTVTAEVDYLHAGKFGEAGTENFAWTPADKKNSVGG |
| 443 |
PF01718 (Orbi_NS1) |
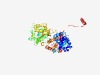 Estimated TM-score=0.77; trivial target. |
>Orbivirus non-structural protein NS1, or hydrophobic tubular protein (Length=548) MERFLRKYNISGDYANATRTFLAISPQWTCSHLKRNCLFNGMCVKQHFERAMIAATDAEEPAKAYKLVELAKEAMYDRET VWLQCFKSFSQPYEEDVEGKMKRCGAQLLEDYRKSGMMDEAVKQSALVNSERIRLDDSLSAMPYIYVPINDGQIVNPTFI SRYRQIAYYFYNPDAADDWIDPNLFGIRGQHNQIKREVERQINTCPYTGYRGRVFQVMFLPIQLINFLRMDDFAKHFNRY ASMAIQQYLRVGYAEEIRYVQQLFGRVPTGEFPLHQMMLMRRDLPTRDRSIVEARVRRSGDESWQSWLLPMIIIREGLDH QDRWEWFIDYMDRKHTCQLCYLKHSKQIPTCSVIDVRASELTGCSPFKMVKIEEHVGNDSVFKTKLVRDEQIGRIGDHYY TTNCYTGAEALITTAIHIHHWIRGSGIWNDEGWQEGIFMLGRVLLRWELTKAQRSALLRLFCFVCYGYAPRADGTIPDWN NLGNFLDIILKGPELSEDEDERAYATMFEMVRCIITLCYAEKVHFAGFAAPACEGGEVINLAAAMSQM |
| 444 |
PF18767 (AID) |
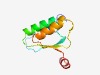 Estimated TM-score=0.76; trivial target. |
>Activation induced deaminase (Length=89) DKDDHAEIKFIDDNKDPFEITDLLIKNSPCHECSEKLLKHFEDLNNKPTIHVGRIWHLYDSDDDNGIIELLKDEFDIIVW EDLHSLICT |
| 445 |
PF18656 (DUF5633) |
 Estimated TM-score=0.76; trivial target. |
>Family of unknown function (DUF5633) (Length=40) GFKTKAEAEDAAKKVLKEDKVNKSFTVTQGANSNYYFSLS |
| 446 |
PF18299 (R2K_2) |
 Estimated TM-score=0.76; easy target. |
>ATP-grasp domain, R2K clade family 2 (Length=146) FLGRKIWKSRINYIAKHPETWNVFIKPTTLTKKFTGRVVRSPKDLISCGDEFEDTEIWCSEPVKFLAEWRCFVRHGEILG VKSYYGDWRVHFDPQVIEQAIAAYTSAPAGYAADFGVTDKGETLLIEVNDGYSIGAYGLFPPDYAR |
| 447 |
PF18182 (mCpol) |
 Estimated TM-score=0.76; trivial target. |
>minimal CRISPR polymerase domain (Length=116) CLGDGDNIGDTIEFHLLSGNIEEASKFSSQVKTAIAKIAELVKDEIGASLVYVAGDDICFVVSAEKNIKDYLVKYSNFFL KTTGKTISFGVGRNSVEALICLRKAKVSGKGHVVTF |
| 448 |
PF17152 (CHASE8) |
 Estimated TM-score=0.76; easy target. |
>Periplasmic sensor domain (Length=101) LRAYADSNQQLIARSISYTVEAAVVFGDAQAAEESLALIASSEEVSSAIVYDRQGQTLASWHRESTGPLHLLEQQLAHWL LSAPTEQPILHDGQKIGSVEV |
| 449 |
PF17149 (CHASE5) |
 Estimated TM-score=0.76; easy target. |
>Periplasmic sensor domain found in signal transduction proteins (Length=107) DYNREFNNVQARHDEIQTIHAELLASSLWNYDLVVLTQRLEGLVNLPNVDYMKITSGDYHFSAGEPVTSMALNSEIALEY TNPDTQVTENIGTLYVESDAQGIYNYL |
| 450 |
PF17138 (DUF5111) |
 Estimated TM-score=0.76; easy target. |
>Domain of unknown function (DUF5111) (Length=121) DAAYSNVCQVKVTPYLIDMSYINILNEKKDQVLTKLYSPNSDGVYSGYMNASSWFHIWGKENDGTIWGNVGQDGHVYEMD NTESAWNIWFPGQTGIYYTVLDTKAKELKPTYIKSMQLNGE |
