

Go to page (83 pages in total): [First page] << < 23 24 25 26 27 28 29 30 31 32 33 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 2701 |
PF10507 (TMEM65) |
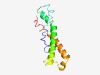 Estimated TM-score=0.60; hard target. |
>Transmembrane protein 65 (Length=107) LAPFIGFGILDNMIMIFAGEYIDHTIGVWLSISTMAAAALGNIISDVGGIGLAHYVEYLVSKAGIRYPILTSEQLVSPCA RMVINVARSIGIVIGCLIGMFPLIFFD |
| 2702 |
PF10497 (zf-4CXXC_R1) |
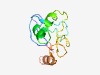 Estimated TM-score=0.60; easy target. |
>Zinc-finger domain of monoamine-oxidase A repressor R1 (Length=99) YDKVLGNTCHQCRQKTIDTKTVCRNQSCGGVRGQFCGPCLRNRYGEDVRSALLDPDWMCPPCRGICNCSYCRKRDGRCAT GILIHLAKFYGYDNVKEYL |
| 2703 |
PF10325 (7TM_GPCR_Srz) |
 Estimated TM-score=0.60; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Srz (Length=264) ICYIVLMLIFFGLLPFYVYVNKVNRVRDESIFIFPITNHFYEMVKKAYFLFLYLVVSFILCFVFGLNDSNYCVPAFCTSC ITCFALYIIVQVFHLLISLLAVQRLVIYFVPSMEKQTIWVQNKLYGNIWYIYIAFGLKESIGIVCLFVCEVAACSPSKQM TFRIFYMSTFFFLNAFLVTSALLYIPITLSIWKLTSLTMNQDKPQKYIFFQTVVIFIFKAACVPALIAFIFCDYSINVSI VFVASMDIIIIPLIIQISYLGCNK |
| 2704 |
PF10192 (GpcrRhopsn4) |
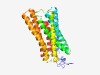 Estimated TM-score=0.60; easy target. |
>Rhodopsin-like GPCR transmembrane domain (Length=258) ELEYDIQLVNGNPNRQSASFFNPLLFHFSFDQQNTLEMYLIFFVIYLVMVPLQIYAVRLQKHPVTRLFTISLLLEFISVC LLLTHTVRYAMNGVGNENLAITGDIFDIFSRTSFMLILLLLAKGWAVTRLQISVSSWILLMVIWIPYCAIHVLLYIWNRT EVDIISDIDEYQTWPGWLVLACRSTMMLWFLWELRITMKYEHSTQKLDFLLHFGASSLVWFIYLPIVAIIAVNVNPLWRY KLLLGITNSADCLAYCVM |
| 2705 |
PF10191 (COG7) |
 Estimated TM-score=0.60; easy target. |
>Golgi complex component 7 (COG7) (Length=765) DFSKFLADDFDVKDWINAAFRAGPKDGAAGKADGHAATLVMKLQLFIQEVNHAVEETSLQALQNMPKVLRDVEALKQEAS FLKEQMILVKEDIKKFEQDTSQSMQVLVEIDQVKSRMQLAAESLQEADKWSTLSADIEETFKTQDIAVISAKLTGMQSSL MMLVDTPDYSEKCVHLEALKNRLEALASPQIVAAFTSQSVDQSKVFVKVFTEIDRMPQLLAYYYKCHKVQLLATWQELCQ RDLPLDRQLTGLYHALLGAWHTQTQWATQVFKNPYEVVTVLLIQTLGALVPSLPMCLSEAVERAGPELELTRLLEFYDTT AHFAKGLEMALLPHLQDHNLVKVVELVDAVYGPYKPYQLKYGDLEESNLLIQISAVPLEHGEVIDCVQELSQSVHKLFGL ASAAVDRCAKFTNGLGTCGLLTALKSLFTKYVSHFTNALQSIRKKCKLDDIPPNSLFQEDWTAFQNSVRIIATCGELLRQ CGDFEQQLANRILSTAGKYLSDSYSPRSLAGFQDSILTDKKSPAKNPWQEYNYLQKDNPAEYANLMEILYTLKEKGSSNH NLLSVSRTALTRLNQQAHQLAFDSVFLRIKQQLLLVSRMDSWNTAGIGETLTDDLPAFSLTPLEYISNIGQYIMSLPLNL EPFVTQEDSALELALHAGKLPFPPEQGEELPELDNMADNWLGSIARATMQTYCDGILQIPAVTPHSTKQLATDIDYLINV MDALGLQPSRTLQNIATLLKAKPEEYRQVSKGLPRRLAATVATMR |
| 2706 |
PF10078 (DUF2316) |
 Estimated TM-score=0.60; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2316) (Length=89) MSLTIIERRNTKKELAANFNLAGVTLEQAAQDLATTPEHVEAVLQLQVDQIEEPWILRNYLNKQLAAQGKTPLPYSRLQG DPAEYWFLN |
| 2707 |
PF10056 (DUF2293) |
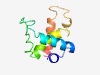 Estimated TM-score=0.60; hard target. |
>Uncharacterized conserved protein (DUF2293) (Length=92) ALRDLFPRIPNTDRQIIIEHAFTRISQRASGKGEQPVGFSDDIPLARRVQLAVLAHIRHSHTRYDDLLKEAGWQAARKAV EELCLDILVKWR |
| 2708 |
PF10048 (DUF2282) |
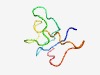 Estimated TM-score=0.60; hard target. |
>Predicted integral membrane protein (DUF2282) (Length=49) WEKCAGIVKVGMNACGTSKHGCGGAAKTDADPEEWIMVPQGTCDKIVGG |
| 2709 |
PF09965 (DUF2199) |
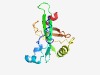 Estimated TM-score=0.60; very hard target. |
>Uncharacterized protein conserved in bacteria (DUF2199) (Length=145) TCRSCGEVHEGIPSFGADAPQPYTALPESEREQRAELSSDQCIIDGKEFYVRGCLEIPVHGQEEPLVWGVWVSVSEQSFQ RMSERWDELGREKEPPSFGWLCTHVPLYPDTLLLKTHVRTRPVGQRPFIELEPTNHPLAVDQREG |
| 2710 |
PF09947 (DUF2180) |
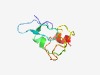 Estimated TM-score=0.60; easy target. |
