

Go to page (83 pages in total): [First page] << < 4 5 6 7 8 9 10 11 12 13 14 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 801 |
PF13114 (RecO_N_2) |
 Estimated TM-score=0.72; trivial target. |
>RecO N terminal (Length=71) MQGYIIHTQKLRDEDLIVDILTKNSLVKTYRFYGARHSNIMLGYKIDFELISNIKFLPQLRSVMHLGFKWL |
| 802 |
PF13020 (DUF3883) |
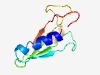 Estimated TM-score=0.72; easy target. |
>Domain of unknown function (DUF3883) (Length=91) GEEYVYQQLCRSLGEQEGVQVKWVNETEESGLPYDITVRSGASQDIEYIEVKSTRTMEKGVFEISMNELDTAGMHGSRYS IYRVFNAGNAA |
| 803 |
PF12812 (PDZ_1) |
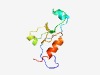 Estimated TM-score=0.72; easy target. |
>PDZ-like domain (Length=80) AITPDKFVSVAGASFHDLSYQQARLYAISLKGAGVYVCEAAGSFRFADGYTSGWLIQEVDNQPTPDLDTFIQVMKKIPDR |
| 804 |
PF12733 (Cadherin-like) |
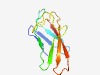 Estimated TM-score=0.72; trivial target. |
>Cadherin-like beta sandwich domain (Length=91) SALSASVGALNPSFSPATKTYVINVPTGTTSTTITPTLNDLKATLTVNGAAHTSGATAGPFPLPVGNTSISIKVTAESDT TISTYTVSIVR |
| 805 |
PF12679 (ABC2_membrane_2) |
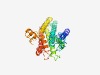 Estimated TM-score=0.72; easy target. |
>ABC-2 family transporter protein (Length=303) WNLALKELYVSVKSRRFIVIVVLYFIIFGLAVYSIKDYLVQMGVPSVDSNELGLWGVSAEIYTTPLAMLFTVNMTIITVL GAVLGVALGADAINREVESGTVRVLLGHPVYRDEIINGKFLGMGLLIAVTYMVSYIVMIAVMLILGIPLDGESLFRSFLA ILVTMLYTMVFLSLGILLSTLSKKPETSMLAGVGLAIFLTVFYGIVVGIVAPKLAGPEPPIGTSAYQIWADELHVWMNRL HSINPAHHYVQLVSYIFAGDRFFNYYIPLSDSLIYGFNNLAALLVMLLLPFALAYARFLTSDL |
| 806 |
PF12390 (Se-cys_synth_N) |
 Estimated TM-score=0.72; easy target. |
>Selenocysteine synthase N terminal (Length=40) LRQLPSVDELLQEPSIREMVQALPRWAVVEAIREVLERWR |
| 807 |
PF12038 (DUF3524) |
 Estimated TM-score=0.72; easy target. |
>Domain of unknown function (DUF3524) (Length=166) VLLIEPFYGGSHKQLMDLLQEELQEDCVLCTLPAKKWHWKARTSALYFMQTVPTSSNYRILFASSVLNLAELAALRPDLG KLKKVLYFHENQLAYPVQKRQERDFQYGYNQILSCLVADVVVFNSAFNMESFLNSIGKFMKLIPDHRPKDLEKIIRPKCQ VLYFPV |
| 808 |
PF11854 (MtrB_PioB) |
 Estimated TM-score=0.72; easy target. |
>Putative outer membrane beta-barrel porin, MtrB/PioB (Length=635) YNDSNDIRSANAFAAEDEFAGKLDADVKYVSESGYQASIEADNLGMENGRLDVNAGKAGQYNLNLNYRQIATYKTDQAMS PYQGAGSDHLTLPDNWQTAGSSQDMTQLYSSLNPLELSLKRKRAGLGFEYQGEELWSTYVNYQREDKTGLKQASGSFFNQ SMMLAEPVDYTTDTIEAGIKLRGDNWFTALNYSGSSFKNQYSQLTFDNAFNPTFGAQTQGTMALDPDNEAHTVSLMGQVT AGKTIMSGRVHYGQMTQDQAFVTSGYGYDMPADSLDGRVDMQGVNLKLVSRISSSVRLNASYDYSDRDNKTNVEEWTQIS IDSTTGQVAYNTPYDITTQRAKLGADYRIARGMRLEAGYDYRQDERSYQDRETTDEHTLWTKFRLNSFDEWDMWIKGSYG ERDGSEYQASEWTSSESNDLLRKYNLADRKRSMVEARVTHTPIDALTIDFGTRYALDDYNDTQIGLTESKDLSYDINASY LINDDMMVNAFYSRQNIESEQAGSTNFSTPNWYGVVEDQVDVIGAGFSYNNLMEKKLRLGIDYTYSDSNSNTQVTQGITG DYGDYYAKAHNVNVYGQYQATEKMALRLDYKMEKYRDNDAGNDIAPDGIWNVVGFGSNSQDYTAHLIMLSMSYKL |
| 809 |
PF11848 (DUF3368) |
 Estimated TM-score=0.72; hard target. |
>Domain of unknown function (DUF3368) (Length=46) VTGTLGVLLRAKNEGLIDSVKPLVDKMIEEGFHVDDKLYNFVLKKA |
| 810 |
PF11350 (DUF3152) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF3152) (Length=206) LPDGGPFTAEGAGTWRVLPGTSPKVGQGTEQVFTYTVEVEDGIDTTGFGGDESFARMVDQTLANPKSWTNDARFAFQRID AGEPTFRISLTSQMTIREACGYDIQLEVSCYNPGIDRVVLNEPRWVRGAVAFQGDIGSYRQYLINHEVGHAVGYQAHQPC EADGGLAPVMMQQTFGTSNNDIAKLDPEGVVPMDGKTCRFNPWPYP |
| 811 |
PF11249 (DUF3047) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF3047) (Length=197) DGWEALEFPGIDSHTRYELVTEDGTRVVRAEADASASGLISRLSVDPGDSLILRWRWKVANVYEKGDARTKAGDDYPARI YVAFEFQPDKAGFFERAKRKTVKVLFGEELPGNALNYIWANTLPVGSVVPNAFTDQTMMIAVNSGADQAGEWVTVERDIV ADYREAFGEEPPRIVGVAIMTDADNTGESATAWYGDV |
| 812 |
PF11042 (DUF2750) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF2750) (Length=101) NHFVNRVCEQEEIWILTDEHGCMMLTTDDDEDCIPVWPHADYAKDWAVGDWKDCTPEAISLNVWLKRWVPGMTDDELLVA VFPTADETGVVVEPYELKDAL |
| 813 |
PF10948 (DUF2635) |
