

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MASSWPPLELQSSNQSQLFPQNATACDNAPEAWDLLHRVLPTFIISICFFGLLGNLFVLLVFLLPRRQLNVAEIYLANLAASDLVFVLGLPFWAENIWNQFNWPFGALLCRVINGVIKANLFISIFLVVAISQDRYRVLVHPMASRRQQRRRQARVTCVLIWVVGGLLSIPTFLLRSIQAVPDLNITACILLLPHEAWHFARIVELNILGFLLPLAAIVFFNYHILASLRTREEVSRTRCGGRKDSKTTALILTLVVAFLVCWAPYHFFAFLEFLFQVQAVRGCFWEDFIDLGLQLANFFAFTNSSLNPVIYVFVGRLFRTKVWELYKQCTPKSLAPISSSHRKEIFQLFWRN | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCC | |
| 99999997778888788888888766777421526897389999999999998899978977625676869899999999999999997699999996689998987789999999999999999999999998046533116533364525687767999999999998999983336718998788847698478999999999999999999999999999999972556775320001000048899999999999983799999999999982688972899999999999999999999898999995188889999999998667877888888848876555789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MASSWPPLELQSSNQSQLFPQNATACDNAPEAWDLLHRVLPTFIISICFFGLLGNLFVLLVFLLPRRQLNVAEIYLANLAASDLVFVLGLPFWAENIWNQFNWPFGALLCRVINGVIKANLFISIFLVVAISQDRYRVLVHPMASRRQQRRRQARVTCVLIWVVGGLLSIPTFLLRSIQAVPDLNITACILLLPHEAWHFARIVELNILGFLLPLAAIVFFNYHILASLRTREEVSRTRCGGRKDSKTTALILTLVVAFLVCWAPYHFFAFLEFLFQVQAVRGCFWEDFIDLGLQLANFFAFTNSSLNPVIYVFVGRLFRTKVWELYKQCTPKSLAPISSSHRKEIFQLFWRN | |
| 45444342333332313323443331543631330010000220120023023212200100012333310000000000100010000000100000353421001001220101133111110000000031030000100302322322101000000110000001000000203437743111010303460011011111113123312200120001001101323444444445443100000000000001103120100000100231422441313200110110010001300031000000004401410140024104433463344344333533468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCC MASSWPPLELQSSNQSQLFPQNATACDNAPEAWDLLHRVLPTFIISICFFGLLGNLFVLLVFLLPRRQLNVAEIYLANLAASDLVFVLGLPFWAENIWNQFNWPFGALLCRVINGVIKANLFISIFLVVAISQDRYRVLVHPMASRRQQRRRQARVTCVLIWVVGGLLSIPTFLLRSIQAVPDLNITACILLLPHEAWHFARIVELNILGFLLPLAAIVFFNYHILASLRTREEVSRTRCGGRKDSKTTALILTLVVAFLVCWAPYHFFAFLEFLFQVQAVRGCFWEDFIDLGLQLANFFAFTNSSLNPVIYVFVGRLFRTKVWELYKQCTPKSLAPISSSHRKEIFQLFWRN | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.28 | 0.26 | 0.83 | 3.30 | Download | -------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG--LHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.28 | 0.26 | 0.84 | 4.27 | Download | --------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG--LHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKYTEEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.24 | 0.85 | 3.91 | Download | ------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG--AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-----DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 4 | 4djh | 0.25 | 0.25 | 0.81 | 1.55 | Download | --------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------ | |||||||||||||||||||
| 5 | 4djh | 0.25 | 0.25 | 0.81 | 1.17 | Download | --------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-----A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.26 | 0.24 | 0.84 | 3.39 | Download | --------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG--AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRR-----DPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 7 | 4djh | 0.25 | 0.25 | 0.80 | 1.71 | Download | -----------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------A-----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.26 | 0.24 | 0.85 | 4.64 | Download | -----------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK-PCG-------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.28 | 0.26 | 0.83 | 3.56 | Download | -------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG--LHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLR------MKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.25 | 0.25 | 0.92 | 3.95 | Download | QKATPPKLEDKSPDSNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
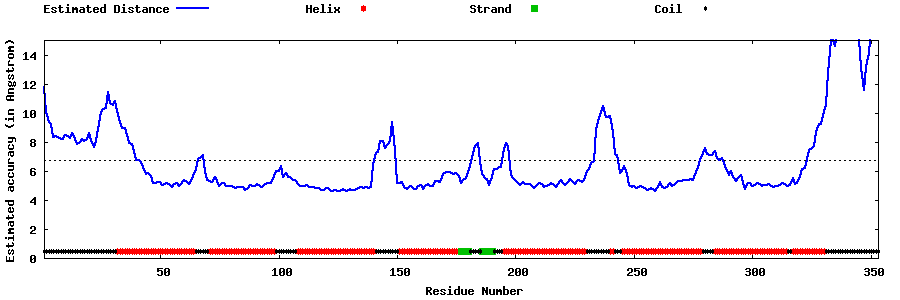
| Generated 3D models | Estimated local accuracy of models | ||
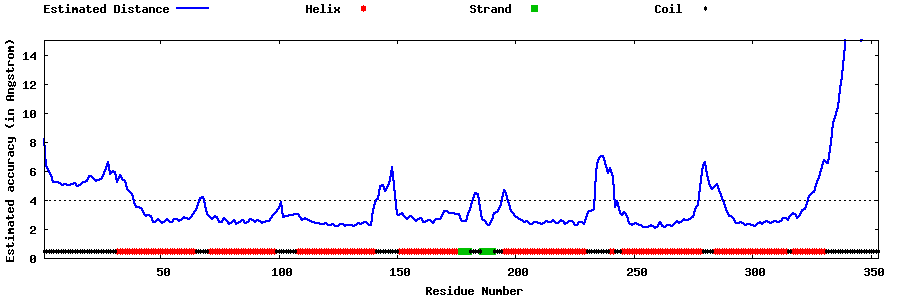 | |||
|
|
|||
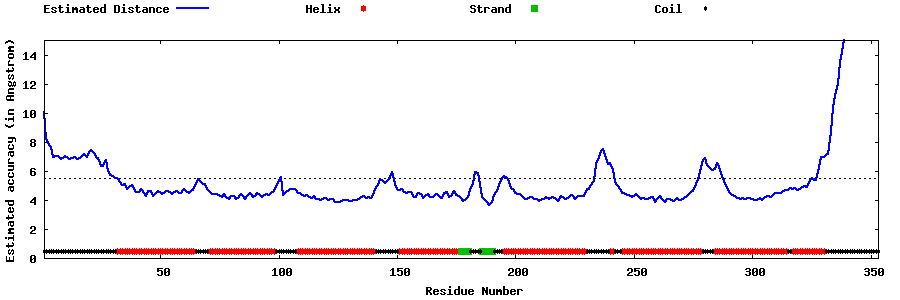 | |||
|
| |||
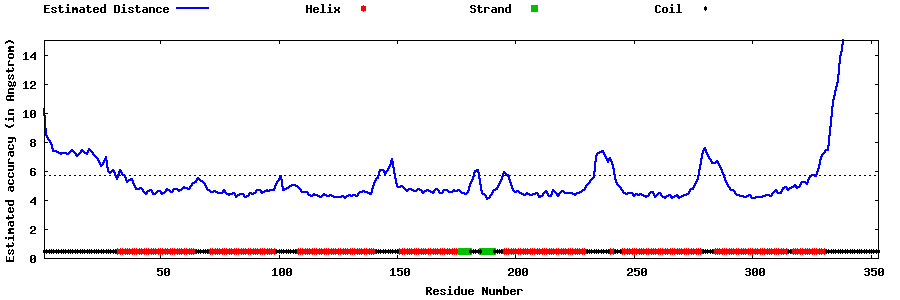 | |||
|
| |||
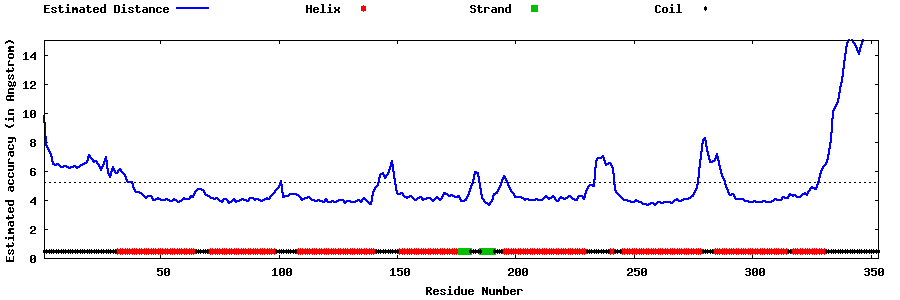 | |||
|
| |||
 | |||
|
| |||