>Uncharacterized protein conserved in archaea (DUF2180) (Length=67) MKCYICAEQGKDTDAVAICIVCGMGVCMDHAIREEIEKWEGGYPFPAKKLKLSIPRILCPPCYNAMK |
| 2711 |
PF09935 (DUF2167) |
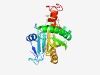 Estimated TM-score=0.60; hard target. |
>Protein of unknown function (DUF2167) (Length=239) PQQGEIKLPNGVATLQVPENFYYLSPEDTETVLVKIWGNPPGGEKTLGMIFPTRVTPYDNTAWGVTVEYVEDGYVSDEDA DKINYDELLSDMKESTANASQERIKQGYDAISLIGWAAKPYYDEKTHKLHWAKEIKFGSQEINTLNYNIRVLGRKGVLVL NFIAGMDQLDMINSKIDTVLKIADFNEGSRYADFNPDIDTVAAYGIGALVAGKVAAKVGILATILLFLKKLWIFLLIGA |
| 2712 |
PF09933 (DUF2165) |
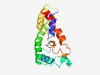 Estimated TM-score=0.60; hard target. |
>Predicted small integral membrane protein (DUF2165) (Length=155) RLSKVALVAAIAFFASLVSFGNITDYGTNYEFVQHVFDMDTIFPNSTIKYRAITSPALHTAGYILIIAAETLTAILCWIG TYAMLTKLGAPAAAFNNAKKWAIAGLTLGFLTWQVGFMSIGGEWFGMWMSHQWNGIDSAFRFFITIIAVLIYVSM |
| 2713 |
PF09894 (DUF2121) |
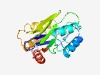 Estimated TM-score=0.60; easy target. |
>Uncharacterized protein conserved in archaea (DUF2121) (Length=198) MSLIITYVGSKGCVMAGDKRSIGFLGDKEQREVLEEEMYSGKIKTTEELLQRAEQLDINLKITDDVEKVRKIGDVLVGEV KLRTTTETKRKRIYGTSGGYHEVELSGSQIKNAKSGQSSIVVFGNKFTKEIANKHLQKYWKSQTSIKEIAKIFQRIMEDV AQSTPSVSPQFDIYTVHPQLNHKQAMEILRTTIVKDVK |
| 2714 |
PF09808 (SNAPc_SNAP43) |
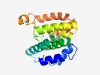 Estimated TM-score=0.60; hard target. |
>Small nuclear RNA activating complex (SNAPc), subunit SNAP43 (Length=185) LQTDCEALLSRFQETDSVRFEDFTELWRSMKFGTIFCGRMRNSEKNMFTKEALALAWQYFLPPYTFQIRVGALYLLYGLY NTQLCQPKQKIRVALKDWDEVLKFQQDLINAQHFDAAYIFRKLRLDKAFHFTAMPKLLSYRMKKKIQRAEVTEEFKDPND RVMKLITSDVLEEMLNVHDHYQNMK |
| 2715 |
PF09791 (Oxidored-like) |
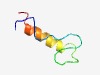 Estimated TM-score=0.60; hard target. |
>Oxidoreductase-like protein, N-terminal (Length=44) IAGVVVPSRPHEPDNCCMSGCVNCVWELYNDDLKEWKTKRKEAA |
| 2716 |
PF09662 (Phenyl_P_gamma) |
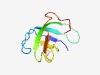 Estimated TM-score=0.60; hard target. |
>Phenylphosphate carboxylase gamma subunit (Phenyl_P_gamma) (Length=83) NQWEVFVMDLAELPEGTELELSVRTLNPGLKKYTYQRVKAELSNALDKFPDRLQVRFGRGQLCSQQFSIRIIEQVQRMPA KYL |
| 2717 |
PF09601 (DUF2459) |
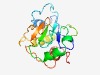 Estimated TM-score=0.60; very hard target. |
>Protein of unknown function (DUF2459) (Length=170) IYIYSNGVHTDIVMPVKNDIVDWSSKIPFTNTKSKETDYNYVGIGWGDKGFYLDTPTWADLKVSTALKAAFWLSESAMHC TYYKTMTEAEDCKKIMISKNQYKELVKFVDAKFDRDQNGNYILIPTNAVYGNNDAFYDAQGRYSFLNTCNTWSNNALKSA GQRAALWTPT |
| 2718 |
PF09571 (RE_XcyI) |
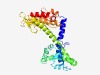 Estimated TM-score=0.60; easy target. |
>XcyI restriction endonuclease (Length=308) YQRMEELKKTTLQEALYKTVEKADIGRLDEELRKYVKNSDLQLLARYGTRGEVMFVVPYVLELNPKLIGYYRLLLGFPQK SFYTGDRGLGFGVFKSMESTGKISKAQQPLIEDLVIAFNKSASLLLNSIEKVSISSSLFRDLTLLTLGPQLRGGRNNDLG IAATKMVFEIIREIVSPAIKESTDTKIVLSNAAGRETTIEFSSDPDIIIREKLKSGKFKNWIAIEIKGGTDLANAHNRLG EAEKSHQKARLAYYTECWTLIGFNIDLELAAKESPTTDKFYHISYLVDKNNAEYEDFKENIIALAGIS |
| 2719 |
PF09501 (Bac_small_YrzI) |
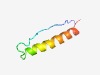 Estimated TM-score=0.60; hard target. |
>Probable sporulation protein (Bac_small_yrzI) (Length=44) MKFKAFFLTITIQKRKLSQNEILHEQHVQNIMDEVKERQASYYS |
| 2720 |
PF09355 (Phage_Gp19) |
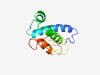 Estimated TM-score=0.60; easy target. |
>Phage protein Gp19/Gp15/Gp42 (Length=116) LWRNLKPDEQERAKALLKTVSDSLRVEARKVGKDLDKLVEADSDYANVVKSVTVDVVARTLMTATDQEPMTQISESALGY SFSGSYLVPGGGLFIKDSELKRLGLKRQRFGVMEIY |
| 2721 |
PF08820 (DUF1803) |
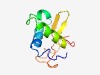 Estimated TM-score=0.60; easy target. |