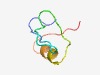 Estimated TM-score=0.72; hard target. |
>Protein of unknown function (DUF2635) (Length=44) KPVTGLTVRDPETMQPLAEKGERKPRTAYWLRRLKDGDVTEVTA |
| 814 |
PF10794 (DUF2606) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF2606) (Length=130) MYSTIFNIGQINKYSKLAIFMSILFLCGCSSQTHSSQKETTIPVTLHVEDAKGLPVEGVQVTIVKAPSSDEEPSTEIGEI LGKTDKNGDIKWDTGRKGDYSVALTKGETSVTHHISLTEDKKDHAIPLVF |
| 815 |
PF10790 (DUF2604) |
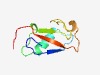 Estimated TM-score=0.72; easy target. |
>Protein of Unknown function (DUF2604) (Length=76) VVVNGQPTQVEANPNQPLHVVRAKALENTQNVAQPAENWEFKDEAGNLLDVDKKVGDFGFANIVTLFLSLKAGVAG |
| 816 |
PF10544 (T5orf172) |
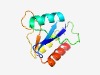 Estimated TM-score=0.72; hard target. |
>T5orf172 domain (Length=105) GFIYVFWYPGGFGHVKIGYTKDVEKRMSEWSKQCGRKPEKYFPSGQADLEPVPHRLRVEQLVHAELARYRREEPRCGKCG GRHVEWFEVDVSLAISVVQKWVNWM |
| 817 |
PF10363 (RTP1_C1) |
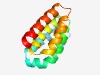 Estimated TM-score=0.72; easy target. |
>Required for nuclear transport of RNA pol II C-terminus 1 (Length=104) KALEDACNELLPVRGHAMIILAKLISQGDSEAKAKKNAILCIFQENIKHEDSYLYLTAIEGLACLATEYPDIVLTTLAEE CVRSEGNVKLKVGESLMRVIRNLG |
| 818 |
PF10354 (DUF2431) |
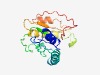 Estimated TM-score=0.72; easy target. |
>Domain of unknown function (DUF2431) (Length=169) LLVGEADFSFAASLAAAFGSSVSNLTATSLEYKRELEKVYSKAVDNIQELKSKGCNVFHAVDATVMASHPSFKNSVFDRI VFNFPFAVISEEEWGNLHLPAVLEKQHSLVREFLENAKKLISENGEIHITHRKDGIFGAWRIKFLASEQGFQVVEAVDFN ICDYPGYNP |
| 819 |
PF10342 (GPI-anchored) |
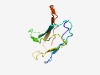 Estimated TM-score=0.72; easy target. |
>Ser-Thr-rich glycosyl-phosphatidyl-inositol-anchored membrane family (Length=85) SPLSTTSLEPGKSYTLEWTDNFRDEVKLELYKDNFLQATISSSTPSDGSFSWTVPTSLTSGSNYNIKMTKVNDSSIYDYS NNFTI |
| 820 |
PF10326 (7TM_GPCR_Str) |
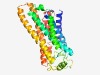 Estimated TM-score=0.72; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Str (Length=306) HIIQYFGFISSQLANSLLIYLILTKARKLFGMYRFVMLGFAFFFLFYSWIEVLTQPVIHIKSPVCIVFMDSSLKYYKSLG FVITCLYCACFALVISLLSAQFFYRFLAVCKPHLLSHGEERLLFYLFVPCFLCFVAWFEFVYYGMANTVEKQLYMKNELS EIYNEDSERVAFIAVMYWSVDQNGQKKWKLCDIALLLACVATIVGGGCFMTIVYCASRIYKKMKDTSCHMSERTMELNRQ LFITLTIQTILPLFMMYIPVGLATSLPIFEIKTGRIANFTAASLAIYPFLEPLIPMMCIKEFKNVL |
| 821 |
PF10127 (RlaP) |
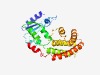 Estimated TM-score=0.72; easy target. |
>RNA repair pathway DNA polymerase beta family (Length=248) MKQRIIDELKRIEQSYGVKIVYAVESGSRAWGFPSQDSDYDVRFIYVPKKEWYFSIEQERDVIEEPIHDLLDISGWELRK TLRLFKKSNPPLLEWLSSDIVYYEAFTTAEQLRKLRTEAFKPEASAYHYINMARRNVKDYLQGQEVKIKKYFYVLRPILA CQWIEKHGTIPPMDFTVLMNELVAEPELKAEMETLLERKRRGEELDLEARIDVIHQFIETEIERIMEAAKELKAEKKDMT SELNRLLL |
| 822 |
PF10030 (DUF2272) |
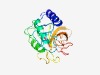 Estimated TM-score=0.72; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2272) (Length=189) ERLPGLWQRVGEYWWIGQDPYETEVGWTGKTDSQGTRFEIEHDGHYAWSAAFISYVMRVAGANDRFPYSPNHSTYINAAA SGQSSGVTAHDPGTYAPKIGDLICVGRGRSKTVTFSMLPTSYGFPAHCGIVSATNQNAQPFGHELSIVGGNVDDAVALTH VPTDANGMIADGSGHSYDNRYPWCAVLEV |
| 823 |
PF09990 (DUF2231) |
 Estimated TM-score=0.72; hard target. |
>Predicted membrane protein (DUF2231) (Length=103) YEVSWWNMLFATISIFIAIIFGQTEAGLAQPYDAVKPILNLHTLIGWSLSAILATITGWRFILRSRDPQKVPVSYLAVSL VLVGIVCFQVYLGDELVWVYGLH |
| 824 |
PF09954 (DUF2188) |
 Estimated TM-score=0.72; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2188) (Length=57) YHITFNKDDRWQVKKVGALKILKTFKTQKEAISFADKLAKNQKAGVVIHRTSGQIRS |
| 825 |
PF09938 (DUF2170) |
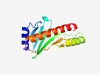 Estimated TM-score=0.72; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2170) (Length=137) WTAQALHEALAATELFQSGQAGIELIQGADASLHITMREYGDLPLFIAVFGEQIIVEALLWPASDVKDAAAFNEEVLRTH KLFPLSSIGLEKMGDGRDCYTMFGALSSSSTLSDVVFEIELLADNVIKATEAYEGFL |
| 826 |
