

You can:
| Name | B1 bradykinin receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | BDKRB1 |
| Synonym | kinin B1 receptor bradykinin receptor BKR1 BK-1 receptor B1R [ Show all ] |
| Disease | Diabetic macular edema Rheumatoid arthritis Pain Osteoarthritis |
| Length | 353 |
| Amino acid sequence | MASSWPPLELQSSNQSQLFPQNATACDNAPEAWDLLHRVLPTFIISICFFGLLGNLFVLLVFLLPRRQLNVAEIYLANLAASDLVFVLGLPFWAENIWNQFNWPFGALLCRVINGVIKANLFISIFLVVAISQDRYRVLVHPMASRRQQRRRQARVTCVLIWVVGGLLSIPTFLLRSIQAVPDLNITACILLLPHEAWHFARIVELNILGFLLPLAAIVFFNYHILASLRTREEVSRTRCGGRKDSKTTALILTLVVAFLVCWAPYHFFAFLEFLFQVQAVRGCFWEDFIDLGLQLANFFAFTNSSLNPVIYVFVGRLFRTKVWELYKQCTPKSLAPISSSHRKEIFQLFWRN |
| UniProt | P46663 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P46663 |
| 3D structure model | This predicted structure model is from GPCR-EXP P46663. |
| BioLiP | N/A |
| Therapeutic Target Database | T58589 |
| ChEMBL | CHEMBL4308 |
| IUPHAR | 41 |
| DrugBank | BE0005831 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 348 | 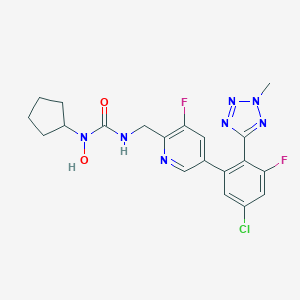 CHEMBL603907 CHEMBL603907 | C20H20ClF2N7O2 | 463.874 | 8 / 2 | 3.1 | Yes |
| 669 | 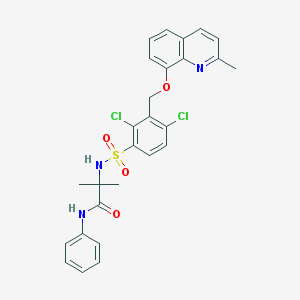 CHEMBL210427 CHEMBL210427 | C27H25Cl2N3O4S | 558.474 | 6 / 2 | 5.8 | No |
| 951 | 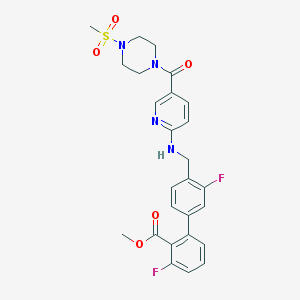 CHEMBL206982 CHEMBL206982 | C26H26F2N4O5S | 544.574 | 10 / 1 | 2.9 | No |
| 2207 | 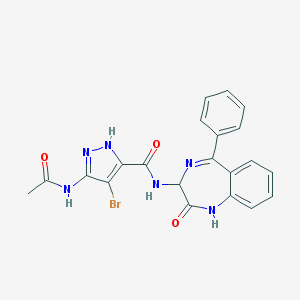 CHEMBL243432 CHEMBL243432 | C21H17BrN6O3 | 481.31 | 5 / 4 | 2.3 | Yes |
| 2986 | 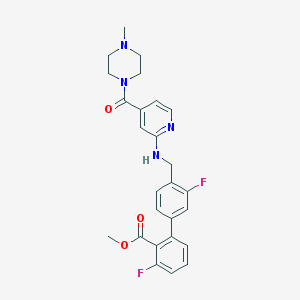 CHEMBL208247 CHEMBL208247 | C26H26F2N4O3 | 480.516 | 8 / 1 | 3.6 | Yes |
