

You can:
| Name | Histamine H4 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HRH4 |
| Synonym | Pfi-013 SP9144 HH4R H4R H4 receptor [ Show all ] |
| Disease | Allergic rhinitis Asthma Inflammatory disease Rheumatoid arthritis |
| Length | 390 |
| Amino acid sequence | MPDTNSTINLSLSTRVTLAFFMSLVAFAIMLGNALVILAFVVDKNLRHRSSYFFLNLAISDFFVGVISIPLYIPHTLFEWDFGKEICVFWLTTDYLLCTASVYNIVLISYDRYLSVSNAVSYRTQHTGVLKIVTLMVAVWVLAFLVNGPMILVSESWKDEGSECEPGFFSEWYILAITSFLEFVIPVILVAYFNMNIYWSLWKRDHLSRCQSHPGLTAVSSNICGHSFRGRLSSRRSLSASTEVPASFHSERQRRKSSLMFSSRTKMNSNTIASKMGSFSQSDSVALHQREHVELLRARRLAKSLAILLGVFAVCWAPYSLFTIVLSFYSSATGPKSVWYRIAFWLQWFNSFVNPLLYPLCHKRFQKAFLKIFCIKKQPLPSQHSRSVSS |
| UniProt | Q9H3N8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9H3N8 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9H3N8. |
| BioLiP | N/A |
| Therapeutic Target Database | T26500 |
| ChEMBL | CHEMBL3759 |
| IUPHAR | 265 |
| DrugBank | BE0000146 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 76 | 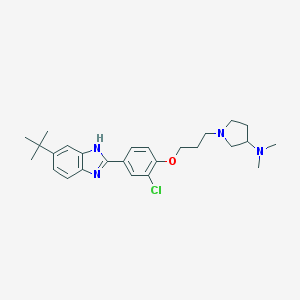 SCHEMBL602163 SCHEMBL602163 | C26H35ClN4O | 455.043 | 4 / 1 | 6.0 | No |
| 229 | 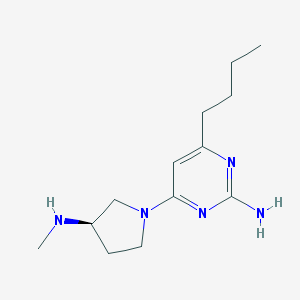 CHEMBL3236571 CHEMBL3236571 | C13H23N5 | 249.362 | 5 / 2 | 1.8 | Yes |
| 375 | 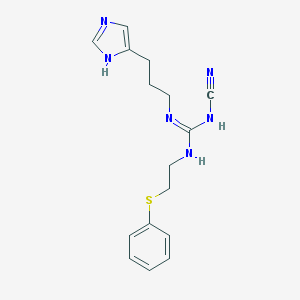 CHEMBL1078529 CHEMBL1078529 | C16H20N6S | 328.438 | 4 / 3 | 2.2 | Yes |