>Domain of unknown function (DUF1803) (Length=89) IINPTRLTRQPFFKDLINYLDQHDDVILRQIKAQFPDQPVDKLMEEYIKAGFILRENKRYTLNLPFLESADRVDLDQEVF VREDSAVYQ |
| 2722 |
PF08611 (DUF1774) |
 Estimated TM-score=0.60; hard target. |
>Fungal protein of unknown function (DUF1774) (Length=95) RIFGNVFIWSILVYGLFFIVVYKDYTMGFSLSVLAASIGVAQFTRKIIAFQWIFAFVIMSVLFVLTLVVAVPAWTGREIK WRRAPEADQERAPLL |
| 2723 |
PF08471 (Ribonuc_red_2_N) |
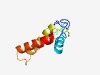 Estimated TM-score=0.60; easy target. |
>Class II vitamin B12-dependent ribonucleotide reductase (Length=101) DGSKVFEMNDVEVPKQWSQVAADILAQKYFRKTGVPQADEKGKPVLDENGKQVTGSEHSVRQVVHRLVGCWKEWGEKHKY FDTPEDAQAFYDEVAFMLLAQ |
| 2724 |
PF08401 (DUF1738) |
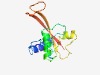 Estimated TM-score=0.60; hard target. |
>Domain of unknown function (DUF1738) (Length=122) DVYARLTAEIVAAIEAGAGEYVMPWHHNGSSTALPVNVASGKAYRGVNTLALWVAAQSAAYPMGLWGTYRQWQALGAQVR KGERSTTVVFWKQIGGDCDEDDSGSDDGAERRRFFARGYSVF |
| 2725 |
PF07959 (Fucokinase) |
 Estimated TM-score=0.60; easy target. |
>L-fucokinase (Length=405) AGGLSQRLPNASALGKIFLALPLGDPVFQMLELKLAIYVDFPLYMKPGIVVTCADDIELYSISETENIAFDRSGFTALAH PSSLSIGSTHGVFVLEPEKKSKICQMEYRSCYRFLHKPSIDEMHSSGAVCNKAAEIVALNDSEFVYTDSTYYVDHTTTVR LLALLKVIEPLCCEIDAYGDFLQALGPGATSTYTQNSANVTEEENNLIQTRQKIFHELQGMPLNVILLNNSKFYHVGTTR EYLFHFTKDHNLRAELGLVSNAFSICPREIKGSCIMHSVVHPSCDIASGSVIEYSRLDANVTVEKGTIISGCQIGNGFRV PSGVFMHSLSVSISGATTFVTVSFAIEDNLKQSLTTPKEINKLHYFGVSLEKCLDHWGISVEELKFSGDRHSFSLWNARI FPQCS |
| 2726 |
PF07751 (Abi_2) |
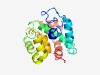 Estimated TM-score=0.60; hard target. |
>Abi-like protein (Length=186) TLSRISFYRLRAYTYPFQNNDDPNHPFIVDVDFETIIKLYKFDRKLRLLVFDAIEKIEIALRTQIIYHFALTHGSHWQTD PNLFRDIARFAKHLETLNKEVERSDETFIKHYKEKYSSPSQPPSWMSLEVASMGTLSKFYQNLKKSPEKETISRSFGLPK FFILENWMFCFSSLRNICAHHGRLWN |
| 2727 |
PF07597 (DUF1560) |
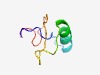 Estimated TM-score=0.60; hard target. |
>Protein of unknown function (DUF1560) (Length=49) CTLGFLHPTSFAKRSRWECRRFEAKHPLSKNISVRSAEISVPLAALAEH |
| 2728 |
PF07569 (Hira) |
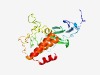 Estimated TM-score=0.60; trivial target. |
>TUP1-like enhancer of split (Length=239) GRRILNGMVLEAQPVIVECRGWWLLCITAVGMCYVWNIKSLSSPHPPVSLAPILEIAMHSLGMHVTSAPGVTSAHLNSNG HIVVTLSNGDGFLYSPNLYCWQRLSEAWWAVGSQYWNSNDSSISSLQSTGVGPDATRNGEVSSSNISAGIIPHLERHTTN EVLLKGRAYTLQRLVKTLLSKEGFESFESSVSIAHLENRVAAALQLGAKEEFRIYLFMYAKRLGAEGLKSKVEELLKSL |
| 2729 |
PF07507 (WavE) |
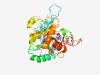 Estimated TM-score=0.60; hard target. |
>WavE lipopolysaccharide synthesis (Length=293) ITVIVQGDVDGEKTPICLQSIRRALPGAFIILSTWEGTDVEGLEYDEVVFSEDPGGFDEFNVPFSPKNNINRQIVSTYHG LLKAKTVYALKLRTDFYLCDDGFKDYFNIYNRECTEYKIFEHKVICCELYSRNPRYIFSDMKRAYHPSDIFFFGYTKDLV RLFDVPLFSKEDEEYMMWKYYPNNSISTFRYDAESYLWMNLVNKGISFKAPDDLEKYTREDIDRSEATFAANLIILSNDD IGLKTLSNSIYRKEAVPQNCYTHEDWEKLYAFYCCNDAKEYIKYLLLIFVKNL |
| 2730 |
PF07482 (DUF1522) |
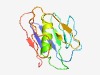 Estimated TM-score=0.60; hard target. |
>Domain of Unknown Function (DUF1522) (Length=114) DTLTVNGKTITFRSGSAPGATGVPTGSGVSGNIVTDGNGNSTVYLGTNGTPAATVNDLLSAIDLASGVKTVSISSGAATI ATSNGQTASTVAAGAVTLRSSTGADLSVTGKADL |
| 2731 |
PF07459 (CTX_RstB) |
 Estimated TM-score=0.60; hard target. |
>CTX phage RstB protein (Length=90) KVMVIGVEHSKGLSKANDKPYDFAQVNYLAPNEGWQSTKGSCKAVGLDKKTIAMSTQPELFAAFAQLDYPVMCELSLDCD PANPQRNHVV |
| 2732 |
PF07441 (BofA) |
 Estimated TM-score=0.60; hard target. |
>SigmaK-factor processing regulatory protein BofA (Length=75) IAIVGLYVIVKVFSWPIKILFKLIINAVLGVVLLVIVNLIGSYFGFSIGINAVTALIAGFLGIPGVIFLIIFKLF |
| 2733 |
PF07438 (DUF1514) |