| 4145 | 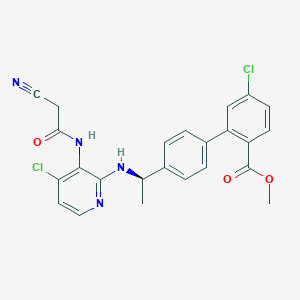 CHEMBL375288 CHEMBL375288 | C24H20Cl2N4O3 | 483.349 | 6 / 2 | 5.4 | No |
| 4254 | 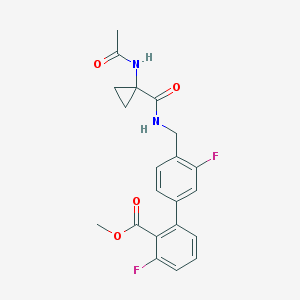 CHEMBL387331 CHEMBL387331 | C21H20F2N2O4 | 402.398 | 6 / 2 | 2.4 | Yes |
| 4770 | 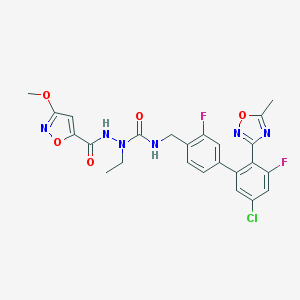 CHEMBL589819 CHEMBL589819 | C24H21ClF2N6O5 | 546.916 | 10 / 2 | 4.4 | No |
| 4950 |  CHEMBL452238 CHEMBL452238 | C33H42ClN3O5S | 628.225 | 7 / 1 | 6.0 | No |
| 5021 |  CHEMBL380260 CHEMBL380260 | C27H32Cl2N4O4S | 579.537 | 7 / 3 | 4.5 | No |
| 5195 |  CHEMBL2311187 CHEMBL2311187 | C47H75N13O11 | 1002.21 | 14 / 11 | -4.1 | No |
| 5196 |  CHEMBL2369827 CHEMBL2369827 | C47H75N13O11 | 998.197 | 14 / 11 | -4.1 | No |
| 5198 |  CHEMBL413382 CHEMBL413382 | C47H75N13O11 | 998.197 | 14 / 11 | -4.1 | No |
| 6222 |  CHEMBL299164 CHEMBL299164 | C34H41N7O2 | 579.749 | 6 / 2 | 5.4 | No |
| 6312 |  CHEMBL591110 CHEMBL591110 | C20H23N5O3 | 381.436 | 5 / 3 | 0.7 | Yes |
| 6327 |  des-Arg10-Kallidin des-Arg10-Kallidin | C50H73N13O11 | 1032.21 | 14 / 11 | -3.8 | No |
| 6599 | 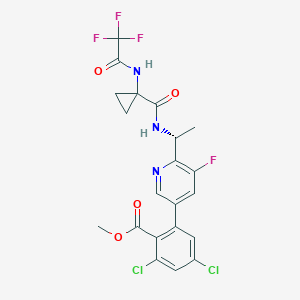 CHEMBL232750 CHEMBL232750 | C21H17Cl2F4N3O4 | 522.278 | 9 / 2 | 4.0 | No |
| 7456 | 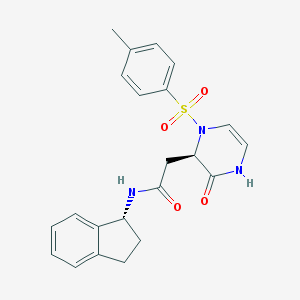 CHEMBL1939763 CHEMBL1939763 | C22H23N3O4S | 425.503 | 5 / 2 | 1.8 | Yes |
| 7610 | 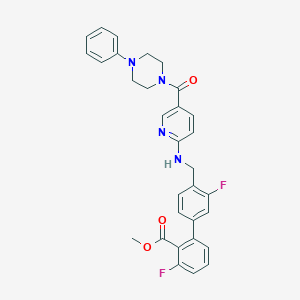 CHEMBL208210 CHEMBL208210 | C31H28F2N4O3 | 542.587 | 8 / 1 | 5.4 | No |
| 8356 | 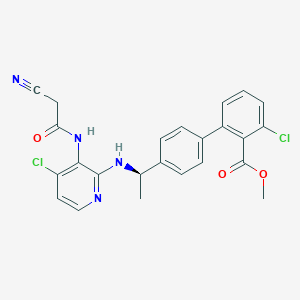 CHEMBL221998 CHEMBL221998 | C24H20Cl2N4O3 | 483.349 | 6 / 2 | 5.4 | No |