| 451 |
PF16961 (OmpA_like) |
 Estimated TM-score=0.76; easy target. |
>Putative OmpA-OmpF-like porin family (Length=199) YEVRALSTNWADKKLMVGFEGGWFFKDQWKLNLGGGVSFTNNPGYPAVPGTIDDSNKNNSADENMGEIPNYRAVADAQSF AYNVSAGVDRYFNIKRVPNLMWYTGIRVGFAYGENEMKYDEETSMGKSIAESWNLRGALTIGVDYFILPALYIGAQIDPF AYTYNKTTYNPQAGLGDLSADSHNYSVLAAPTFKIGFKF |
| 452 |
PF16828 (GAGBD) |
 Estimated TM-score=0.76; trivial target. |
>GAG-binding domain on surface antigen (Length=145) KKTPLQHLKELVEELKTDLEKLKEYPEYDSTKYPVQKMLWDDNLSFGEREMGRVSSLKEEDSAVSEIPQRLFDFRNHIIW ERLTRQVDQYRKKYPDNKEIEEEYKQHLEQTDSKNYASQEASNLQASVNEAEQAFEKIKEIAKEI |
| 453 |
PF16323 (DUF4959) |
 Estimated TM-score=0.76; easy target. |
>Domain of unknown function (DUF4959) (Length=105) FGCEEIEREPITKDSVPPGVITSLVVENIPGGAVITYALPKETDLLYVKAEYELSNGKKVETRSSIYASSVQVEGFGDTR EREVKLYAVDRSENISAPVTVKVQP |
| 454 |
PF16268 (DUF4921) |
 Estimated TM-score=0.76; easy target. |
>Domain of unknown function (DUF4921) (Length=428) MADGTIKQVNPFSGTQVWTVPGRANRPLTHLPADIHKLTYQDHVSTCAFCSDRQIETPPEKARMVQSSVKKNSDWEILRG VGAEDLYATTAAFRRVPNLFEIVSYDYWHDNYGYSLDSETQARMERYIASDLGRKHVLEIIKTKRKAAGNAVSTDYPENE LLKDASAFFGGGHDVIIARRHYTDDATHSDQLASAGTLTYEEHEAFIRFTIDSLRDLYSRNRYAPYVAVFQNWLKPAGAS FDHLHKQLVAVDERGVHSEMEVQKLRQNPNMYNEWAVDFAAMHNLVIAENDHAICFAGFGHRFPTLEVFSKSAVSEPWLQ SSEEIRDMSDLVHACHAGAGPQVPCNEEWHHKPVDVDIPMPWRIMIKWRVSTLAGFEGGTKIYLNTLSPWDVRDRIVHEL YSLRDQGRISPNIRIATECAVQRNSLKY |
| 455 |
PF16161 (DUF4867) |
 Estimated TM-score=0.76; easy target. |
>Domain of unknown function (DUF4867) (Length=200) IQKVTDPAFRKYGQVLEGYDFTSLIKEMKHTPVPEDVIYVPSVEELEALDVMKKLQNKGYGGLPIQIGYCNGHNKKLNAV EYHRNSEINVAVTDLVLLIGHQQDIEPDHTYDTSKIEAFLVPAGTGIEVYATTLHYAPCHVNEGGFQCVVVLPKGTNTEL TFPTEKTGEDSLMTAKNKWLIAHEDAKIEGAFNGLKGENI |
| 456 |
PF14626 (RNase_Zc3h12a_2) |
 Estimated TM-score=0.76; trivial target. |
>Zc3h12a-like Ribonuclease NYN domain (Length=124) YEPELNLPVKPLTDIILLFLIRGHKTTVYLPKYYQEFLSDCGVSKVDDLVAFQKLIDLGFIKFLEITNPFNFKWFNVVAD LADRCGAVFVSSEDYRQRKMKINYSKPSGRIITPCFLNAEDRLL |
| 457 |
PF14473 (RD3) |
 Estimated TM-score=0.76; trivial target. |
>RD3 protein (Length=129) GWMKWPKNYSYKPTQYPGSEVVTKTLLRELKWHLKERERLVQEIENEQRVQKTGVDYNWLKNYQNPQATIPATEQRQLEA LCSQIQPCQTGTVLSRFREVLAENDVLPWEIVYIFKQVLKDFLTTIERE |
| 458 |
PF14416 (PMR5N) |
 Estimated TM-score=0.76; trivial target. |