PF09848 (DUF2075) |
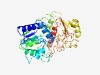 Estimated TM-score=0.72; easy target. |
>Uncharacterized conserved protein (DUF2075) (Length=363) MFVVQGDAGTGKSVILNSLFNDVQKLANNNPDTTDILKGTRNYLIVNHPEMLKLYHRMSTNYTYINKASLERPTSLINNL QKRKEMADVVIIDEAHLLATSKDAFKRFYGENHLKELMELAKVLVLVYDEKQALRMGCYWDENTENGANLQTFYDEIPES KRGWYNLKQQFRVAAPPDVLDWINDISVEGKIPKYPKSAKGSLEPKFDFQIWDDCNEMYMKLKEKNETFGQCRMLSTYDF PYRLDGKTYYVTCGDFKLRWDMYAPKALLPWSEREDTIDEVGSVYTVQGFDLNYAGVILGRSVGYDKANDCIKLRPELYD DHAGFTKKKNISDADTVKQKIIMNSINVLLTRGTKGLYVYAWD |
| 827 |
PF09559 (Cas6) |
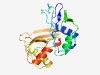 Estimated TM-score=0.72; easy target. |
>Cas6 Crispr (Length=195) VDLNFQVLGNILPADHGYGLYAALTHWNPIIHDLEGLSIQTIAGIPDKQGKIYLTERSRFRIRLPHDQVPLVYGLAGKTL TIGKHTIRLGIPQIYLLQPASKLRSRLVVIKGYQEPKIFLEAVQRQLEKLRINGIVAISTANSKPDRKTIKIKRFTVVGF GLEMSNLSDEDSLLLQEVGLGGKRRMGCGVFVPCQ |
| 828 |
PF08856 (DUF1826) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF1826) (Length=199) EELSGLAGIYQDDVNLMVWQRNLDSETAQSVSAVLQTDRPLSLNQIVTPENVNQSLERGLPQVEGRDALIRDIALLVDAY CCLFDLETVGLRLTQVDRAMCPRFHVDQVPCRLITTFAGPATQWLEENSLNRSKLGRGSNGQPDSSSGLIQANANIQQIT VGDVALLKGERWEGNEGRGIVHRSPGVEQGQQRLLLTLD |
| 829 |
PF08480 (Disaggr_assoc) |
 Estimated TM-score=0.72; easy target. |
>Disaggregatase related (Length=202) MDDVEIYNNLFHNTYGPGIALVGYNNGGGIYPLTEACNIYIHNNIFYDCASQQTYNWMGGVTACGFDNVVIENNTFDGCY NVGVFVHQPTVGVSAPGTGYRIYAKNNIIANTRLRAYDPSGTGNGVQNDMPTTHTLTSEYNCFHGNAGSNYKNVTASSTD LSSDPLFYDQPNRDYHLKSLYGRWNGSSWETDTVHSPCIDAG |
| 830 |
PF08400 (phage_tail_N) |
 Estimated TM-score=0.72; easy target. |
>Prophage tail fibre N-terminal (Length=127) XSGVLKDGAGKPIQNCTIQLKAKRNSTTVLVNTVASENPDEAGRYSMDVEYGQYSVILLVEGFPPSHAGTITVYEGSRPG TLNDFLGAMTEDDVMPEALRRFEEMVEEAARNAEAASQSAAAAKKSE |
| 831 |
PF08386 (Abhydrolase_4) |
 Estimated TM-score=0.72; trivial target. |
>TAP-like protein (Length=104) SPTWGPIWSGIRMNCIDWPKFPKTNFRGPFNATTSHPLLVIGNTADPVTPLWAAKKMSAGFTDSVVLTQDSPGHCSLAAP SVCTQSHIRRYFIDGTLPEPGTVC |
| 832 |
PF08366 (LLGL) |
 Estimated TM-score=0.72; trivial target. |
>LLGL2 (Length=100) SPFGPFPCKSITKIATHSSRRHNEDYLFFIGGMPRAAYSDRNVISVQLGEKQVVFDFTSKVLDFSIFTADRETGCTEPEA LFVLCEEEFVAIDLLSDDWP |
| 833 |
PF08348 (PAS_6) |
 Estimated TM-score=0.72; easy target. |
>YheO-like PAS domain (Length=114) LKQYIPMVHFIAEIMGENCEVVLHDVTNPDHSIIEIANGHVSGRKINSPITDLALKILKEESYKDQDYICNYKSSSKTNK TLRSSSYFIKDENNKIIGMICVNMDITDFINARN |
| 834 |
PF08220 (HTH_DeoR) |
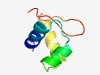 Estimated TM-score=0.72; easy target. |
>DeoR-like helix-turn-helix domain (Length=57) RREQIIQRLRQQGSVQVNDLSALYGVSTVTIRNDLAFLEKQGIAVRAYGGALICDST |
| 835 |
PF07857 (TMEM144) |
 Estimated TM-score=0.72; easy target. |
>Transmembrane family, TMEM144 of transporters (Length=355) VGLAACGVSSVFFGSMFVPIRKFDAADGIFAQWVMATAILLIGFVVFCILGFPGFYPLAMLGGMSWTIGNATAIPIISRL GMALGMLIWNTTNCLTGWAGGRFGLFGMKAHPPASSILNYGGLVCVIIGGILFSRIRSESAPEKEKDPKIRMASVESKAN GTVEKSLLSSEEGSTEPDEDSVSDRDRAVKKAGVQRILLVYLCGFVAALIAGVFYGMTFVPVIYMMDNPDLFVGYPDDGL AYVFSHFFGIFLTATVIFIGYAAVKRNRPIVPSFIFLPSLASGMLWGIAQSSFFIANQHLSQAVTFPIISMLPGCVASAW SIFYFHEIRGKRNFTILAVAMAVTITGAIMVGLSK |
| 836 |
PF07802 (GCK) |
 Estimated TM-score=0.72; easy target. |
>GCK domain (Length=72) GECGFCLFMKGGGCKDTFINWEDCVKEAEDKNEDLVEKCAEVTAKLKQCMDEHSDYYEPILRAEKHAEEQVT |
| 837 |
PF07608 (DUF1571) |
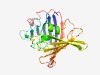 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF1571) (Length=230) YPGHRAQFKRLERIHGQLGPWQTIELKVLHNPLSVYMKFLEPDDGKEVLYSQGRFDNHLIAHPGGMARLLTPRLKLDPNG PLAMAENRHPITSAGLLNLVRRLIHFRRLDLNHPTSHLRLDRIETPDGQMVFRSCHRHLKRTDQHPYSRSEIWYDSATCL PIRVENHDWPAGIILGHTISSPADLGDPFAIFDAPDNGETLVEIYEYAHLDFETPLTPLDFDPANPAYGF |
| 838 |
PF07344 (Amastin) |
 Estimated TM-score=0.72; easy target. |