| 9244 | 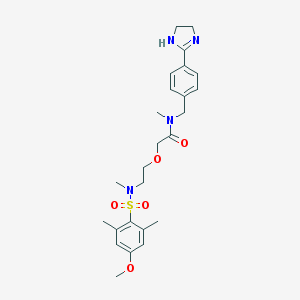 Safotibant Safotibant | C25H34N4O5S | 502.63 | 7 / 1 | 1.6 | No |
| 10067 | 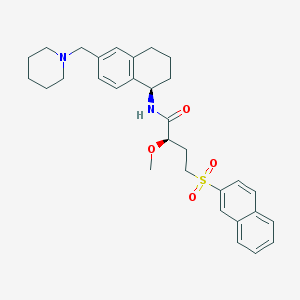 CHEMBL453338 CHEMBL453338 | C31H38N2O4S | 534.715 | 5 / 1 | 5.0 | No |
| 557578 | 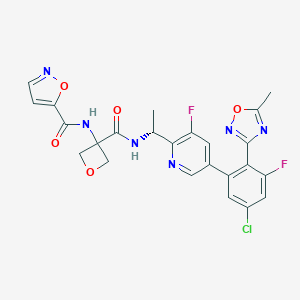 SCHEMBL10147804 SCHEMBL10147804 | C24H19ClF2N6O5 | 544.9 | 11 / 2 | 2.4 | No |
| 548001 | 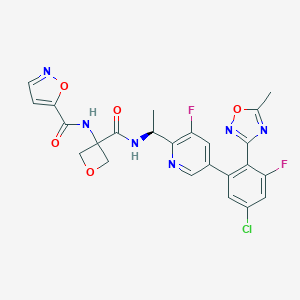 CHEMBL3980627 CHEMBL3980627 | C24H19ClF2N6O5 | 544.9 | 11 / 2 | 2.4 | No |
| 10810 | 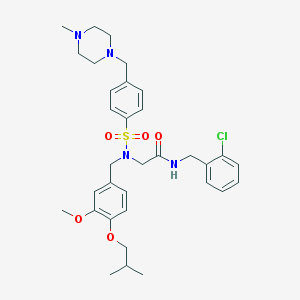 CHEMBL447392 CHEMBL447392 | C33H43ClN4O5S | 643.24 | 8 / 1 | 5.0 | No |
| 10940 | 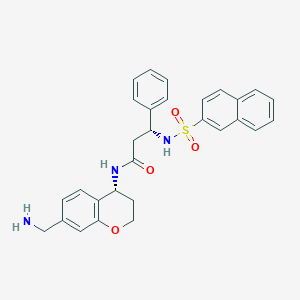 CHEMBL227828 CHEMBL227828 | C29H29N3O4S | 515.628 | 6 / 3 | 3.4 | No |
| 12010 | 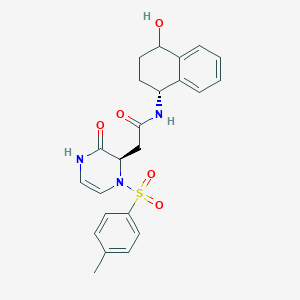 CHEMBL1939920 CHEMBL1939920 | C23H25N3O5S | 455.529 | 6 / 3 | 1.0 | Yes |
| 12024 | 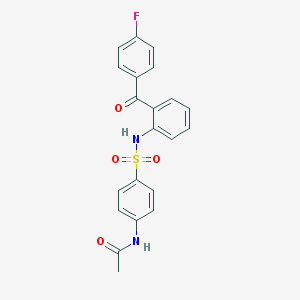 CHEMBL516783 CHEMBL516783 | C21H17FN2O4S | 412.435 | 6 / 2 | 3.6 | Yes |
| 12075 | 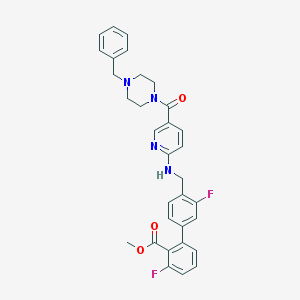 CHEMBL426681 CHEMBL426681 | C32H30F2N4O3 | 556.614 | 8 / 1 | 5.1 | No |
| 12216 | 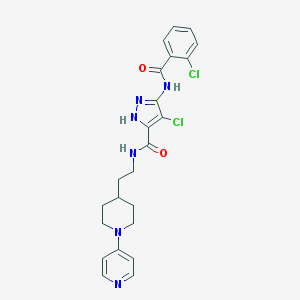 CHEMBL390916 CHEMBL390916 | C23H24Cl2N6O2 | 487.385 | 5 / 3 | 4.1 | Yes |
| 12902 | 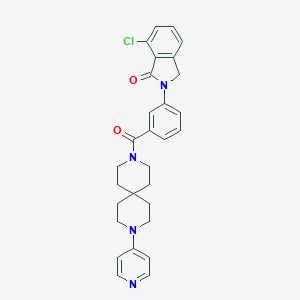 913064-47-8 913064-47-8 | C29H29ClN4O2 | 501.027 | 4 / 0 | 4.4 | No |
| 12967 | 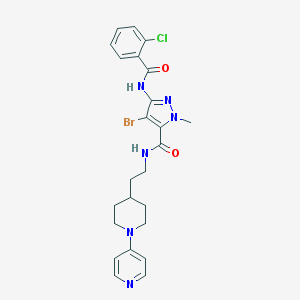 CHEMBL241935 CHEMBL241935 | C24H26BrClN6O2 | 545.866 | 5 / 2 | 4.2 | No |
| 13275 | 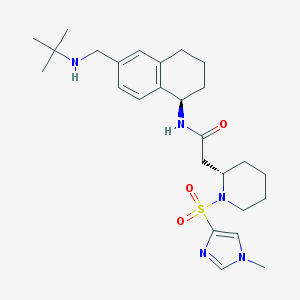 CHEMBL206246 CHEMBL206246 | C26H39N5O3S | 501.69 | 6 / 2 | 2.3 | No |