 Estimated TM-score=0.60; hard target. |
>Protein of unknown function (DUF1514) (Length=62) MWIVISIVLSIFLLVLLSSISHKMKTIEALEYMNAYLFKQLVKNNGVEGLEDYENEVERIRK |
| 2734 |
PF07283 (TrbH) |
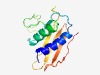 Estimated TM-score=0.60; hard target. |
>Conjugal transfer protein TrbH (Length=129) LDQQKLATDAVQQLATLYAPARTRLELQQPTPDPFGQALVKSLRDKGYALLEYAPAGASAQAPASAASQPSAPATTAAPG GLPLRYVLDQAGDSNLYRLTLMVGNQSITRPYLAQNGTFAPAGYWVRKE |
| 2735 |
PF07280 (Ac110_PIF) |
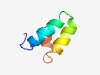 Estimated TM-score=0.60; hard target. |
>Per os infectivity factor AC110 (Length=43) VLMVVVFVTCVFILITLRLNKFQLQELLYYQYNYIPESLLSVV |
| 2736 |
PF07059 (DUF1336) |
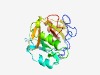 Estimated TM-score=0.60; very hard target. |
>Protein of unknown function (DUF1336) (Length=213) WSQPDWMQFKLRSKTYLQNKIKETGAPPLWFESFRGTHEELMHVCSSKKSFASKALAKFGNDMPQLFVVTLIIPGSPLVA TVQYFARTKSSGSEDPTEAEKLWERFLDADDAFRSSRFKLIPTIVDGPWIIRKSVGTTPCIIGKAIKTTYFKGPNYLEVH VDISSDTVAKHITSLCRSQSTHFTVDMGFVIEGQSEEELPEALLGCVQYARMD |
| 2737 |
PF07035 (Mic1) |
 Estimated TM-score=0.60; hard target. |
>Colon cancer-associated protein Mic1-like (Length=160) DQFLVQVVTAYVASLCRNSLVIQPVTSELMVRALVTRGAAARLRAALRRGALGDARALACALLSLGHLDPAAAQLALDML WRLKAYEEVVEVLLSWEEPVGAAGAARQAGTTWLSLPPRKLLAAARAHAQAHGRPTAFTAIYHALQNRNERTRGSPHFLK |
| 2738 |
PF06985 (HET) |
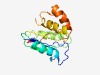 Estimated TM-score=0.60; very hard target. |
>Heterokaryon incompatibility protein (HET) (Length=138) YDALSYRWGDPNDAVKILVNDAELKVTRNLSIALQNLRHQDRELVIWTDAICINQMDIEERQVQVTLMGVIYSKADCVQI WLGEASLDTEAGMDLANSCGNTDAAEVVKKICADDRGSAGLTELLERPYWSRIWMFQE |
| 2739 |
PF06868 (DUF1257) |
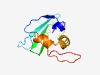 Estimated TM-score=0.60; very hard target. |
>Protein of unknown function (DUF1257) (Length=103) ALTDLGIDWKAGPEQVRGYQGQTRNAEVAIEQENGYDIGFSWNGSEYELVADLQFWQQAWSVDRFLQKVTQRYAFHTVMS ETAKQGFQVTEQEQNQDGSIRLV |
| 2740 |
PF06569 (DUF1128) |
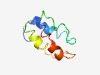 Estimated TM-score=0.60; hard target. |
>Protein of unknown function (DUF1128) (Length=71) LTQPSQENVEYMIEQIKEKLRMVNVDAMKSEHFDDSQYEDLHDLYAMVMKRDTFSPSEMQAIASELGSLRK |
| 2741 |
PF06454 (DUF1084) |
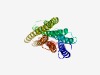 Estimated TM-score=0.60; easy target. |
>Protein of unknown function (DUF1084) (Length=272) VSDWWDEINESTQWQDGIFYALCAAYALVSSVALIQLIRIELRVPEYGWTTQKVFHLMNFIVNGVRAIVFGFHKQVFVLH PKVLTLVLLDLPGLLFFSTYTLLVLFWAEIYHQARSLPTDKLRIFYMSINGVIYFIQVCIWVYLWIDDNSVVEFIGKIFI AVVSFIAALGFLLYGGRLFFMLRRFPIESKGRRKKLHEVGSVTAICFTCFLIRCFVVVLSAFDSDASLDVLDHPVLNLIY YMVVEILPSALVLYILRKLPPKRISAQYHPIR |
| 2742 |
PF06291 (Lambda_Bor) |
 Estimated TM-score=0.60; hard target. |
>Bor protein (Length=78) QQTFVMAPQESEATTPTTEKSHHFFINGLAQKKQIDAAKVCNGIDNVAKVEVQETFVNGLLAAVTFGIYTPREARVYC |
| 2743 |
PF06196 (DUF997) |
 Estimated TM-score=0.60; hard target. |
>Protein of unknown function (DUF997) (Length=77) QAHKEARIGVGLVLFNFLWWYGFAYGLGSRPAKQYHYIFGMPSWFFYSCIAGYIVVTALVIFIVKFFFKDIPFDDEE |
| 2744 |
PF06119 (NIDO) |
 Estimated TM-score=0.60; very hard target. |
>Nidogen-like (Length=78) NTFQTVLSTDGNQTFLLYNYGNIQWPSMNWDGPLALAGLNSGYDTGHYTLPGSLTLSVANLSSTSNVNVTGRWAFKVD |
| 2745 |
PF05715 (zf-piccolo) |
 Estimated TM-score=0.60; easy target. |
>Piccolo Zn-finger (Length=57) LCPVCKNTELNLKSKDSLNHNTCTQCKSEVCNMCGFSPPDATAKEWLCLNCQMQRAL |
| 2746 |
PF05656 (DUF805) |
 Estimated TM-score=0.60; hard target. |
>Protein of unknown function (DUF805) (Length=110) YARFDGRATRSEYWLFVLVQWVLLVGVIAGLAAALPREAASAPFMVMGVATVLITLLLPNLAVTSRRLHDSGLSFAWWLI SFIPLIGGFVNLVLMCLGGTRGGNQYGPDP |
| 2747 |
PF05611 (DUF780) |
 Estimated TM-score=0.60; hard target. |