| 532 | 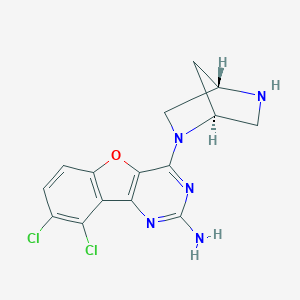 CHEMBL1915037 CHEMBL1915037 | C15H13Cl2N5O | 350.203 | 6 / 2 | 3.0 | Yes |
| 1011 | 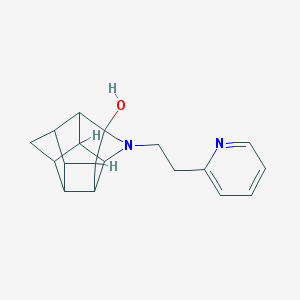 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 521482 | 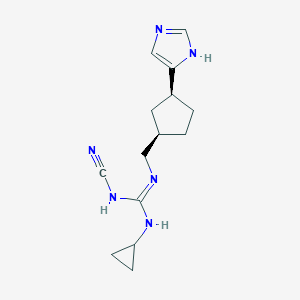 CHEMBL3806146 CHEMBL3806146 | C14H20N6 | 272.356 | 3 / 3 | 1.7 | Yes |
| 521485 | 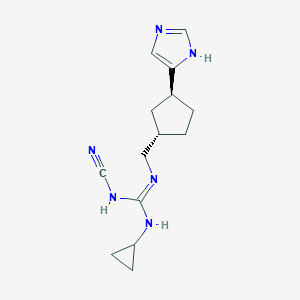 CHEMBL3804837 CHEMBL3804837 | C14H20N6 | 272.356 | 3 / 3 | 1.7 | Yes |
| 441729 | 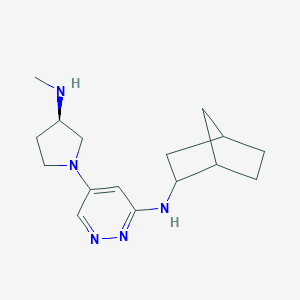 CHEMBL3393556 CHEMBL3393556 | C16H25N5 | 287.411 | 5 / 2 | 1.8 | Yes |
| 441730 | 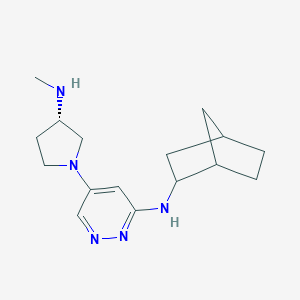 CHEMBL3393557 CHEMBL3393557 | C16H25N5 | 287.411 | 5 / 2 | 1.8 | Yes |
| 1787 | 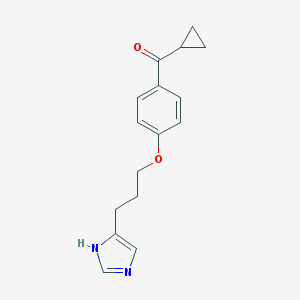 ciproxifan ciproxifan | C16H18N2O2 | 270.332 | 3 / 1 | 2.5 | Yes |
| 1805 | 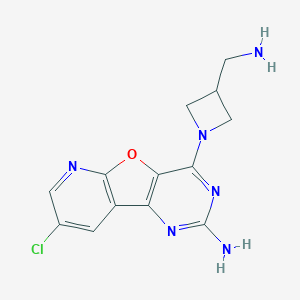 CHEMBL1915051 CHEMBL1915051 | C13H13ClN6O | 304.738 | 7 / 2 | 1.1 | Yes |
| 2285 | 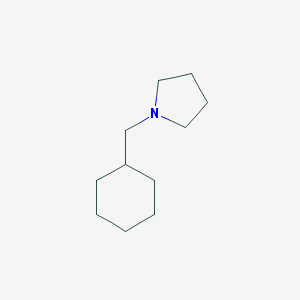 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2432 | 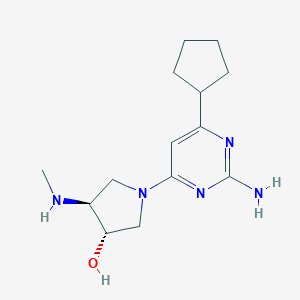 CHEMBL3236565 CHEMBL3236565 | C14H23N5O | 277.372 | 6 / 3 | 0.8 | Yes |
| 3023 | 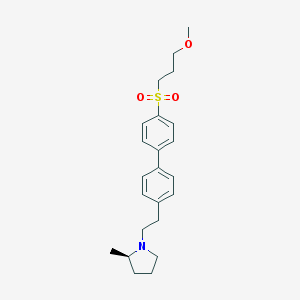 UNII-10521U3EQA UNII-10521U3EQA | C23H31NO3S | 401.565 | 4 / 0 | 4.2 | Yes |