| 14884 | 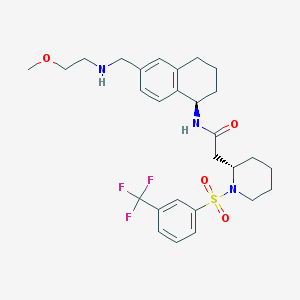 CHEMBL381372 CHEMBL381372 | C28H36F3N3O4S | 567.668 | 9 / 2 | 3.6 | No |
| 15060 | 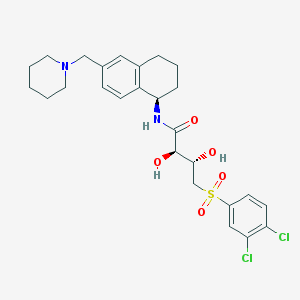 CHEMBL507168 CHEMBL507168 | C26H32Cl2N2O5S | 555.511 | 6 / 3 | 4.0 | No |
| 15124 | 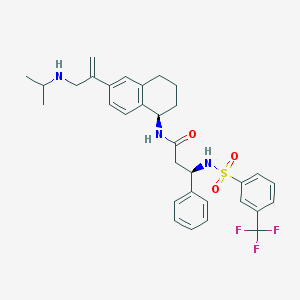 CHEMBL1210753 CHEMBL1210753 | C32H36F3N3O3S | 599.713 | 8 / 3 | 6.0 | No |
| 15171 | 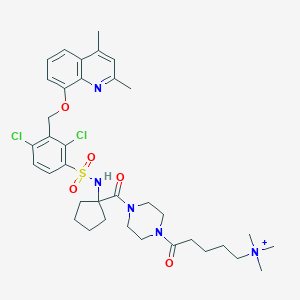 CHEMBL386794 CHEMBL386794 | C36H48Cl2N5O5S+ | 733.77 | 7 / 1 | 5.2 | No |
| 15226 | 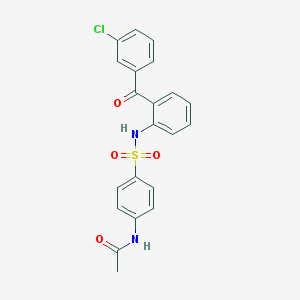 CHEMBL470693 CHEMBL470693 | C21H17ClN2O4S | 428.887 | 5 / 2 | 4.1 | Yes |
| 15850 | 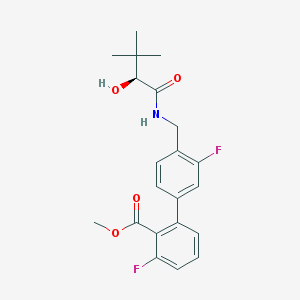 CHEMBL272491 CHEMBL272491 | C21H23F2NO4 | 391.415 | 6 / 2 | 3.8 | Yes |
| 15976 | 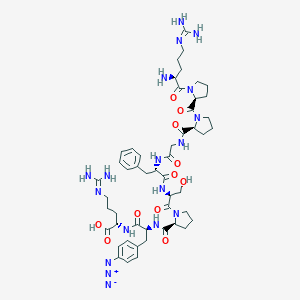 CHEMBL2372168 CHEMBL2372168 | C50H72N18O11 | 1101.24 | 16 / 12 | -3.8 | No |
| 15982 | 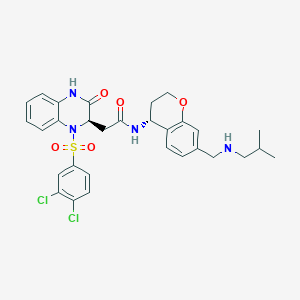 CHEMBL444431 CHEMBL444431 | C30H32Cl2N4O5S | 631.569 | 7 / 3 | 4.5 | No |
| 16465 | 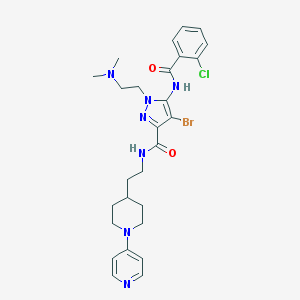 CHEMBL244284 CHEMBL244284 | C27H33BrClN7O2 | 602.962 | 6 / 2 | 4.2 | No |
| 17135 | 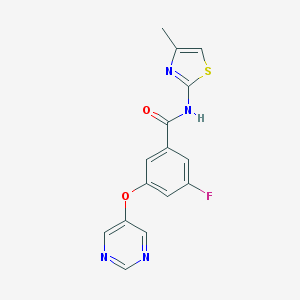 CHEMBL2440659 CHEMBL2440659 | C15H11FN4O2S | 330.337 | 7 / 1 | 2.4 | Yes |
| 17172 | 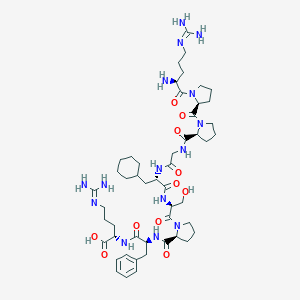 CHEMBL2372142 CHEMBL2372142 | C50H79N15O11 | 1066.28 | 14 / 12 | -3.6 | No |
| 17629 | 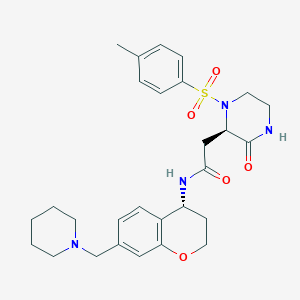 CHEMBL1777968 CHEMBL1777968 | C28H36N4O5S | 540.679 | 7 / 2 | 2.1 | No |