>Caenorhabditis elegans protein of unknown function (DUF780) (Length=72) MADNSAYMSAGGCSFGYMGSKTSSSGYAREEYASGGSGGEACINQNQRSGRNTNPGQQVFKSRIDKSCYLEP |
| 2748 |
PF05531 (NPV_P10) |
 Estimated TM-score=0.60; easy target. |
>Nucleopolyhedrovirus P10 protein (Length=77) MSQNILLLIRADIQAVSQKVDVLQSSVDDVRANLPDVTELNEKLDAQSASLATLQTAVDAITDILNPEIPEIPEVPD |
| 2749 |
PF05404 (TRAP-delta) |
 Estimated TM-score=0.60; hard target. |
>Translocon-associated protein, delta subunit precursor (TRAP-delta) (Length=155) ALFSGANGQNCQNPKVEAASFTSLDATIVTQIAYITEFTLVCDNPLPPNYALYAEVDGKPLTAARIGENKYQVSWTEEPA AARSGMHEVRVLDEEGWASLRRARRADPAATVAPLLAIQLHHPGSYSGPWVNSEVLATALSVLIAYIALHNKSKI |
| 2750 |
PF05177 (RCSD) |
 Estimated TM-score=0.60; very hard target. |
>RCSD region (Length=90) ETTIEKTETTMTTEMTHESEESRTSVKKEKTPEKVDEKPKSPTKKDKSPEKSITEEIKSPVKKEKSPEKVEEKPASPTKK EKSPEKPASP |
| 2751 |
PF04991 (LicD) |
 Estimated TM-score=0.60; hard target. |
>LicD family (Length=224) CRENDLTCYFCGGGCIGAIRHKGFIPWDDDLDFFMPRKDYEVFVKKWKDYEPGKRYILSDTTLDYVDRNNFATLRDSETT QIKPYQKDLRIPHGVALDVIPLDGYPDGKWKRKFQCMWALIYQLFRAQTVPEKHGGLMAVGSRFLLSVFRGKKIRYKIWN VAKRHMTQYPISECNYITELCSGPFYMKKKYKKEWFEKAVFVPFEGENMPIPVGYDGYLKEAFG |
| 2752 |
PF04927 (SMP) |
 Estimated TM-score=0.60; hard target. |
>Seed maturation protein (Length=54) KYGDCFNVKGDLADQPIAPQDAAMMQSAEAMVFGKNQKGGPASVMQAAAAKNER |
| 2753 |
PF04884 (DUF647) |
 Estimated TM-score=0.60; easy target. |
>Vitamin B6 photo-protection and homoeostasis (Length=238) DSLVSTAKDVFLPQGYPDSVSADYLTYQIWDTAQAFCSSLTSALASRAVLTGYGVGDQGASVAAATLAWLLRDGCGMVGR ILFAWLYGTALDWDCKRYRLLADVLNDLAILIQLLCPLAGPPGSPAVVAVLCIASVTLALVTVCGGATRAAFTMHQARQH NMADVSAKDASQETLVNLVGLVVNLTFVPLITGPVAVPVFIIFTSGHIFGNWCAVRSVAMETLNPSRLHLVVVSFVAS |
| 2754 |
PF04859 (DUF641) |
 Estimated TM-score=0.60; easy target. |
>Plant protein of unknown function (DUF641) (Length=125) RAATEAFVAKLFASVSSVKAAYAQLQYAQSPYDPDGIQTADEMVVSELKNVSELKQCYLKKQIDESSPETTQILAEIQEQ KSLLKTYEIMGKKLDSQLKLKDSELTFLREKLEEANKENKLLEKR |
| 2755 |
PF04819 (DUF716) |
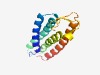 Estimated TM-score=0.60; hard target. |
>Family of unknown function (DUF716) (Length=136) LTQLLGAIAFAQQLLLFHLHSADHMGPEGQYHLLLQILVFVSVSTALIGIVLPHSFLVNFVRSVSIFFQGLWLIVMGFML WTPSLIPKGCYMNDEDGHMVVRCSSHEALHRAISLVNIEFSWFIIGVTVFAVSLYL |
| 2756 |
PF04457 (MJ1316) |
 Estimated TM-score=0.60; very hard target. |
>MJ1316 RNA cyclic group end recognition domain (Length=74) MKTAGDVRHRIQWDENLHEEYFTVGYMDRFSGVVEKPFTSFHWEHLALVDDDQLAIPQHRIQYFKYKGTKVWDK |
| 2757 |
PF04418 (DUF543) |
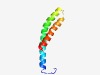 Estimated TM-score=0.60; hard target. |
>Domain of unknown function (DUF543) (Length=72) TQSPSTKVVPSENILNDKWDVVLSNSLVKTGLGFGFGVITSVLFFKRRAWPVWLGVGFGAGRGYAEGDAIFR |
| 2758 |
PF04411 (PDDEXK_7) |
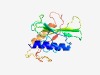 Estimated TM-score=0.60; hard target. |
>PD-(D/E)XK nuclease superfamily (Length=156) TLYEYWTFLKLGQILDKKYQLVSQDIIQVNREGLFVNLQTNRSATRKYKHPHTKEEIILTYQKYESGLPTTPQKPDTMLS IAKKGKGFTYNYVFDAKYRIDYAQEGSYYQNRYKQPGPLEDDINTMHRYRDSIVAYNNGPYERTAFGAYVLFPWFN |
| 2759 |
PF04400 (NqrM) |
 Estimated TM-score=0.60; hard target. |
>(Na+)-NQR maturation NqrM (Length=40) VMFKRKAIQGSCGGLNTVGVDKVCNCETACSEHKLYQIAE |
| 2760 |
PF04366 (Ysc84) |
 Estimated TM-score=0.60; very hard target. |
>Las17-binding protein actin regulator (Length=125) GGGFGGQIGFELTDFVFILNDANAVKTFAQAGSLTLGGNVSLAAGPVGRNAEAAGAASLRSVAGIFSYSKTKGLFAGVSL EGSGIIERRDANEKLYGTRYTAQQILSGGVAPPPQSQALMAILNG |
| 2761 |
PF04219 (DUF413) |
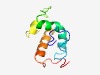 Estimated TM-score=0.60; hard target. |