| 3131 | 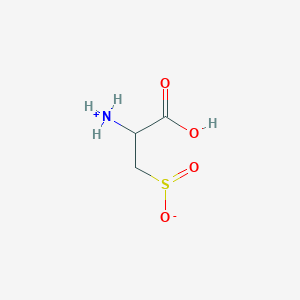 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3328 | 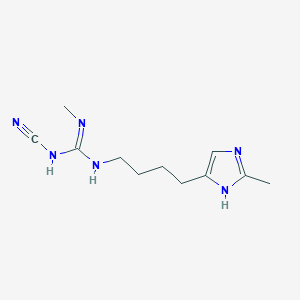 CHEMBL3220637 CHEMBL3220637 | C11H18N6 | 234.307 | 3 / 3 | 0.8 | Yes |
| 3453 | 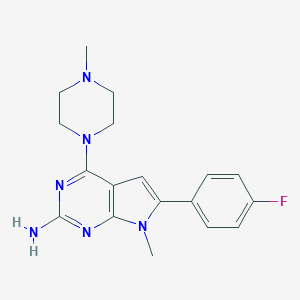 CHEMBL2314768 CHEMBL2314768 | C18H21FN6 | 340.406 | 6 / 1 | 2.3 | Yes |
| 3699 | 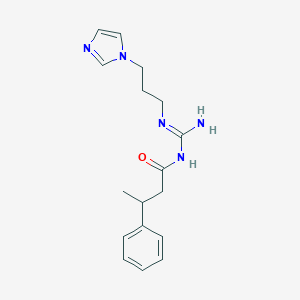 CHEMBL3220892 CHEMBL3220892 | C17H23N5O | 313.405 | 3 / 2 | 1.7 | Yes |
| 3728 | 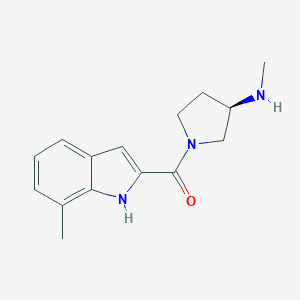 CHEMBL2047444 CHEMBL2047444 | C15H19N3O | 257.337 | 2 / 2 | 2.0 | Yes |
| 4218 | 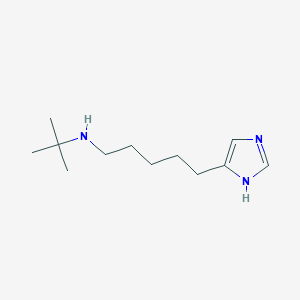 CHEMBL202638 CHEMBL202638 | C12H23N3 | 209.337 | 2 / 2 | 2.0 | Yes |
| 4636 | 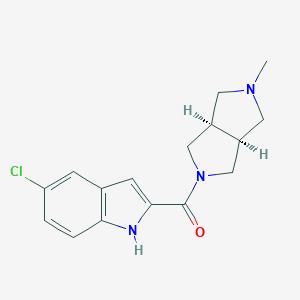 CHEMBL1915345 CHEMBL1915345 | C16H18ClN3O | 303.79 | 2 / 1 | 2.6 | Yes |
| 5094 | 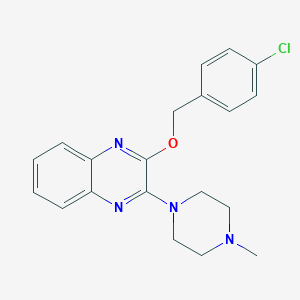 CHEMBL260456 CHEMBL260456 | C20H21ClN4O | 368.865 | 5 / 0 | 3.9 | Yes |
| 5403 | 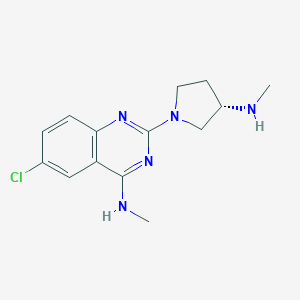 CHEMBL3085393 CHEMBL3085393 | C14H18ClN5 | 291.783 | 5 / 2 | 2.7 | Yes |
| 5404 | 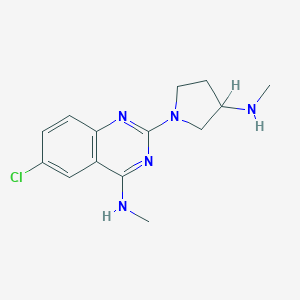 CHEMBL435439 CHEMBL435439 | C14H18ClN5 | 291.783 | 5 / 2 | 2.7 | Yes |
| 5513 | 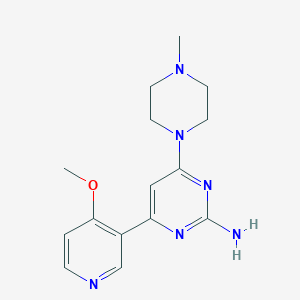 CHEMBL522019 CHEMBL522019 | C15H20N6O | 300.366 | 7 / 1 | 0.7 | Yes |