| 17793 | 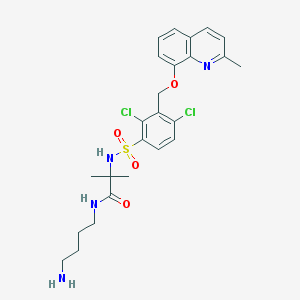 CHEMBL379259 CHEMBL379259 | C25H30Cl2N4O4S | 553.499 | 7 / 3 | 4.0 | No |
| 18255 | 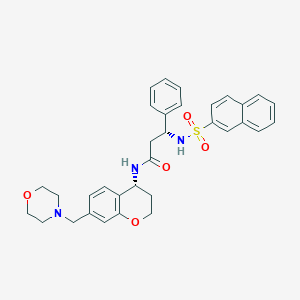 CHEMBL412762 CHEMBL412762 | C33H35N3O5S | 585.719 | 7 / 2 | 4.0 | No |
| 18264 | 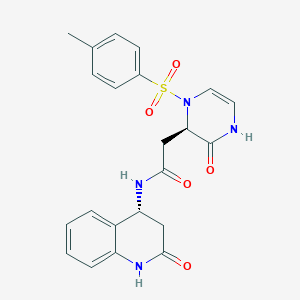 CHEMBL1939770 CHEMBL1939770 | C22H22N4O5S | 454.501 | 6 / 3 | 0.3 | Yes |
| 19229 |  CHEMBL1835757 CHEMBL1835757 | C32H32N2O3 | 492.619 | 4 / 1 | 5.4 | No |
| 19231 |  CHEMBL1934259 CHEMBL1934259 | C32H32N2O3 | 492.619 | 4 / 1 | 5.4 | No |
| 19573 |  CHEMBL216618 CHEMBL216618 | C49H69N15O12S | 1092.24 | 16 / 13 | -5.6 | No |
| 20926 |  CHEMBL1956869 CHEMBL1956869 | C34H46Cl2N5O5S+ | 707.732 | 8 / 1 | 4.6 | No |
| 20967 | 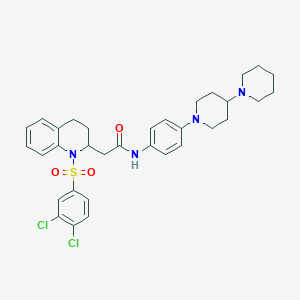 CHEMBL2016601 CHEMBL2016601 | C33H38Cl2N4O3S | 641.652 | 6 / 1 | 6.7 | No |
| 21799 | 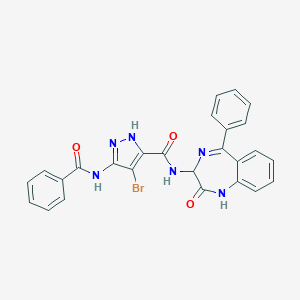 CHEMBL390272 CHEMBL390272 | C26H19BrN6O3 | 543.381 | 5 / 4 | 4.0 | No |
| 22647 | 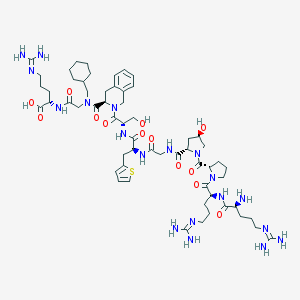 CHEMBL3038098 CHEMBL3038098 | C59H91N19O13S | 1306.56 | 18 / 15 | -5.9 | No |
| 22698 | 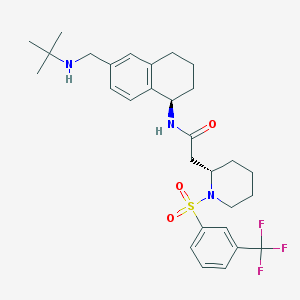 CHEMBL381366 CHEMBL381366 | C29H38F3N3O3S | 565.696 | 8 / 2 | 4.7 | No |
| 22794 | 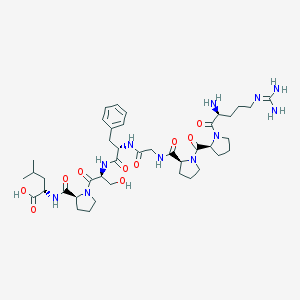 8-Leu-9-des-arg-BK 8-Leu-9-des-arg-BK | C41H63N11O10 | 870.022 | 12 / 9 | -3.6 | No |
| 23320 | 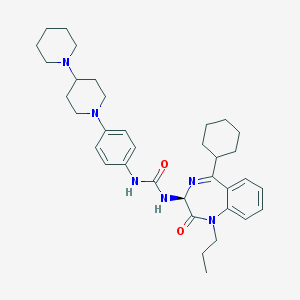 CHEMBL1788318 CHEMBL1788318 | C35H48N6O2 | 584.809 | 5 / 2 | 6.3 | No |
| 23321 | 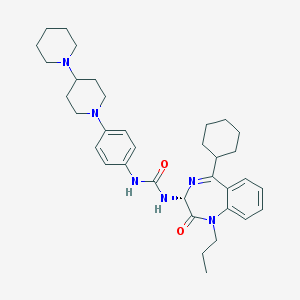 CHEMBL1744082 CHEMBL1744082 | C35H48N6O2 | 584.809 | 5 / 2 | 6.3 | No |