>Amastin surface glycoprotein (Length=158) FVAFMFVLVATPIDMFRIRSRDSQVLPDRCYTLWGLKTNCDSFSYQFLSDDLWESCPPRRDRFRVAQAFAVMSIFVYLAA VVLGVIMLFCCRYLRWVCLALNCFGAVTVCIVWACMAVTYNRNEGLNCLPLKRTHIYGAGFVLLVLAWLLDILNIPVL |
| 839 |
PF07223 (DUF1421) |
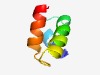 Estimated TM-score=0.72; easy target. |
>UBA-like domain (DUF1421) (Length=45) YNELIEKLVSMGYRGDHVVNVIQSLEESGQPVDFNAILDRLNGHS |
| 840 |
PF06450 (NhaB) |
 Estimated TM-score=0.72; easy target. |
>Bacterial Na+/H+ antiporter B (NhaB) (Length=520) MPMTMSQAFIGNFLGNSPKWYKIAILSFLIINPILFFYVSPFVAGWVLVLEFIFTLAMALKCYPLQPGGLLAIEAVAIGM TSASQVLHEIEANLEVLLLLVFMVAGIYFMKQLLLFAFTKMITKVRSKILVSLLFCLASAFLSAFLDALTVIAVIITVAV GFYSIYHKVASGKDFSADHDHTSENKNEEGEDQLNEAELESFRGFLRNLLMHAGVGTALGGVCTMVGEPQNLIIAAQANW QFGEFAIRMSPVTVPVFIAGILTCYIVEKFRIFGYGAQLPDAVHKILCDYDAHEDARRTNKDKMKLIVQAFIGVWLIAGL ALHLASVGLIGLSVIILATAFNGITDEHALGKAFEEALPFTALLAVFFSVVAVIIDQQLFAPVIQWALNHEGNTQLVIFY IANGLLSMVSDNVFVGTVYINEVKAALLNGQITRDQFDLLAVAINTGTNLPSVATPNGQAAFLFLLTSALAPLIRLSYGR MVWMALPYTIVLSIVGVMAIQIGFLEQMTQYFYDSHAILH |
| 841 |
PF06381 (DUF1073) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF1073) (Length=355) QLEWMYRGSWLVGKAVDVVAEDMTRAGIDLNGTSNPDDIETLHRAINRLQIWQSINDTLKWARLYGGAIAVMLIDGQDVS TPLRIETVTKDQFKGLLVLDRWMVQPSLERPVKEYGPYLGKPEFYTINQNAPALINKEVHHSRVIRIEGYELPFWQQQTE NGWGLSVVERLYDRLVAFDSTTQGAAQLVYKAHLRTYKVAGLRKIISAGGAVMDALVKQMELIRKYQSNEGLTLMDAEDQ FETHQYAFSGLADITLQFGQQLSGAIDIPLVRLFGQSPSGMNATGESDLRNYYDSINAQQERRLRLPMTTLLDVLYRSVL GSAPPSSFSFRFNPLWQMSEKERADIANVIATAVA |
| 842 |
PF06258 (Mito_fiss_Elm1) |
 Estimated TM-score=0.72; easy target. |
>Mitochondrial fission ELM1 (Length=321) PGSENQSLGLVRALGLSDNHILYRVTRPRGGINEWLHWLPVSFHKKDGPLLVVASGRDTISIASSIKRLASQNVFVVQIQ HPRSNLNRFDLVITPHHDYYPLTPHAQEQVPRFLRKWITPHEPPDRHVVLTVGALHQIDSAALRSAASAWHDEFAPLPKP LLVVNIGGPTRHCRYGADLAKQLTTYLFSVLESCGSVRISFSDTTPEKVSNIVAKELGNNPKVYIWDGEEPNPQMGHLAW ADAFVVTADSISLISEACSTGKPVYVMGAERCRWKLSEFHKSLSERGVVRPFTGSEDISESWSYPPLNDTAQAARRVHEA L |
| 843 |
PF06239 (ECSIT) |
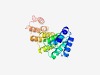 Estimated TM-score=0.72; easy target. |
>Evolutionarily conserved signalling intermediate in Toll pathway (Length=218) KSKLSLVPRPPESQRRPVKALELHEELFRQAPAGERDKASFVRTVQNFEQHNVHKRGHVDFIYLALRKMREYGVERDLAI YNRLLDVFPKEVFRPRNVIQRIFIHYPRQQECGIAVLEQMENHGVIPNKETEFLLIQIFGRKSYPMLKYLRMKLWLPRFK NISPFPVPQELPQDPVDLAKLGLRHMEPDLSARVTIYQMPLPRDSLGAADPTQPHIVG |
| 844 |
PF06115 (DUF956) |
 Estimated TM-score=0.72; easy target. |
>Domain of unknown function (DUF956) (Length=116) MVQSLNTKVDLTIKGTSYMGMSDYGQIMVGDNAFEFYNDRDANKFIQIPWEEVDYVVASVMFKGKWIPRYAIQTKRNGTY SFASKEPKKVLRAIREYVDPDRMVQSLSFFDVVKRG |
| 845 |
PF05525 (Branch_AA_trans) |
 Estimated TM-score=0.72; easy target. |
>Branched-chain amino acid transport protein (Length=427) SYIIIIGLMLFALFFGAGNLIFPPMLGQLAGKNVWIANAGFLVTGVGLPLLAITAFVFSGKQNLQSLASRVHPVFGIVFT TILYLAIGPFFAIPRSGNVSFEIGVKPFLSNDASPVSLIIFTILFFALACLLSLNPSKIIDIVGKFLTPIKLTFIGLLVV VALIRPIGTIQAPSKGYTSQAFFKGFQEGYLTLDALVAFVFGIIIVNALKEQGASTKKQLMGVCAKAAAIAAVLLAVMYT ALSYMGASSVEELGILENGAEVLAKVSSYYFGSYGSILLGLMITVACLTTSVGLITACSSFFHELFPNISYKKIAVVLSV FSTLVANIGLTQLIKVSMPVLLTMYPIAISLIFLTFLHSVFKGKTEVYQGSLLFAFIISLFDGLKEAGIKIEVVNRIFTQ ILPMYNIGLGWLIPAIAGGICGYILSI |
| 846 |
PF05506 (DUF756) |
 Estimated TM-score=0.72; easy target. |
>Domain of unknown function (DUF756) (Length=83) PEVRVCYDIANGNVYLQMRNDGKAPCKFTVRAKAYRDDGPWNATVNGGAQAEQHWELAASGQWYDFAVTCDSDPAFYRRF AGR |
| 847 |
PF05428 (CRF-BP) |
 Estimated TM-score=0.72; easy target. |
>Corticotropin-releasing factor binding protein (CRF-BP) (Length=307) MAPTFKLQCHFILICLMALRGESRYLELREAVDHDPFLLFSANLKRELAGEQLYRRALRCLDMLSLQGQFTFTADRPQLH CAAFFIAEPEEFITIHYDRVSIDCQSGDFLKVFDGWILKGEKFPSSQDHPLPTSERYIDFCESGLSRRSIRSSQNVAMIF FRVHEPGNGFTLTIQTDPNLFPCNVISQTPNGRFTLVVPHQHRNCSFSIIYPVVIKISDLTLGHLNGLQLKKSSASCGGI GDFVELLGGTGLDPSKMMPLADLCYPFHGPAQMKIGCDNTVVRMVSSGKHINRVTFEYRQLEPYELE |
| 848 |
PF05275 (CopB) |
 Estimated TM-score=0.72; easy target. |