>PMR5 N terminal Domain (Length=53) GCDVAQGEWVYDEAARPWYAEEECPYIQPQLTCQAHGRPDTAYQHWRWQPRGC |
| 459 |
PF14378 (PAP2_3) |
 Estimated TM-score=0.76; easy target. |
>PAP2 superfamily (Length=176) RWDLAIGFDWSAWHAALLNQPTLNRLLMVVYNTLFAQFLLAVIYFSKRDRSARIEELMLLMCATVAPTALISALWPTLGP FAVHGVGDVTYLRDVLTLRAAGPWHFESLAMQGIVPMPSFHTVLAVLFTYAFRGTGLLGYGIATLNLAMLLSIPPIGGHY LVDVLAGAALALGAIA |
| 460 |
PF14125 (DUF4292) |
 Estimated TM-score=0.76; easy target. |
>Domain of unknown function (DUF4292) (Length=209) VSFETLAGRLKVRYQDEKQTQNVSVSIRMEKDKAIWMSANILGIPLAKVLITPNSVKYYEKISGSYFDGDFRLLSDVLGT ELDFNKVQNILLGQTIYDLRQEPYILETSDRGYVLQPKKQFDLFTRLFLLNASTYKTEAQQLVNNNENQSAIVTYPTYQT ISGQVFPNEINIVANQNNKNTLIDIEYRSVEFNVDVSFPFSIPSGYDEI |
| 461 |
PF13933 (HRXXH) |
 Estimated TM-score=0.76; easy target. |
>Putative peptidase family (Length=238) ILSSAAIAAALPTPETSEGNQSQPFSGLAPWNAGAVNQFPIHSSCNATQRRQIELGLNETITLASHAKDHILRWRNESEI YKKYFGDRPSMEAIGAFDIVVNADKKDILFRCDNPDGNCDNESWAGHWRGENGTDETVICDLSYQTRRSLSTMCGLGYTV SESETNTFWAADLLHRLYHVSAIGQNWIDHFADGYEEVIDQAKENATLSTRDSETLQYFALEVYAYDIAVPGIGCPGE |
| 462 |
PF13679 (Methyltransf_32) |
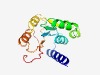 Estimated TM-score=0.76; easy target. |
>Methyltransferase domain (Length=148) KKQHEVKLASSLISWLCRDSKASVVIDAGDGKGYLSSCLAFEHNLHVIGLEANAAYQESAQKRVSKLKKKWKKTEDGGNY KSLAVRVTPNLRIGTICKEAFPEIEQPDCCLSGLHTCGDLAVSCLEIFTQDPSVRYLCNIGCCYNLLT |
| 463 |
PF13629 (T2SS-T3SS_pil_N) |
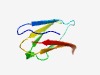 Estimated TM-score=0.76; easy target. |
>Pilus formation protein N terminal region (Length=68) GIEVTMNQAKIVKLTRPADTIVVGNPAIADASVQDASTIVLTGKGFGVTNLVVLDHDGSPIVDEQVTV |
| 464 |
PF13625 (Helicase_C_3) |
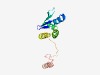 Estimated TM-score=0.76; trivial target. |
>Helicase conserved C-terminal domain (Length=123) LLIQADLTAVAPGPLVPRVAAELARMADIESAGAATVYRFTESSLRRALDAGSSAEDLHDLLGQLARGGVPQTLTYLIDD TARRHGRLRSGPAASYLRCDDTALLTEVVASRHTQALAMRRVA |
| 465 |
PF13550 (Phage-tail_3) |
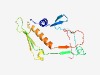 Estimated TM-score=0.76; easy target. |
>Putative phage tail protein (Length=157) TTQRKAWDVLSDFCSAMRCMPVWNGQTLTFVQDRPSDKVWTYNRSNVVMPDDGAPFRYSFSALKDRHNAVEVNWIDPNNG WETATELVEDTQAIARYGRNVTKMDAFGCTSRGQAHRAGLWLIKTELLETQTVDFSVGAEGLRHVPGDVIEICDDDY |
| 466 |
PF13528 (Glyco_trans_1_3) |
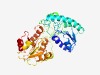 Estimated TM-score=0.76; easy target. |