| 24310 | 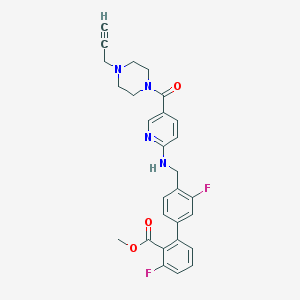 CHEMBL207000 CHEMBL207000 | C28H26F2N4O3 | 504.538 | 8 / 1 | 3.7 | No |
| 24698 |  CHEMBL1956722 CHEMBL1956722 | C36H48Cl2N5O6S+ | 749.769 | 8 / 1 | 4.3 | No |
| 25113 |  CHEMBL2087036 CHEMBL2087036 | C23H26Cl2N4O3S | 509.446 | 5 / 1 | 2.9 | No |
| 25540 |  CHEMBL2087415 CHEMBL2087415 | C23H29ClN4O4S | 493.019 | 6 / 1 | 1.9 | Yes |
| 26205 |  RS 39604 RS 39604 | C26H36ClN3O6S | 554.099 | 9 / 2 | 3.1 | No |
| 27059 |  CHEMBL1834749 CHEMBL1834749 | C31H33N5O2 | 507.638 | 5 / 1 | 4.3 | No |
| 28179 |  CHEMBL234296 CHEMBL234296 | C22H22F4N4O | 434.439 | 8 / 2 | 4.2 | Yes |
| 28776 |  CHEMBL1956857 CHEMBL1956857 | C36H45Cl2F3N6O8S | 849.745 | 15 / 4 | N/A | No |
| 29429 |  CHEMBL2372164 CHEMBL2372164 | C50H72N17O11+ | 1087.23 | 15 / 12 | -4.1 | No |
| 29656 | 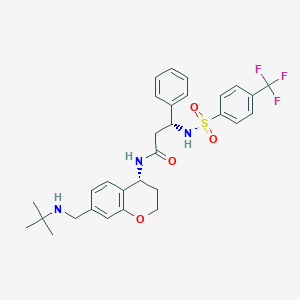 CHEMBL389054 CHEMBL389054 | C30H34F3N3O4S | 589.674 | 9 / 3 | 4.5 | No |
| 30313 | 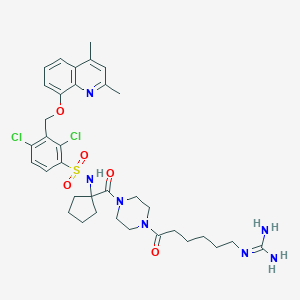 CHEMBL222156 CHEMBL222156 | C35H45Cl2N7O5S | 746.749 | 8 / 3 | 4.0 | No |
| 558220 | 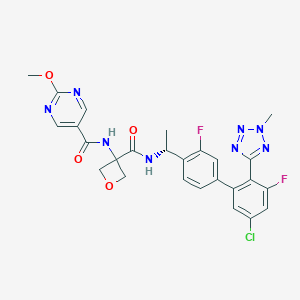 SCHEMBL10182621 SCHEMBL10182621 | C26H23ClF2N8O4 | 584.969 | 11 / 2 | 2.8 | No |
| 548222 | 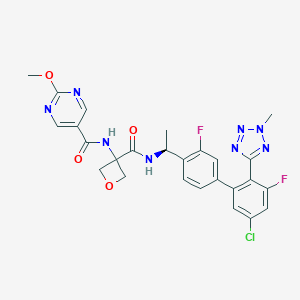 CHEMBL3892284 CHEMBL3892284 | C26H23ClF2N8O4 | 584.969 | 11 / 2 | 2.8 | No |
| 31306 | 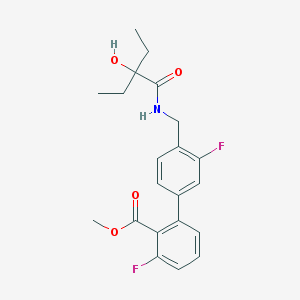 CHEMBL256793 CHEMBL256793 | C21H23F2NO4 | 391.415 | 6 / 2 | 3.7 | Yes |
| 31314 | 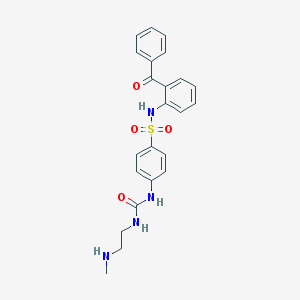 CHEMBL487444 CHEMBL487444 | C23H24N4O4S | 452.529 | 6 / 4 | 2.9 | Yes |