>Copper resistance protein B precursor (CopB) (Length=207) AMIFADRLEYQSNEGDSVLLWDAQGWVGGDINKLWIKTEGEYLSNEGGFEEAEVQALYSRAISPYFDLQLGYRYDIEPDP ERHFAVFGVQGLAPYWFEVDAAAFMSDDGDVSARLEAEYELFLTQKLILQPRAELDFAVQDVPELGIGSGLSTAEAGLRL RYEFRREFAPYIGVSWRRAFGETADFARADGEDVEAVSFVAGIRLWF |
| 849 |
PF04796 (RepA_C) |
 Estimated TM-score=0.72; easy target. |
>Plasmid encoded RepA protein (Length=157) VPYGSRARMILLYLQTRALQTDSREVELGRSMHDWLDRMGLSVGGQTYRDIREQASRIAACNLTFAWEDANSLGFEKDSI VKGGIHFSSEHNSPQGTLWEDRVLLSEAFFRELKAHPVPIWEPALRHIQNNSTSIDIYIWLAYRLHSLNKSTTITWP |
| 850 |
PF04407 (DUF531) |
 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF531) (Length=169) MLTMGIINTYDKIKVLDAHYRAIARAAPICHAFGFRLALYDFPFSMTAEELVEYVTDKTTIGESGKYLRLLHEKGHFFVF DLPKKGFQPQFGTAVVTTSKPEKKRAINPEEIASGILRKSSYLLLVGLGRKGLPKEMFEQAPLHLDITSAGISLETCTAI GCIPAYIMG |
| 851 |
PF04319 (NifZ) |
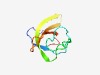 Estimated TM-score=0.72; easy target. |
>NifZ domain (Length=71) PRFRIGERVISRSSIRNDGTYTGKDMGELLVSKGDVGYVTGIDTFLQRFYIYAVHFVESDHRVGMRAKELC |
| 852 |
PF04311 (DUF459) |
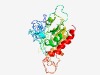 Estimated TM-score=0.72; easy target. |
>Protein of unknown function (DUF459) (Length=322) RHKSVKNIYLAFVDPLMHTAEQWGIDTVFPLLRESFLHATGLIQHPEWEDTFYHCEQRSYEPAAALAESVPSPVLQEAVA VSPPGVAVNDSVAERKTTAPARVFSRTALRILMCGDSQMRYLTGGALQVLGTSSHVQIQEVTVSSSGFVRTDYYHWPRKF LALLDTHTQQEPYAAVIMAFGMNDYQNFYDADGSLCVTKTARWERAYEQKMRACLNIILHTVPKVYLLGMPETRNKQLNE KLVYIEHVQKKVVAQYDPQRVRYYSLKPIVPGVHGTYASAIRDTHGRWVHVMHKDGIHYTIEGGAYVMETLLPLILADLE RS |
| 853 |
PF04165 (DUF401) |
 Estimated TM-score=0.72; trivial target. |
>Protein of unknown function (DUF401) (Length=389) ASSSLALLFSIAVVLILIRKVNIGLAIFVGSAILAYLTLGADGLLVMLKSLANPQTIEIVVIVILAFTLGYSMEYFGMLE KVTGGLSESFGVYSFLLIPLLIGLLPMPGGALISAVMLGPLTKMYRISPEKATLLNYWFRHVWVTFWPLYPSVIIGAAVV NVDYFHFAAATFPIGVFALTAGLTLTRGMERRFRVSRNLFDSLINLYPIAILLILSVLLRLDLLLALVVAIAAVSIHNRA KLEDFREIFRRTVDKKIILLVFAVMVYKSVIQNSGAAESLFTDISLGFPAPAAAFLLTLLVGFATGIEMSYSSIALPLLT SFTGSGIVVAKNLMLVIAAGYIGVMLSPMHLCYILTAEYFGTEIGKTYRQLLAIAGITAALVWIAYLVI |
| 854 |
PF03741 (TerC) |
 Estimated TM-score=0.72; hard target. |
>Integral membrane protein TerC family (Length=172) SLIVLEGLLSADNALVLAIMVKHLPKEQQKKALFYGILGAYLFRFIAIGVGTYLIKIWWIKAAGALYLLWMAIQFFMKKN KEEDEAAVPETPHGFWRTVLAVEIMDIAFSIDSVLAAFGVSDQVWVLYLGGIFGVMMMRGVAQIFLTLIERYPELETTAY IIIGIIGAKMMA |
| 855 |
PF03217 (SlpA) |
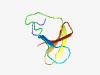 Estimated TM-score=0.72; hard target. |
>Surface later protein A domain (Length=52) KIMSKSYVYTSAGKRTGAYKSAYTYVYVIGGIVKIGSNNYYKIGTNQYIKVN |
| 856 |
PF02035 (Coagulin) |
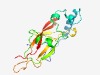 Estimated TM-score=0.72; trivial target. |
>Coagulin (Length=173) PNVPTCLCEEPTLLGRKVIVSQETKDKIEEAVQAITDKDEISGRGFSIFGGHPAFKECGKYECRTVTSEDSRCYNFFPFH HFPSECPVSVSACEPTFGYTTSNELRIIVQAPKAGFRQCVWQHKCRAYGSNFCQRTGRCTQQRSVVRLVTYDLEKGVFFC ENVRTCCGCPCRS |
| 857 |
PF02030 (Lipoprotein_8) |
 Estimated TM-score=0.72; easy target. |
>Hypothetical lipoprotein (MG045 family) (Length=484) MKKQLKYWFSLMGLTMSVGLTACSSNQFVFANFESYVSPLLLERAEAKQPMTFLTYPTNEKLINGFANNTYTVAVASSYA VSELQQQGHLLPIDWAKFNLKKTQNGSNQATIQNKEDAKELFTKEIGDISGELLNWGVPYFLQDLVFVYRGEKIQELEGQ DVTWSTIIKAIVNHKDRFNNNRLALIDDARTIFSLANVVHHEVKNTTVDVNPTGSTLNYFGNVYESFANLGLKRDNLNTL FVNSDSNIIINELANGRRQGGIVYNGDAVYAALGGDLRDEINENNLPNGDNFHIVQPKHSPVALDFLIINQQQTHFRDAA HQLIYQLALEGADQTAEELLKTDEEKGTSDEDYYTYGAMQNFSYVNYVSPLKNISDETTGIVFKENKQADTKQVVKQQSQ SEQQSESAEKEETEQDDFYTATLKSLLKADSLDDKAKKLVDTIKKTYKIKKADNINWANLIEKPITPLQRSNLTLSWLDF KQRF |
| 858 |
PF01925 (TauE) |
 Estimated TM-score=0.72; easy target. |
>Sulfite exporter TauE/SafE (Length=235) AMVGVLSGLMSGLFGIGGGTIIVPALVWLGLTQRHAVATSLAAIIPTAIAGVISYAQSGHVDWIAALLLACGTIVGAQLG SWLLSRLPEMVLRWAFAAFLVFVIVSQIVGTPSRDGTIHMTFLTGALLMLLGIFVGTLSGLLGIGGGAIVVPALSLAFGA SDLVARGTSLLMMIPNAIAGSVANVRRGLVNLRDGLIIGVVAACLTPVGAWIAGSISARLGANLFAVYLCVIGVR |