>Glycosyl transferase family 1 (Length=323) MRILYGVVGEGMGHATRSRVLLEALTKEHEVHIVVSGRAQHYLAQRFQNVHGIWGLTIAYEGNSVKKWQTVLQNLQGAVA GWPQNVRQYFDLVEGFKPDVVVSDFETFSYLFAKRHMLPVISVDNMQIINRCTHDPALLAGHEESFEASRAIVKAKLPGA FHYLTTTFFYPPVRKRRTTLAPSILRPEILAAKPEAGEHLLVYQTATTNTHLPEILKQSGVPCRVYGLRRDLKEDVVDGN LTYRPFSEAGFIDDLRTARAVVAGGGYTLMSEAVYLRKPLLSIPVGGQFEQVLNALYLERLGYGMYVKELSLDALKTFLS RVP |
| 467 |
PF13524 (Glyco_trans_1_2) |
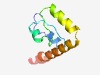 Estimated TM-score=0.76; trivial target. |
>Glycosyl transferases group 1 (Length=85) AENYANNMRLYEATGCGALLITDYKDNLNEIFEVGKEVVAYRSPEECVDLIDYYMKHPKEAEQIARAGQERTLQDHTYTE RMNQI |
| 468 |
PF13115 (YtkA) |
 Estimated TM-score=0.76; easy target. |
>YtkA-like (Length=82) AIGDLRLRISTQPEPAKVGENALRIEVKDARGQPVTHAALAVEYTMDMPGMPIDKAEVRHIGDGVYETQVRFTMAGPWGV TV |
| 469 |
PF12481 (DUF3700) |
 Estimated TM-score=0.76; trivial target. |
>Aluminium induced protein (Length=229) LAIFHKAFANPPEELNSPASHKSTKKPKQPEETLNDFLSHHPHNTFSMSFGNAAVLAYVRHERAASVHQRLFCGFDDIYC LFLGSLNNLFMLNRQYGLSKGSNEAMFVIEAYRTLRDRGPYPADQVVKDLEGSFAFVIYDSKTGTVFAALGSDGGVKLYW GIAADGSVVISDDLDVIKEGCAKSFAPFPTGFMFHSEGGLMSFEHPMNKIRAMPRVDSEGVICGANFKV |
| 470 |
PF12261 (T_hemolysin) |
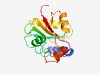 Estimated TM-score=0.76; easy target. |
>Thermostable hemolysin (Length=172) PQQHADRESLEWFVRECFATVHHADIRHFMPTLMALRDSHGTFCAAAGVRTADSGALFLEQYLQQPIERVVSRLAGAPVE RQQIVEVGNLAARSGGSARLIIVVMTWLLAHRKLDWVVFTGAASLINSFQRLGLNPLLLGDADPQRLGSEQHSWGSYYAQ RPQVFTGSIRAG |
| 471 |
PF11910 (NdhO) |
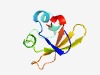 Estimated TM-score=0.76; trivial target. |
>Cyanobacterial and plant NDH-1 subunit O (Length=66) IKKGEMVRVVREKLENSVEAKASDPRFPSYIFETQGEVVDLKGDYAFVKFGKVPTPNIWLRQDQLE |
| 472 |
PF11775 (CobT_C) |
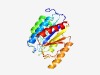 Estimated TM-score=0.76; easy target. |
>Cobalamin biosynthesis protein CobT VWA domain (Length=219) SFMHEKEATFRDTVVTLLLDNSGSMRGRPITVAATCADILARTLERCGVKVEILGFTTRAWKGGQSREAWLAAGKPANPG RLNDLRHIIYKSADAPWRRARKNLGLMMREGLLKENIDGEALDWAHKRLLGRPEQRKILMMISDGAPVDDSTLSVNAGNY LERHLRHIIEEIETRSPVELIAIGIGHDVTRYYRRAVTIVDAEELGGAITEKLAELFSE |
| 473 |
PF11265 (Med25_VWA) |
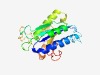 Estimated TM-score=0.76; easy target. |