| 31967 | 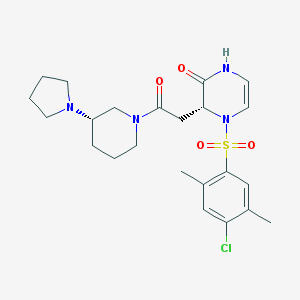 CHEMBL1939949 CHEMBL1939949 | C23H31ClN4O4S | 495.035 | 6 / 1 | 2.1 | Yes |
| 31997 | 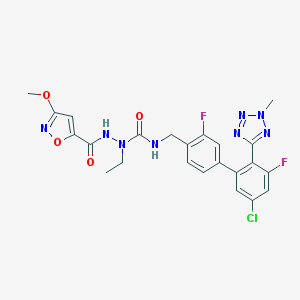 CHEMBL609157 CHEMBL609157 | C23H21ClF2N8O4 | 546.92 | 10 / 2 | 4.0 | No |
| 33118 | 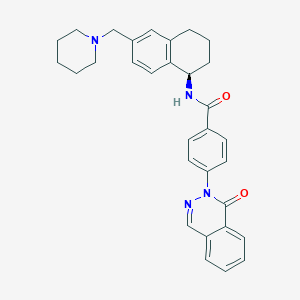 CHEMBL1835766 CHEMBL1835766 | C31H32N4O2 | 492.623 | 4 / 1 | 4.7 | Yes |
| 33284 | 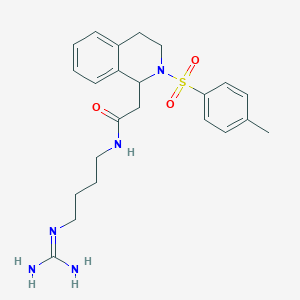 CHEMBL498393 CHEMBL498393 | C23H31N5O3S | 457.593 | 5 / 3 | 1.4 | Yes |
| 33447 | 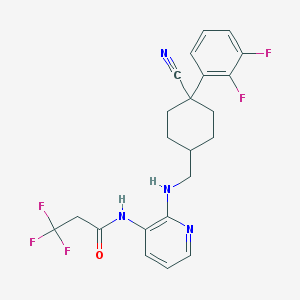 CHEMBL388031 CHEMBL388031 | C22H21F5N4O | 452.429 | 9 / 2 | 4.3 | Yes |
| 33987 | 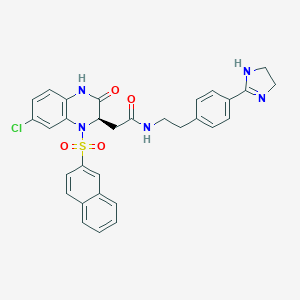 CHEMBL160547 CHEMBL160547 | C31H28ClN5O4S | 602.106 | 6 / 3 | 3.7 | No |
| 34186 | 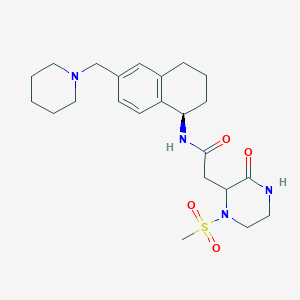 CHEMBL1777880 CHEMBL1777880 | C23H34N4O4S | 462.609 | 6 / 2 | 0.9 | Yes |
| 35950 | 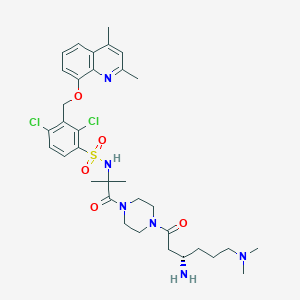 CHEMBL205093 CHEMBL205093 | C34H46Cl2N6O5S | 721.739 | 9 / 2 | 3.8 | No |
| 36614 | 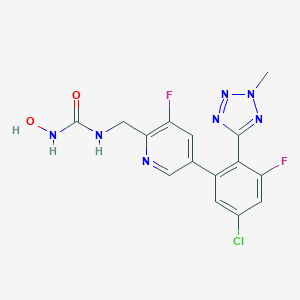 CHEMBL598868 CHEMBL598868 | C15H12ClF2N7O2 | 395.755 | 8 / 3 | 1.7 | Yes |
| 37354 | 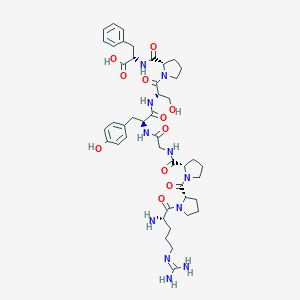 CHEMBL2372151 CHEMBL2372151 | C44H61N11O11 | 920.038 | 13 / 10 | -3.7 | No |
| 558415 | 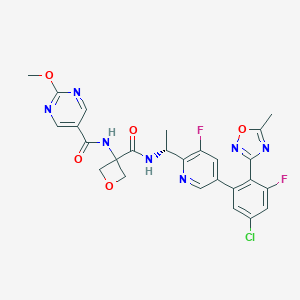 SCHEMBL10147835 SCHEMBL10147835 | C26H22ClF2N7O5 | 585.953 | 12 / 2 | 2.2 | No |
| 548322 | 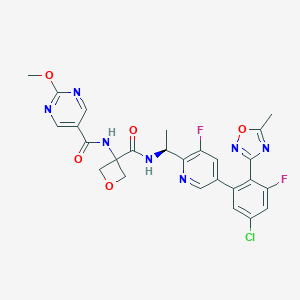 CHEMBL3987084 CHEMBL3987084 | C26H22ClF2N7O5 | 585.953 | 12 / 2 | 2.2 | No |