>Protein of unknown function, DUF (Length=89) RFFDNKHYPRGFSRHGDFTIKEAQLLEQHGYALNELELGKREPVTEDEKQFVSVCRGEREPATEMERVWLKYMARIKRPK RFHTLSGGK |
| 2762 |
PF03981 (Ubiq_cyt_C_chap) |
 Estimated TM-score=0.60; hard target. |
>Ubiquinol-cytochrome C chaperone (Length=145) FSLEKTFRTNFSMLVVHMWLCLRRLKAEGKEGIELGQYVYEIYNHDLEERVSKAGVNLLLSKWMRELEKVFYGNIVAFDT AMLPEAKPDDLQTAIWKNVFAEDGSSKLDAAALPAVMAFTRYVRRECTCLSLTDKEAMMSGNFLF |
| 2763 |
PF03977 (OAD_beta) |
 Estimated TM-score=0.60; easy target. |
>Na+-transporting oxaloacetate decarboxylase beta subunit (Length=367) EVIMLCISFILLYLAIKKEYEPLLLVPIAFGMLLANLPLGGLMDPPIVEVVEKSGVLVPETKQVGGLLYYLYQGVKLGIY PPLIFLGVGAMTDFGPLIANPRSILLGAAAQFGIFFTFIGAILLGFTGPEAASIGIIGGADGPTALYLTSRLAPHLLGPI AVAAYSYMALVPIIQPPIMRALTTKEQRQVVMEQLRPVSKKEKIIFPIMVTVLVSLLLPSAAPLIGMLMLGNLFRESGVV DRLSSTAQNELVNIITIFLGTTVGATANAQTFLSPSTLKIILLGLIAFSLGTATGVLFGKLMYRTSKGKVNPLIGAAGVS AVPMAARVVQKVGQEENPRNFLLMHAMGPNVAGVIGSAVAAGLLLMF |
| 2764 |
PF03772 (Competence) |
 Estimated TM-score=0.60; hard target. |
>Competence protein (Length=271) LLLGQKKDISKDMYDDYASAGVIHILAVSGLHVGIILMLLQLILSPLFRFKHGNVFKTVLIVLFLWCFAIIAGLSPSVIR AVTMFSFLAVALNLKRKTSTLNTLFLSALVILLVEPQFLFSVGFQLSYLAVFSIILLQPKLSKLIPRPKYYISRILWGVF TVTIAAQIGVLPLSLFYFHQFPGLFFVTNLIIIPFLGIILGYGLLCISLALLGILPSFIAEGFRLVIGFMNSVISWVAQQ NSFVFKEIPFSEIQTIVTYVLIGTILISIQR |
| 2765 |
PF03609 (EII-Sor) |
 Estimated TM-score=0.60; hard target. |
>PTS system sorbose-specific iic component (Length=232) LVQAILVGIWAGIAGVDKLVLQTHIHRPIVTGLIVGLILGDVNTGLITGATLELVWIGAVAIGGAQPPNVVIGGVIGTAL AIITKSDPQVTVGLAVPFAVAAQALITLLYTAMSPIMHKFDAYAKKADTRGIERMSSFLPIVLFICYFFVVFLCIFFGAD KASSFISILPEWSIHGLSVAGGMMPAVGFAMLLKTMWETSYVPFFIFGFVMAAYLGLDTLAVAGLAVAIALY |
| 2766 |
PF03428 (RP-C) |
 Estimated TM-score=0.60; easy target. |
>Replication protein C N-terminal domain (Length=174) RSMTLGMLASQVAAGEIKPDQSVDKWKLFRTLCEAKPLLGITDRALAVLNALLSFYPKNELAQDNGLIVFPSNVQLSLRT HGMAEQTVRRHLAALVDAGLLVRKDSPNGKRYVRRDRAGEIDEAFGFSLAPLLARAEEIEQLAAAVMAERLHVQRLRERI TLCRRDIAKLIEAA |
| 2767 |
PF03406 (Phage_fiber_2) |
 Estimated TM-score=0.60; easy target. |
>Phage tail fibre repeat (Length=41) STTQKGLVQLSSATNSTSEVLAATPKAVKAAYDLANGKYTA |
| 2768 |
PF03323 (GerA) |
 Estimated TM-score=0.60; easy target. |
>Bacillus/Clostridium GerA spore germination protein (Length=469) RIKQITGNSNDIVIREFRAGVEQNIHLAIVYTDGLADEDFIHNYILESLMLNIRESKLNRTDAKIFHPVDFIASAFLTVG DLKSVDDFDTALNHILSGNTIILIDGSHSCFACDTRGWNDRSIEEPSAQTVIRGPKDSFTETLRTNTALIRRRVKDPNLR IEGKVIGKRTKTDVSIVYIKGIADSKVVEEVHKRLDRINIDSILESGYIEELIEDGTYSPFPTVYHTERPDSTAASLLEG RVAILVDGTPFVLLVPAIFMQFLQSAEDYYQRFDIGTFIRLLRHITFFLALLTPAVYIAVTTFHQEMLPTQLLVSLAAQR EAIPFPAIVEAVLMEVTFEILREAGVRMPRAVGPAISIVGALVLGEAAVNAGLVSSAMVIVVSITAIASFVFPAYNLAIA VRILRFLFMLLASMFGLFGIILGLIVMVLHLCSLRSFGVPYLSPLGPYYKKDIKDTAVRFPWWAMTTRP |
| 2769 |
PF03208 (PRA1) |
 Estimated TM-score=0.60; hard target. |
>PRA1 family protein (Length=143) SLSDRRPWTELIDRSAFSRPDSLSDATSRLRRNLAYFRVNYAAVVAFALGASLLAHPFSLLVLLGLLAAWCFLYLFRGSD QPVVLFGRTFSDRETLLGLVAASFVAFFFTSVASLIISGLLVGGAIVAVHGACRMPEDLFLDD |
| 2770 |
PF03155 (Alg6_Alg8) |
 Estimated TM-score=0.60; easy target. |
>ALG6, ALG8 glycosyltransferase family (Length=478) ATAFKVLLFPAYKSTDFEVHRNWLAITNSLPVRDWYYEKTSEWTLDYPPFFAYFEWILSQFAKLVDPEMLRVYNLQYDSW QTVYFQRATVIITELVLLYALHLFVQSAPPALKRPSHAAALSILLSPGLLIIDHIHFQYNGFLYGILILSLVLARKKSTT LASGLLFAALLCCKHIYLYLAPAYFVYLLRAYCLGPKSIFHIKFGNCLKLGTGIIAVFGAAFGPFVYWEQIPQLLSRLFP FSRGLCHAYWAPNIWAMYSFTDRILIYVAPRLGLAVNQNAINSVTRGLVGDTSFAVLPEITPRTTFILTLFFQILPLIKL FFDPTWDTFVGAVTLCGYASFLFGWHVHEKAILLVIIPFSLLALKDRRYLGAFRPLAVAGHVSLFPLLFTAAEFPIKTVY TIFWLVFFLMAFDRLAPASKRPRIFLLDRFSLLYIAVSIPLILYCSLVHGVVFGDKLEFLPLMFMSSYSAVGVVGSWV |