>Mediator complex subunit 25 von Willebrand factor type A (Length=214) DHTVQADIVFVIEGTAINGAYLNDLKTNYLLPTLEYFSQGNLEDRDYVCEGTSTLYGLVVYHAADILPKSSCDCYGPYSS PQKLLMTLDKLELIGGKGESHANIAEGLATALQAFEDLQQKREGNNIITQKHCILVCNSPPYFMPVMEASTYSGHTIDQL AAILNEQNINLSILSPRKIPAIFKLFEKAGGDLTQSQNKNYAKDPRHLVLLKGF |
| 474 |
PF11215 (DUF3010) |
 Estimated TM-score=0.76; easy target. |
>Protein of unknown function (DUF3010) (Length=136) MKICGVELKGNDAVICLLDHSDNVMHLPDCRVAKISIGNANSTEEVKKFQFSFAKLVEDYKVEKVVIRQRQTKGKFAGGA VGFKLEAAIQLINDLQVEVVSPTQIKESLKKNPLAISFKETGLKQYQEQAFTTAYA |
| 475 |
PF11142 (DUF2917) |
 Estimated TM-score=0.76; easy target. |
>Protein of unknown function (DUF2917) (Length=58) FEVQPGATMSWRVDANTELLVQGARVWLTRAASPYDHWLQIGETFRLHRGERVWISAD |
| 476 |
PF11086 (DUF2878) |
 Estimated TM-score=0.76; easy target. |
>Protein of unknown function (DUF2878) (Length=155) ITNLIAFQIGWFACVLGGANGLPWIGVGVVALVAGLHLSQSRHPRRESALLLTIGLLGAGWDSLLVRLGFLDYPSGMLLP WLAPVWIIAMWIAFATTLNVSLGWLKDRWFLATVLGAVGGPLAYYAGHKLGGVQFPDVFVGMAVLAGGWSFLMPL |
| 477 |
PF11071 (Nuc_deoxyri_tr3) |
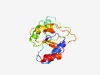 Estimated TM-score=0.76; easy target. |
>Nucleoside 2-deoxyribosyltransferase YtoQ (Length=140) YLSGEIHTDWRERIEAGADEAGLDITFSAPVTDHEASDDCGVAILGQEDKPFWKDHKGAKLNAIRTRTLIEQADVVVVRF GDKYKQWNAAFDAGYAAALGKPLIVLHPQEHTHALKEVDAAALAVAETPEQVVEILRYVI |
| 478 |
PF10881 (DUF2726) |
 Estimated TM-score=0.76; easy target. |
>Protein of unknown function (DUF2726) (Length=124) RYLLSKAERSFYGVLTQAVGDKVLIFSKVRVADVLTPQKGLNRSNWQRAFNQISAKHVDFLLCDPQDCAVKLAIELDDAS HGSAKRQKRDAFLEQACESAGLPLLRIRAARGYGVADIRQQIET |
| 479 |
PF10686 (YAcAr) |
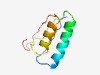 Estimated TM-score=0.76; easy target. |
>YspA, cpYpsA-related SLOG family (Length=67) PKVAVTGGLDFNDHHLIWAKLDQVHAKHPDMVLLHGGSPKGAERIAARWADHRNVPQIAFKPDWTRH |
| 480 |
PF10662 (PduV-EutP) |
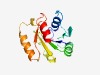 Estimated TM-score=0.76; easy target. |
>Ethanolamine utilisation - propanediol utilisation (Length=142) MGRVIFMGKTGCGKTTLCQALNNLEIKYKKTQAIELYDNAIDTPGEYMENRGYYTALIVTAADADIIALVYDCTSEENYI APGFASMFCKEVIGIITKINLAKDKSEIEVAEERLNLAGVSKIFKVDTVDDVGIKELFNYLN |
| 481 |
PF10137 (TIR-like) |
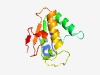 Estimated TM-score=0.76; easy target. |
>Predicted nucleotide-binding protein containing TIR-like domain (Length=119) KIFIVHGHNNQIKSEVARVLEKLNLDPIILHEQADNGMTIIEKFEQHSNDVNFAIILLTADDFGNVKTEENTKPRARQNV IFEMGYFYAKLGRKNVFLLKENEVENPSDLSGIIYTPYD |