| 859 |
PF01827 (FTH) |
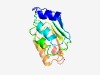 Estimated TM-score=0.72; easy target. |
>FTH domain (Length=133) LEDTRAATGTKSLLKKKSLTLRNLMAPDVFSILPYFDSNCMENIHLTGEYEEAQPLNGIVELPLWNHLKNVTLDHFFIGN IPQNITHLETFTGNVREITAEDVIQIKNMMLRSGQLKCCYMSSNWNQDESFQQ |
| 860 |
PF01697 (Glyco_transf_92) |
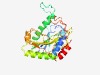 Estimated TM-score=0.72; easy target. |
>Glycosyltransferase family 92 (Length=242) KGITLCLQPVYYYSQWQNIVLYIEAWIAQGATRFIVFYHSSTKDTRKVLDYYQNLGLLEIRSWPNFGDLPIKAADKYPKI DESAFIFSYFLAMNICVLDIKTSVGSIADFDEIMVPRNGTTLEYALKEMVDTDVGALSFENNYVAMEPSIYSSDFSGVSK PIFFERGGPRKYIFNASVIDLCQVHWVRSFIDQSKKSKNADGALMHLRFNAKDFKEKRVSKPFQFFPSNTSQHIQNMKTT IR |
| 861 |
PF01050 (MannoseP_isomer) |
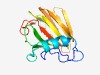 Estimated TM-score=0.72; easy target. |
>Mannose-6-phosphate isomerase (Length=151) VGIDNLVIVDTKDALLVATKDKVQDVKKIVEQLKAEGRTEFKNHREVYRPWGKYDSIDNGSRYQVKRITVKPGEKLSIQM HHHRAEHWIVVSGTAKVTNGDNTFLVTENESTYIPIGVVHALENPGQLPLEIIEVQSGSYLGEDDIVRFED |
| 862 |
PF18855 (baeRF_family11) |
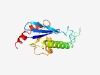 Estimated TM-score=0.71; easy target. |
>Bacterial archaeo-eukaryotic release factor family 11 (Length=138) QTAFVLALSQNAARLVEIVPDTAAETIKVPDLPTDAASHVGKSSIKDRAPSRRLQGSEGQKVRLRQYARGVDQALRPLLT GSGIPLILATTQPLDGIYRSVNSYPGLAPRSIEGNPEELSDQQLAEAARPILDEIYAE |
| 863 |
PF18761 (Heliorhodopsin) |
 Estimated TM-score=0.71; easy target. |
>Heliorhodopsin (Length=244) LRRFNLIMGFLHLIQGVLMIVLSNDTTYPIFTNYLTFNPESLSLVPDPKLFYELRFGPAVAAFLLLSAIAHFSLASFGYN WYVKNLKKGMNPVRFYEYALSSSLMIVLIGMLVGMWDLGALILIFGINAMMNLFGIMMEYHNQYTEKTNWTSFIYGSIAG IIPWIVIVLYFLGSINSSDAKPPAFVYAIIPTIFVFFNIFAVNMYLQYKKVGKWADYLYGERVYIILSLVAKTLLAWQIF AGTL |
| 864 |
PF18742 (DpnII-MboI) |
 Estimated TM-score=0.71; easy target. |
>REase_DpnII-MboI (Length=147) IIENICYKFHSVARQLQSRHSDRSTLEIEDEYDVQDLLHALLKLHFDDIREEEYTPSYAGSSSRVDFLLKDENIIIEVKK TRKNLKAKEVGEQLIIDKAHYTAHPNCKSLICFVYDPDARISNPNGLEKDLERNNNDLNVKVIIAPK |
| 865 |
PF18481 (DUF5616) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF5616) (Length=138) NQLVGREIMIDGFNLLIVLESALSGAYLFKGMDNCYRDLSGVHGSYKRVQQTEKALLLIGDFLKECAVGKVTWVFDKPVS NSGRLKTRVRELAEAHGFNWEVILDNNPDKLLAESNKIAISSDAWILDRAKRWFNLAG |
| 866 |
PF18208 (NES_C_h) |
 Estimated TM-score=0.71; easy target. |
>Nicking enzyme C-terminal middle helical domain (Length=109) HYSFDEKRMVSQLSKELKTFVSLENLDEKQRMLFNWKNSTMIKQAIGQDMTKQLITINQQENSLAEANQLLDKVVNRTVR KLYPDIDFERVTMAEKRELIKETDSEQKV |
| 867 |
PF17312 (Helveticin_J) |
 Estimated TM-score=0.71; easy target. |
>Bacteriocin helveticin-J (Length=312) LMYELENLHHVVVQATNLVNNQIFALQLLHKQTDIIVYQAEANRKHIFFDEDNPIVYLSGMAGGHTQTWVYSGKPGNYFI GTKPKKHGHILWDTQIARVNINGGSVQYDSNTEMPRLSYLNRAGSGFGDGSVAYPGKELERLEAAISPDYKRLLIASIDL NHVGHFALYDLNEVNEALDNAEASASDVNIQNLHCLVAFTIPSFNQTISSIQGYDIDQDNNVYVSCQLGPKANFLGMAKE GHPRQIVKIPWGSSNPDEWEVANLDHDHTVDTLGYVTEFESVQVVSPNEIYLTVAYHRKNDLKTLKNRVYKI |
| 868 |
PF17194 (AbiEi_3_N) |
 Estimated TM-score=0.71; easy target. |
>Transcriptional regulator, AbiEi antitoxin N-terminal domain (Length=92) EQKIKKLLDQYKPGTVCLASWLEELGISYDLQKRYRRSGWLSSIGTGAFVRPGDEVTWQGGLYALQTQANLPIHAGAITA LAMQDMSHYLRL |
| 869 |
PF17161 (DUF5123) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF5123) (Length=118) VIANNTFANITGGNSNGFARVRAVVAGGITISNNLFLNMMSSNYPFVHSNTTSENTVNLSNNFFYNFHEEWFSDMDEATA TANGGAVLTASPVADIDGGDFTLTNAVLMAKKAGDPRW |
| 870 |
PF17008 (DUF5088) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF5088) (Length=198) GQVILKSRLFTFSSGGSLPLFFSFILLTSAGLVKVYVWKLAVRKFFCIKEGDYIFIRGFKIKRRPGALLIADRTNTDSDT KYASIPEISVNLTEPTGHIMEMEMLHGLPKVPYDPEFTTVIGIVEYVSSLFRYREGKARMKFREYVYLKVSGIIIKLLAN DTESIIDIKTGTSIEVRHLRKGTIGPFEFYISSLYTQF |
| 871 |
PF16460 (Phage_TTP_11) |
 Estimated TM-score=0.71; easy target. |