| 2771 |
PF02992 (Transposase_21) |
 Estimated TM-score=0.60; easy target. |
>Transposase family tnp2 (Length=211) DGKLRHPVDYVTWDHMNAKYPTFADEARNLRLGLSTDGFNPFNMKNSMYSCWPVLLVNYNLPPDLCMKKENIMLSLLIPA PHQPGNSIDVYLEPLIDDLNTLWSIGEVTYDALTRSAFTLKAMLLWTISDFPAYGNLAGCKVKGKMGCPLCGKNTESMWL KFSRKHVYMGHRKGLPPTHSFRGKKKWFDGKVEQGRRGRILTGRDISQNLR |
| 2772 |
PF02413 (Caudo_TAP) |
 Estimated TM-score=0.60; easy target. |
>Caudovirales tail fibre assembly protein, lambda gpK (Length=134) KTNAFYPVELEQNYIAAGSLPDDIIEVSHDIYQEYAANHAPEGKHRVANKNGLPEWADIPPPTKDELRQHAEFQKQQLIA EATNQIAPLQDAVDLGIATEEEKIALLAWKEYRVILNRIDVSQAMDIDWPEKPE |
| 2773 |
PF01976 (DUF116) |
 Estimated TM-score=0.60; hard target. |
>Protein of unknown function DUF116 (Length=154) FMILMGRLVGVSKEKVRQSFIELNNHLVRSNHHRTRPNKLLILLPHCIQDFDCEIKITGNVKNCKGCGKCEIKDLNELSD QYQVKIAVATGGTLARRIIVDNRPEAIVAVACELDLTSGIQDSYPIPVIGILNERPNGPCINTKVDIQKVRDAI |
| 2774 |
PF01889 (DUF63) |
 Estimated TM-score=0.60; hard target. |
>Membrane protein of unknown function DUF63 (Length=276) ILQFVNKYYIEPIIYDSGYNPVNTLTWALILGVCVFGVIKLLDRMKVEVDERFIYSIIPFVLAGSSLRVLEDANAFSAPL KYLIITPNIYFVVFVVTLACLVVSKKLYDLGVAGDWKKTFAAAGGLWFLANLTALLYLEDIVRPDALVLILVIGTLVAFS IYGLARWQGIGLITDRLNFTILLAHLMDASSTFIGIDMLGYYEKHVVPAYLIDLTGTALVMYPLKLAIFIPVIYVLDTQF NDSEESISLRTFVKMVIIVLGLSPATRNTLRMALGI |
| 2775 |
PF00839 (Cys_rich_FGFR) |
 Estimated TM-score=0.60; hard target. |
>Cysteine rich repeat (Length=58) PECHHFLWDYKKNLTKDIRFDQAAYEVCKEPLAQLPECSKLETGQGLVISCLMDHKGN |
| 2776 |
PF00379 (Chitin_bind_4) |
 Estimated TM-score=0.60; hard target. |
>Insect cuticle protein (Length=57) YQFNYEQSDGQKREEKGELKPVEGSDEPALSVTGSYEYTDPNGQRYRVDYVADERGY |
| 2777 |
PF18866 (CxC7) |
 Estimated TM-score=0.59; hard target. |
>CxC7 like cysteine cluster associated with KDZ transposases (Length=64) NWSRFSEDYKKEFRVPYSLTECPECGDIFKDCPPGYVGDGRKGVLRGFYSTIDFPGYCSAECHF |
| 2778 |
PF18834 (LPD22) |
 Estimated TM-score=0.59; hard target. |
>Large polyvalent protein associated domain 22 (Length=93) RTALSYVADTNPDEQANIQRLAELTGLPPEMVSRNRPEIERKAKLDAVDYDALLSRAPVTNKFLTDPNNAGVAHDDIDAL SNVEGAFDSFVKE |
| 2779 |
PF18796 (LPD1) |
 Estimated TM-score=0.59; hard target. |
>Large polyvalent protein-associated domain 1 (Length=71) DKKTKGYYFSKPEEMCARAFEAFVQDAPIKNNFLVKGTKQSEEAKLGLYPQGEHRLRINAAFKQYFHTLGH |
| 2780 |
PF18188 (PPL4) |
 Estimated TM-score=0.59; easy target. |
>Prim-pol 4 (Length=162) PQDELEKWLLKHNEKNLVLCLEHPTHVSQRGGHYHIIFRSQQMYNEYLSAWKKTILDKSKMYIEEMRTKHAFRLYVDIDL KVEIEKPCDIVELGWVDSIMQFTKQYFPKKDISMVLTQCHGKWEDAVTTTAKYKSGYRLYFHEIIVDYDTFCDYTHQLCM MI |
| 2781 |
PF17986 (EKAL) |
 Estimated TM-score=0.59; hard target. |
>EMP3-KAHRP-like N-terminal domain (Length=53) VTEFKSGGLKEYQENYETKHYKLKENVVDGNKECDEKYEAAQYGFKEKCPYDV |
| 2782 |
PF17445 (Mfa1) |
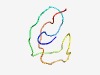 Estimated TM-score=0.59; very hard target. |
>Mating factor A1 (Length=42) MFSIFAQPAQTSVSETQESPANHGANPGKSGSGLGYSTCVVA |
| 2783 |
PF17431 (YpmT) |
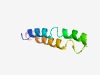 Estimated TM-score=0.59; hard target. |
>Uncharacterized YmpT-like (Length=62) MKRIYQYFSLLSFTFSLYFGWLAYHHLAAEDMDQMYLNISYCALFLSVMVFTFGMRDRKKTD |
| 2784 |
PF17273 (DUF5338) |
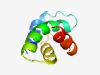 Estimated TM-score=0.59; hard target. |
>Family of unknown function (DUF5338) (Length=68) DELAEWVKKRDKTRTRQDKNVVAFLAVRDDVKEAIAAGYAIKTIWEHMHEQGKIAYRYETFLKHVKRH |
| 2785 |
PF17270 (DUF5336) |
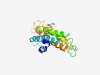 Estimated TM-score=0.59; hard target. |
>Family of unknown function (DUF5336) (Length=173) HGYQPGQQPTTQFSAPTQQFARVNEPGPGAEGPSKLPEYLSAATAGLGLLAYLSYFAPQFTVESSDFPGLGELSGSSVGL GIAVVASLVGALVAGVGLLPKQKNRVALAAVASVLGFLLVISEVLNKPSGTSVAWGLWLVTAFTLLQAAVAVVVLLFDSG ILTAPTPRPKYEQ |