| 482 |
PF09989 (DUF2229) |
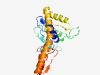 Estimated TM-score=0.76; easy target. |
>CoA enzyme activase uncharacterised domain (DUF2229) (Length=220) TVGIPRVLNIYENYPFWFTFFTELKFRVELSPRSSKKIYEMGIETIPSESVCYPAKITHGHIMSLVNKGIKFIFYPCIPY EQKEKEDADNHYNCPIVTSYSEVIRTNMDILKEKDVKFMNPFLALDDKEKLKKRLFEEFKTFNITKNEIENAVEKAFKEQ ENFKSDIRKKGEEVLEYLNKTGKKGIVLSGRPYHIDPEINHGIPNLITEFDMAVLTEDSI |
| 483 |
PF09863 (DUF2090) |
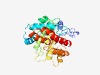 Estimated TM-score=0.76; trivial target. |
>Uncharacterized protein conserved in bacteria (DUF2090) (Length=308) ELDDYLSRANDVQRPDIDARLNHLHRVTTRKPNWEELCVLAFDHRVQFVDMVREAGADFDSIKPLKKLILQASQEVAKEA GLQGKAGILCDGTFGQDVLNEVTGSGWWIGRPIELPGSRPLCLEHGNIGSQLVNWPLEHVVKCLAFFHPDDQHPLRLEQE SMIKEVYAACCQSGHELLLEVILPADMPSSDDLYLRAVQRFYNLGVKPDWWKLPPQSVEGWNNISAVITERDTYCRGVVI LGLDAPEAELKAGFTAAANQPLVKGFAVGRTIFGQPSRDWLAGNIDDRALIEKIKANYHNLITLWRQR |
| 484 |
PF09844 (DUF2071) |
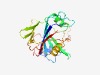 Estimated TM-score=0.76; easy target. |
>Uncharacterized conserved protein (COG2071) (Length=213) KQTWRNLLFAHWPVPADELRAYVPKELPLDTFEGSAWLAVVPFRMTDIRLRGLPPIPGTSRFPELNVRTYVTLHDKPGVY FFSLDAAHTLAVKLARWLYHLPYYRADMGVTVQNETVFYRSERRQAGPAFAARYRPVSAPYTAEPGSLEHWLTERYCLYT LNGRRDVMRCDIHHRPWPLQKAEAELYENTMLTGNGLPFAPDVPPLLHYAEAL |
| 485 |
PF08937 (DUF1863) |
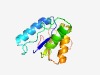 Estimated TM-score=0.76; easy target. |
>MTH538 TIR-like domain (DUF1863) (Length=118) RRTFFSFHYGPDVWRAWNVRNSWVVREANKEDRGFFDSSVLEASKKESDNTLKNFLRKGLDNTSVTCVLAGTQTWERRWV RYEIARSVIKGNGLLTVYIHGVQNKDKETSTKGANPLA |
| 486 |
PF08218 (Citrate_ly_lig) |
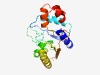 Estimated TM-score=0.76; easy target. |
>Citrate lyase ligase C-terminal domain (Length=175) QGHRYLIQQAAAQCDWLHLFLVKEDSSRFPYEDRLDLVLKGTADIPHLTVHRGSEYIISRATFPCYFIKEQSVINHCYTE IDLKIFRQYLAPALGVTHRFVGTEPFCRVTAQYNQDMRYWLETPTISAPPIELVEIERLRYQEMPISASRVRQLLAKNDL TAIAPLVPAVTLHYL |
| 487 |
PF07550 (DUF1533) |
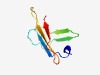 Estimated TM-score=0.76; trivial target. |
>Protein of unknown function (DUF1533) (Length=57) VWRSAITGITVDGALLEPIQYTVTAGNISIAPGVLNTDANHSIVISATSYQDTTVVQ |
| 488 |
PF07302 (AroM) |
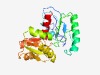 Estimated TM-score=0.76; easy target. |