>Phage tail tube, TTP, lambda-like (Length=137) VLEIEGVTSFNPGGNPADQIEDPCLSDTSRKYKKGLRTPGQATLGINADPRLASHVRLFQLSEKDGETSVKWAIGWSDGI DVKPTVSTEGDDFVLPPARTWFTFEGYVSDFPFDFASNTLVATQATIQRSGAGKWTP |
| 872 |
PF16288 (DUF4934) |
 Estimated TM-score=0.71; trivial target. |
>Domain of unknown function (DUF4934) (Length=102) LKVSDLGKTIRYIPLETTDSCLIGDFPNIKLLDDKIMVYNGKQCLLFDKETGKFICSVGHRGDDPEAYSSTCGYLNPQNQ LLYFNRDPNQLVKYNQKGNFAG |
| 873 |
PF16261 (DUF4915) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF4915) (Length=319) RQFISWLSEQRISLAFTTYQTGKLFLIGVASDRRLSVFERTFERCMGLWASPDRLYTSSLYQVWRFENALESGQLYNGGY DRLYVPQVGYTTGDIDIHDIHVDSQGRVIFVNTLFSCLATISDRYSFIPLWQPSFISRLAAEDRCHLNGLAMENGQPRYV TAVSQSDVSDGWRDRRRNGGCVIDVPSNEIVVSGLSMPHSPRVYRDRVWVLNSGTGYFGTVDINAGKFNPIAFCPGYLRG LALWGDYAVVGLSKMRGNKTFSGLPLDENLAAKDVEPRCGLQVIDLRSGDIVHWLRIEGMVEELYDVAVIPGVRQPMAL |
| 874 |
PF16065 (DUF4807) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF4807) (Length=120) IDYEWNDRMNYLIWTRVEIENAFSWLSTLGGAYSALGDYFDHCAEEAGRISLRQYKLSRLLGDDGLAARSKLYSSIAYAQ RGKLKLARYIVRDIAAFARQTHDRRLNRMCQGVWAKLKYL |
| 875 |
PF16030 (GD_N) |
 Estimated TM-score=0.71; easy target. |
>Serine protease gd N-terminus (Length=113) SPCPAVFSYDERDDSYDTWFGTIRLKSNVPLYGIFVDIIFSSPVLSFGTFLRQYNTDDNVHFHIMDKSYRVRAGEVVIIK FFVRYAPDQPVPLLRQIRFNGQNICTETRQNPV |
| 876 |
PF15976 (CooC_C) |
 Estimated TM-score=0.71; easy target. |
>CS1-pili formation C-terminal (Length=93) LYPGNVAVIEPEVKQMVTVSGRIRAEDGTLLANARINNHIGRTRTDENGEFVMDVDKKYPTIDFSYGGNKTCEVALELSQ ARGAVWVGDVVCS |
| 877 |
PF15405 (PH_5) |
 Estimated TM-score=0.71; trivial target. |
>Pleckstrin homology domain (Length=128) IVFKSQLRKTPHDSSPANEITSYLFDHAVLLVRVKQAGKAEEIKAYRRPIPLELLAIREMDEVIPQQGVKRTSSSLLPLG GNNAKDGKKSEGWPITFRHLGKAGYELTLYAANQAGRDKWLQFIDKAQ |
| 878 |
PF14966 (DNA_repr_REX1B) |
 Estimated TM-score=0.71; trivial target. |
>DNA repair REX1-B (Length=97) LVQRFYALHAERVEAYRLFEEGHEAYLRTAPHYDFQRYRQLVHEITLAFSGISKELIEMKERLHEECERPDLSEHIEKLQ EKERHKLELTAKLQLAK |
| 879 |
PF14900 (DUF4493) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF4493) (Length=223) ETGNISLSLTASTGFSANTRSVNEADYKNVNQYTIQILNSAGVEKVNYLYDDLPSKIKLENGSYTLKAFYGEDKDASRNG FYVEGSSTFTVQGADQTLNVDCAPVCGKVAVNFDESMDKYFSDYSVVYETSALTAANTTATWVKADTEPWYLKLNKDGET VKATIHVTRTSDGKSITTEKTYNLERNKSWTLNIAPKDDNGSMTITISIDESTEDHEIDIVVP |
| 880 |
PF14891 (Peptidase_M91) |
 Estimated TM-score=0.71; easy target. |
>Effector protein (Length=172) AQFIKIEGSDEFRERVLADLDMYRASPTGQQMLANLEDKRDPNFLFGENTLTIKELNEENGYASRGHKFVNSDSVIQYNP AFTFDSERGRPSVVLYHELGHVYDYWNDTFDGDTIAGGPDDGIKKAERQATGLPFDHDDDPSTPDRLDPDHPIQYTENGL RREMGMRERDTY |
| 881 |
PF14500 (MMS19_N) |
 Estimated TM-score=0.71; easy target. |
>Dos2-interacting transcription regulator of RNA-Pol-II (Length=261) VQVLGEFLTNEDGTIRAKGTGLLSTVLADCPQEQVNNAAINVLVDFYCERLSDTASVPELLQGIESLAHFNSFHNENSLK VAKSIFASVNVQSFQQTTRHYVFQIFDRLLKHHLNALKQMRQDFVTGFIQAMDGEKDPRNLLLAFSLIKLIIHEFDISQH VELLFEVTFCYFPITFKPPPDDPYGITADDLKIALRECLSATPYFAELAMPLLLEKLTSSSGNAKKDSMETLTACAPVYG AKSIVPYLEDLWDYLKDEIVN |
| 882 |
PF14452 (Multi_ubiq) |
 Estimated TM-score=0.71; hard target. |
>Multiubiquitin (Length=63) LNFLSRQVSDPVPLGRQLLEAAALDPQDGYSLFAVLPSGDFEDVRLDEPFDLREHGTERFVTF |
| 883 |
PF14387 (DUF4418) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4418) (Length=117) FIILGLLIAVGPQSIFAVCGPMEDGKFMKCHWTAQAELGIGLAIVFLAAGILFFKSEQIRAGLSIAAFALSIVSVLVPTK LIGVCGGEHMQCHSLTKPVLIILGIAGAVFALANTFF |
| 884 |
PF14373 (Imm_superinfect) |
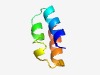 Estimated TM-score=0.71; hard target. |
>Superinfection immunity protein (Length=42) LYFLPMMIAFKRHHENYTAIFMLNLLLGWTGIVWVVCLIWAF |
| 885 |
PF14289 (DUF4369) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4369) (Length=84) FTIKGYVAGEKETKKVYLRYGQQFDSTLMKDGRFEFSGYVSDPLRATIWCNRNNSIDFYLDPADVQLSSMNSLSGAVVNG GPTE |
| 886 |
PF14109 (GldH_lipo) |
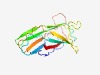 Estimated TM-score=0.71; hard target. |