| 2786 |
PF17264 (DUF5330) |
 Estimated TM-score=0.59; hard target. |
>Family of unknown function (DUF5330) (Length=64) PQVGASQAVSAATAAVSDMSQFCKRQPQACEVGGQAATVIGHRAQEGARKLYQIITDKKAPDHT |
| 2787 |
PF17208 (RBR) |
 Estimated TM-score=0.59; very hard target. |
>RNA binding Region (Length=58) RPGTGPGRTRGRLPANWTQTMALQQAQTQAQTQAQPIKIPKKRGPKPGSKRKPRVVPN |
| 2788 |
PF17140 (DUF5113) |
 Estimated TM-score=0.59; easy target. |
>Domain of unknown function (DUF5113) (Length=162) YNDLYQIAGAYVSIGKYMNEHGRYTEALDTLAKALDCVNQHHMLYYHHAADTLDKLHVFVEGDTTYTGVPWIMQEDVRTV PEWISRIREQLSVSYAGLGMKYASDYNRNIYLDILNYTRQDKELESRYLSLEADSRQMTLVLSLVIVGLVLVVILWWFFN KR |
| 2789 |
PF16784 (HNHc_6) |
 Estimated TM-score=0.59; easy target. |
>Putative HNHc nuclease (Length=200) GVELEQSHIDLLENGYPLKAEVEVPDNKKLSIEQRKKIFAMCRDIELHWGEPVESTRKLLQTELEIMKGYEEISLRDCSM KVARELIELIIAFMFHHQIPMSVETSKLLSEDKALLYWATINRNCVICGKPHADLAHYEAVGRGMNRNKMNHYDKHVLAL CRQHHNEQHAIGVKSFDDKYHLHDSWIKVDERLNKMLKGG |
| 2790 |
PF16676 (FOXO-TAD) |
 Estimated TM-score=0.59; easy target. |
>Transactivation domain of FOXO protein family (Length=41) QERFPSDLDLDMFNGSLECDVESIIRSELMDADGLDFNFDS |
| 2791 |
PF16478 (DUF5055) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF5055) (Length=105) MAKQLNFTYDGKDYTLEFTRRTVAEMEKKGFIASDITEKPMTTLPALFAGAFLAHHRFVKEDIINDIYSKLTKKEDLIGK LSEMYNEPILALVEEPEKAEGNLDW |
| 2792 |
PF16352 (DUF4980) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF4980) (Length=110) SSNHCLYRIQQKDKCLLLPVQESAEMSNIKVIAGNKQMKSLNVRLAMNKVDYYVPLYLDEFNEEKTLALDIHVNGNYRND GGISTFTCWKNIKNAESFDTTNREQYRPLY |
| 2793 |
PF16169 (DUF4872) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF4872) (Length=173) ELEKVRFAKGAFAPKGHMYYPIAFPETLSLEKAIVKGIKHTCRDMTAPVPIVGVRAMRWVAKNIRKWPQKLGAKKANHYL GQIVRMQEEIGTGGGGFRYIYAAFLQEAGKVLKNEKLKDFSKEMTEIGDQWRDFALEASRVYKNRSVQTDAYNKVADQLL DLAKREEAFFQAL |
| 2794 |
PF16064 (DUF4806) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF4806) (Length=89) VTGFSFPLKEKHDIERLEFAIQGDKNIRAQYINYLQKCMPKGMKIQNFLSSLFSDDALYAYNFHGCNASGKFRIPMKNYS VFFDCFLEA |
| 2795 |
PF16060 (DUF4802) |
 Estimated TM-score=0.59; very hard target. |
>Domain of unknown function (DUF4802) (Length=64) KSSKEFYKAMAKQWGITCKMSDHCRCLDCQSHYFDCDYEKDEQEKGDGGLSAGTPMFISEVMHG |
| 2796 |
PF16029 (DUF4787) |
 Estimated TM-score=0.59; very hard target. |
>Domain of unknown function (DUF4787) (Length=62) DMTFREFESACEQSSACSHLSGLIKTRCIRECVSPSCYRELYQADPLEEGEIDVRLNSFKGC |
| 2797 |
PF15983 (DUF4767) |
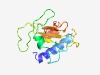 Estimated TM-score=0.59; very hard target. |
>Domain of unknown function (DUF4767) (Length=134) AWTSEKQQALEQFMIRWQAEMGQEYIAATPNNPANMYGITFPEQLTEGLLAVNDQKVAATWNPNGELLPDTYNIVAAYYQ VNGPEMAGHFYLMTIYNGQSVVLHSQQNQGLPDGLVHFTPTENIALANAFTDIA |
| 2798 |
PF15937 (PrlF_antitoxin) |
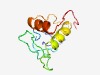 Estimated TM-score=0.59; hard target. |
>prlF antitoxin for toxin YhaV_toxin (Length=97) ESKVTIRGQTTIPAPVREALKLKPGQDSIHYEILPGGQVFMCRQGDEQEDHTMNAFLRFLDADIQNNPQKTRPFNIQQGK KLVAGMDVNIDDEIGDD |
| 2799 |
PF15911 (WD40_3) |
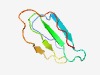 Estimated TM-score=0.59; easy target. |
>WD domain, G-beta repeat (Length=57) VNEYRHVMGVRNIHADASGTRLVIVDDKTDGFIYNPVNDQIIQIVNFPATVRGILWD |
| 2800 |
PF15796 (KELK) |
 Estimated TM-score=0.59; easy target. |
>KELK-motif containing domain of MRCK Ser/Thr protein kinase (Length=80) KIRTMERENRSYKMEKDDLERVFKEANEKIAAQQKELKEAQNQRKLAMAEFSELNDKLADMRSQKTKLSRTVREKEEELE |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218