>AroM protein (Length=218) IGLITIGQSPRTDVTPDLEPIFGPDVQLLQAGALDGSTAQEIAAFAPEEGDYVLISKLRDGTSAVFAEKHILPRLQQCIR DLEDQGAELIMFLCTGSFPAFESRVPLVFPCAVLNGTVPALAGRSRIGVITPKEEQIDQCAHKWKDYVQSVHVVAGSPYG EPEELDRAAKEMAGMDVDLVVLDCIGYTSAMKERVRQLSGKPVVLSRTIAARVVMELL |
| 489 |
PF07288 (DUF1447) |
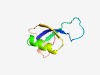 Estimated TM-score=0.76; trivial target. |
>Protein of unknown function (DUF1447) (Length=68) IYKVFYQETKTRNPKREETHSLYIEAADEVTARQRVEENTPYNIEFIQPLEGSHLEYEQENADFKLTE |
| 490 |
PF06862 (UTP25) |
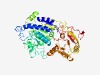 Estimated TM-score=0.76; easy target. |
>Utp25, U3 small nucleolar RNA-associated SSU processome protein 25 (Length=475) TSLQNRLFHQMNNYRDVMYCNRNMSNAKEIRNAYLLHAVNHITKTRDRVLKNNAKLSKAQKEGKDLGEMRDQGFTRAKVL LLLPFRNTVVDVVDTLIKLSGTEQQENKNRFYEQFNLREEEAIDTSKPADFLQNFQGNIDDHFRLSVKFSKKSMKFYCNF YDADMIIASPLGLRTLIGSEGDKKRDFDFLSSIELVILDQTNHFLMQNWEHVDHIFQHMNLIPTDSHGCDISRIKSWYLD GKAKYLRQTLVFADFMTPEFNALFTKHFKNVSGKLKIKQTYAEGSIVDVIPQVQQGFTRIDVPSLKSVNEARYRYFIEKT LPTLRKSAIMQSHTLVFIPSYFDFVRVRNYFEDNKYSYEACSEYASGPAITRARAQFFHGRADFLLYTERLHFFRRYNIR GAFHIVFYGLPEDPLYYTEVVNFLGLKFNEASAAEEATFSCTALFTKYDFLKLERIVGTERAKKMCTAQKNVFMF |
| 491 |
PF06730 (FAM92) |
 Estimated TM-score=0.76; easy target. |
>FAM92 protein (Length=222) MFRRGKLSFLNTKDDRVKIINERINITERHLMEMCSSFALVTRKMAKYRDSFDELAKSVKSYADDEEINESLCQGLKSFT NAVTIMGDYMDINVHRLEHKIVNELAQFEQLCKSTRDNLRLAVIARDKEVLRQRQMLELKSKFSANNSAADSELFKAKME VQRTNKEIDDIIGNFEQRKLRDLKQIISDFILIAMKQHTKALEILSASYYDIGTIDERDDFI |
| 492 |
PF06151 (Trehalose_recp) |
 Estimated TM-score=0.76; easy target. |
>Trehalose receptor (Length=384) KENTTQNCLYFMLVFAQCFGMLPVTGISEENCKNLKFKWSSKRTFYSLTFALVATANTVFFLIKMIRGKRNEFQEWVALL FFFVTVITVVIFLDVAKKWPQLMKKWTEVDTAMNSYGFPIALSKKLKTTTLLVISAAIVEHGLFVAVASAPCEGKNWSEL FNNYFKMRFDYVFDVIPYHFFWGIMFQVLNIFSTVSWNFMDLFIILISLCLSERFKQVATRTNQLAAKEVTDERIWEQLR EDYTRLTILSHTVDKTLANTVVLSFANNLFVILVQLFYSLQAPPRFGILNKLYYVYSFAFLIIRMVAVALNAATINNESL KPKYCLNTLPHFLWNVEIDRFIYQIKYSPVVLTGHKLFKITNALVLQITVAIVTYELIMIQYPS |
| 493 |
PF05978 (UNC-93) |
 Estimated TM-score=0.76; easy target. |
>Ion channel regulatory protein UNC-93 (Length=146) LTFGQLCIMTGYDSQSFILESVIHSIHEKEPERISPYAGYYGQAVCYLAYVTACLFSPSFLYATSAKTTLLISSICFTSF PLGFLFTNSYYYYFSSALNGIGFAFYYTGNGGYITSHSTRQTIESNVSLSMIVGSIIMAIITSVTQ |
| 494 |
PF05973 (Gp49) |
 Estimated TM-score=0.76; easy target. |
>Phage derived protein Gp49-like (DUF891) (Length=89) DFLSSLPPKHQAKALREIDLLAEFGNALKEPYVKHIKGDIWELRIRFSSDISRVFYFTWQADTIVLLHGFVKKTQKTPPQ EIETAQNRM |