| 38018 | 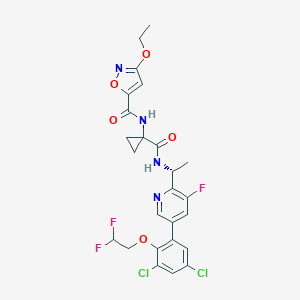 CHEMBL404280 CHEMBL404280 | C25H23Cl2F3N4O5 | 587.377 | 10 / 2 | 5.0 | No |
| 38057 | 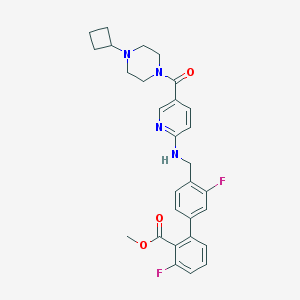 CHEMBL204562 CHEMBL204562 | C29H30F2N4O3 | 520.581 | 8 / 1 | 4.5 | No |
| 38376 |  CHEMBL196494 CHEMBL196494 | C24H23F3N4O2 | 456.469 | 7 / 3 | 4.2 | Yes |
| 38588 |  CHEMBL208386 CHEMBL208386 | C28H39N3O3S | 497.698 | 5 / 2 | 3.9 | Yes |
| 41168 |  CHEMBL218605 CHEMBL218605 | C23H19F5N4O3 | 494.422 | 10 / 2 | 4.4 | Yes |
| 42232 |  CHEMBL525092 CHEMBL525092 | C30H34N4O5S | 562.685 | 7 / 3 | 2.9 | No |
| 558605 | 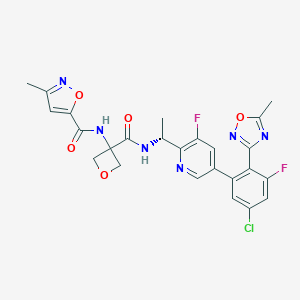 SCHEMBL10147810 SCHEMBL10147810 | C25H21ClF2N6O5 | 558.927 | 11 / 2 | 2.8 | No |
| 548383 | 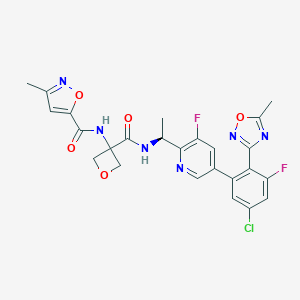 CHEMBL3910554 CHEMBL3910554 | C25H21ClF2N6O5 | 558.927 | 11 / 2 | 2.8 | No |
| 43927 | 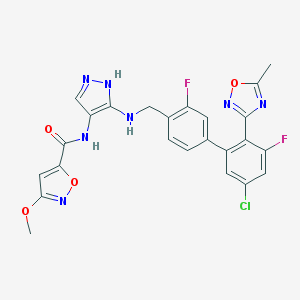 CHEMBL604999 CHEMBL604999 | C24H18ClF2N7O4 | 541.9 | 11 / 3 | 4.7 | No |
| 44231 | 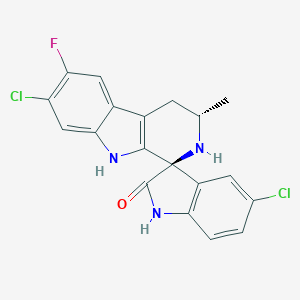 Cipargamin Cipargamin | C19H14Cl2FN3O | 390.239 | 3 / 3 | 3.9 | Yes |
| 44700 | 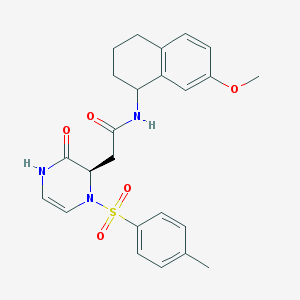 CHEMBL1939764 CHEMBL1939764 | C24H27N3O5S | 469.556 | 6 / 2 | 2.1 | Yes |
| 44732 | 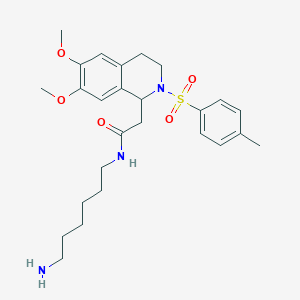 CHEMBL496341 CHEMBL496341 | C26H37N3O5S | 503.658 | 7 / 2 | 2.7 | No |
| 45091 | 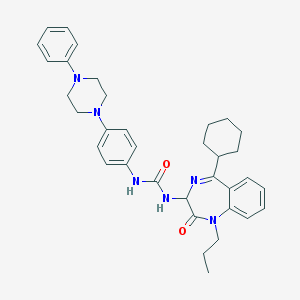 CHEMBL298158 CHEMBL298158 | C35H42N6O2 | 578.761 | 5 / 2 | 6.5 | No |
| 45817 | 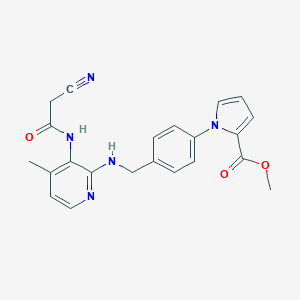 CHEMBL360500 CHEMBL360500 | C22H21N5O3 | 403.442 | 6 / 2 | 3.2 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417