>GldH lipoprotein (Length=136) RVYDEYQSVPERGWDKDSMITFKVEKIDTLKPYNLFINLRNNNNYNYNNIFLITEMNFPNGKVISDTLEYEMAYPNGDWM GQGFGDVYESKLWYKENVRFPETGTYQFKIRQAMRKNGNVNGISELQGITDVGFRI |
| 887 |
PF13711 (DUF4160) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4160) (Length=63) GIIIYMYFNEHNPPHFHAEYNEYKASISIKTLGLMEGKLPSKVMSLVVEWAQEHQDELLENWN |
| 888 |
PF13613 (HTH_Tnp_4) |
 Estimated TM-score=0.71; trivial target. |
>Helix-turn-helix of DDE superfamily endonuclease (Length=51) LTKFQEFCMVLVKLRLSVPHLDLAYRLKISTSSVSRILVTWITVMDIRLTP |
| 889 |
PF13446 (RPT) |
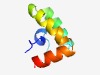 Estimated TM-score=0.71; trivial target. |
>A repeated domain in UCH-protein (Length=59) TVEQALVFLGVDEQTSDDFIITMYTAKISDSPSSKDLARRAVELIAEARKSDTLKHFLK |
| 890 |
PF13376 (OmdA) |
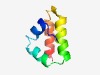 Estimated TM-score=0.71; hard target. |
>Bacteriocin-protection, YdeI or OmpD-Associated (Length=59) PRVPTDLGKALAATPTAKVQWNDLTPIARRDFITWIDSAKQPETRRRRIERACSMLAAG |
| 891 |
PF13335 (Mg_chelatase_C) |
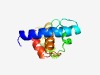 Estimated TM-score=0.71; easy target. |
>Magnesium chelatase, subunit ChlI C-terminal (Length=97) EKSSAVRERVIAARKIQEERFKDSQGVFTNAQMNSKLLRKHCQIDEAGNELLKNAMEKLGLSARAYDRILKVSRTIADLA GHENINNEHLSEAIQYR |
| 892 |
PF13100 (OstA_2) |
 Estimated TM-score=0.71; easy target. |
>OstA-like protein (Length=158) SKVYLLHSDVLKKSPLNPDPDAQILIGNVAFRHDSIYMYCDSACFYEKTNSLEAFDNVKMVQGDTLFLYGDYLFYDGNTQ IAQVRYNVRMENKNTTLLTDSLNYDRIYNLGYYFEGGTLMDEENVLTSEWGEYSPATKISVFNYDVKLVNPKFTLTSD |
| 893 |
PF13058 (DUF3920) |
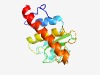 Estimated TM-score=0.71; easy target. |
>Protein of unknown function (DUF3920) (Length=126) VLDSEFPWDIQRVKNDIFSLIEKREIPVIFCDTCDTNNVLVNLGEEEEEFLFPLSGFYHKERQMIFVCMWEQYEQVLKTL LHEFRHSMQHEKNVLYIGKEAYEARWIEKDARAFAERKMNEYIRRN |
| 894 |
PF13005 (zf-IS66) |
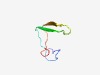 Estimated TM-score=0.71; easy target. |
>zinc-finger binding domain of transposase IS66 (Length=46) PTACHCCGGHRLRRLGEDITETLEVVPRQWKVIQHVREKFPCRDCE |
| 895 |
PF12953 (DUF3842) |
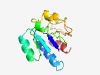 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF3842) (Length=129) RIAVVDGQGGGIGKAIVEKFRKEFEDTIEIIALGTNSLATSLMLKAGANEGATGENAICYNVSKVDLILGPIGIVCANSL LGELTPQMAKAISESPAKKVLIPLNRCNVIVAGVENKPLPHYIDDAIQI |
| 896 |
PF12852 (Cupin_6) |
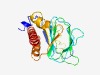 Estimated TM-score=0.71; easy target. |
>Cupin (Length=190) DVLAGLLDGPRARGAFLLRSILNPPWSLRIQDRAPLTLVAMMRGQAWVIPDHGETVRLRPGDVAIMRGPDPYTVADDPAT PPQVVIHPGQHCTTPEGESLAQEMDLGVRTWGNGPDGSTVMITGTYQMHGEISRRLLAALPALLVLPEDAWDCPLIPLLG AEIVKDEPGQEAVLDRLLDLLLIAVLRAWF |
| 897 |
PF12637 (TSCPD) |
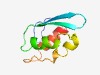 Estimated TM-score=0.71; easy target. |
>TSCPD domain (Length=74) YKTKGVCSQQIHFEIEDNKIHNVSFVGGCNGNLQGISHLVEGMDVDEAISRLEGIQCGFKTTSCPDQLAKALKE |
| 898 |
PF12388 (Peptidase_M57) |
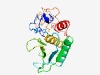 Estimated TM-score=0.71; easy target. |
>Dual-action HEIGH metallo-peptidase (Length=206) VYVGRDAHVTLEASREMLQPATKETQEQYRTTNLVGTSITKICVNPTSGFNSYSRLSQGLDLAIQNYNQRGLSFTMARGP TTGCSANITATTMSGTGGSAGFPSGGRPYGTINIGTGLQSYSVDVNEHVITHELGHAIGFRHTDYYNRNISCGSGGNEGT AGVGAIHIPGTPTTATVGGSIMNSCFRSTETGEFTSSDITALNYLY |
| 899 |
PF12385 (Peptidase_C70) |
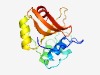 Estimated TM-score=0.71; trivial target. |
>Papain-like cysteine protease AvrRpt2 (Length=140) KLVVPYIPQNGTRGCWEATMNMLRAYFGQPPLDKGQLAYLYDAEGVPKELNGPLDYATRNRKAKLKNIPVPAGKSWDADK IRALLDQHGPLEARIESPNDELVAHAIVLTGIDEDGDVICHDPELGPGQKVSIAMLNAAF |
| 900 |
PF12096 (DUF3572) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF3572) (Length=83) QESAETIALQALGWLAGNEELLPVFLGATGASEADLRSRATEPDFLGSVLDFLMMDDAWVVEFCDSLGLSYETPMMARAA LPG |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218