| 495 |
PF05940 (NnrS) |
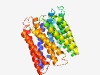 Estimated TM-score=0.76; easy target. |
>NnrS protein (Length=372) FALGFRPFFLAAGLYAVLMMALWLLVLNGAVIPGELLPLVWHGHEMVFGFAVSVIAGFLLTAAQNWTGLRTPSGRPLAAL FLLWLAGRLGFLIPGLPTGLVALIDLAFLPVLAVTLAVPILRTKQLHNYPFPILLLALAAANALVHADALHWMSGTARPG LHLGVYVVIMMITAMGGRVIPSFTDNKLRTRARRWTIIEWLVPAATLLALIAALVAPTSPVTALLAAVAAAAHAIRLAGW YTPKFWSVPLLWILHLGYAWIPLGFALLALSAAGWNAAAGSALHAFTAGAIGALTLGMMARVSLGHTGRPLEPPPIMTLA FIAINLAALARVALPLLFPAAYTQVMTVAGLVWIAAFGLFVAMYAPMLLRPR |
| 496 |
PF05673 (DUF815) |
 Estimated TM-score=0.76; easy target. |
>Protein of unknown function (DUF815) (Length=252) ASIAFRWRKRGAQGYLQPVTHVHRIALKDLQGVDRQKALVEQNTRQFVNGVPANNVLLTGARGTGKSSLVKAVLNKYAPK GLRVIEVEKQDLLDLPEIVDLVSARPERFILFCDDLSFEAEEAGYKALKAVLDGSVAAPSENVLVYATSNRRHLMPEFMQ ENLEARRVGDEIHPGETVEEKVSLSERFGLWVSFYPFSQDEYLSIVFHWLEYFQAPIPNRETVRQEALQWALQRGSRSGR VAWQFARDWAGR |
| 497 |
PF05228 (CHASE4) |
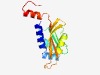 Estimated TM-score=0.76; easy target. |
>CHASE4 domain (Length=144) IAYDIAVLDDQVFDWAVWDDTYQFVVDGNREYIASNLEETIFISPKINFIIFFDNEGKLIFGKGYDLERRRELPLPPSLL TLGQEKRPVKGIVMLPEGPLLVVVEPILDSSGNGPARGMLLFGRYLDDDRIKRLADTVQLSFTV |
| 498 |
PF04727 (ELMO_CED12) |
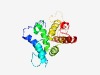 Estimated TM-score=0.76; trivial target. |
>ELMO/CED-12 family (Length=185) LLRKWRNVPVDLEKPDHRRALKGIHLASNPEKATDKSSENGDDTRRSRRHNPEKWRRLGFETETPATEFQEMGFLGMMDL TDYVRKHQDEFQKMLLEHSTKPPEQRCPIARASLAITSVLYEHFEVDKCVTEDAKSYLVLESRSNLDKVFKPLLLHWSRL HVAGLHAFFRLWKATGADVNDFDKI |
| 499 |
PF04450 (BSP) |
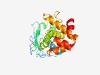 Estimated TM-score=0.76; easy target. |
>Peptidase of plants and bacteria (Length=198) PKFNLRIEDLSHEGVSIFLNAVNPNVALQEAVVASFTWLYNSETIPTNVKQIVLVLRPMSGVAHTLGSKSHKEIHFSLDH IRNSQKRARDEIMGILVHEVVHCYQFDAKGTCPGGLIEGIADYVRLKDGRAPPHWTKKPGETWDAGYEKTAFFLAWIEDR YGDGTICELNALLNDKKYHRRIFKEVTGRPVRKLWALY |
| 500 |
PF04305 (DUF455) |
 Estimated TM-score=0.76; easy target. |
>Protein of unknown function (DUF455) (Length=247) TAAPLEKVRLTREYAAGWRDGRIAAFGTLAPPERPARPERPPLMLPRDMPKRGRGGSAQNRVALLHALAHIELNAIDLAW DIVCRFVGEGMPKGFTDDWVQVADDEARHFQMLEERLNQLGSGYGELPAHDGLWQAATTTAHDLAARLAVVPMVLEARGL DVTPETVRRLREFGDAESAALLQVIHDEEITHVAAGRRWFGHLCAKRGVDPVETWQDLVKRYFRGGLKKPFNVTSRQAAD FGPEFYE |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218