| 5682 | 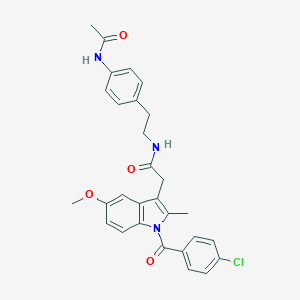 AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 5866 | 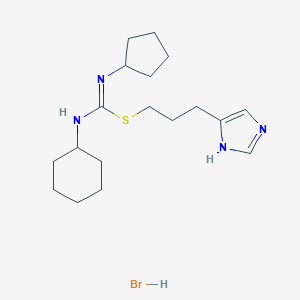 CHEMBL1923030 CHEMBL1923030 | C18H31BrN4S | 415.438 | 3 / 3 | N/A | N/A |
| 6410 | 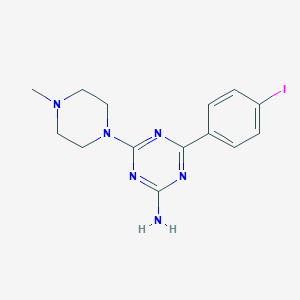 CHEMBL3290579 CHEMBL3290579 | C14H17IN6 | 396.236 | 6 / 1 | 2.2 | Yes |
| 557458 | 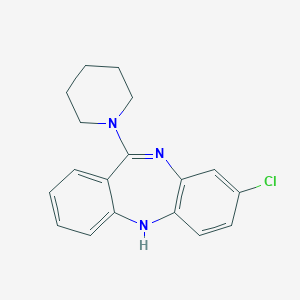 CHEMBL377765 CHEMBL377765 | C18H18ClN3 | 311.813 | 2 / 1 | 4.1 | Yes |
| 463726 | 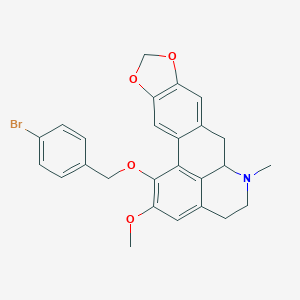 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 7233 | 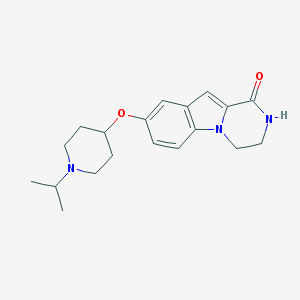 CHEMBL1258754 CHEMBL1258754 | C19H25N3O2 | 327.428 | 3 / 1 | 2.7 | Yes |
| 7355 | 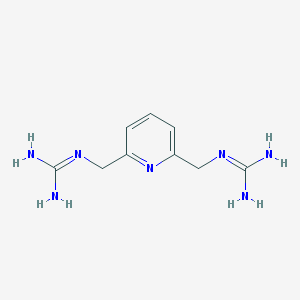 2,6-Bis(guanidinomethyl)pyridine 2,6-Bis(guanidinomethyl)pyridine | C9H15N7 | 221.268 | 3 / 4 | -2.2 | Yes |
| 7356 | 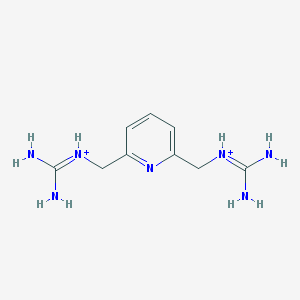 AC1O3VDW AC1O3VDW | C9H17N7+2 | 223.284 | 1 / 6 | -2.2 | No |
| 7455 | 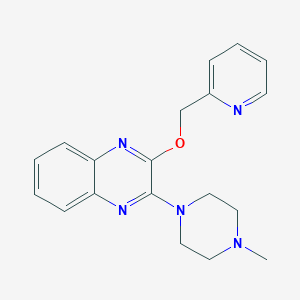 CHEMBL259957 CHEMBL259957 | C19H21N5O | 335.411 | 6 / 0 | 2.2 | Yes |
| 7670 | 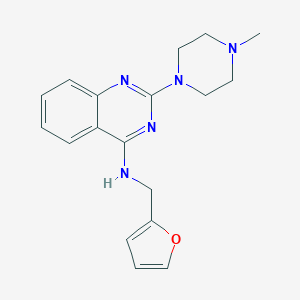 CHEMBL455133 CHEMBL455133 | C18H21N5O | 323.4 | 6 / 1 | 2.7 | Yes |
| 7842 | 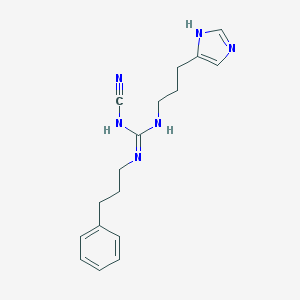 CHEMBL1078210 CHEMBL1078210 | C17H22N6 | 310.405 | 3 / 3 | 2.3 | Yes |
| 8020 | 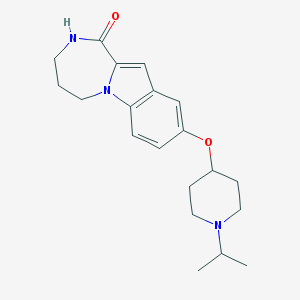 CHEMBL1258755 CHEMBL1258755 | C20H27N3O2 | 341.455 | 3 / 1 | 3.0 | Yes |
| 8102 | 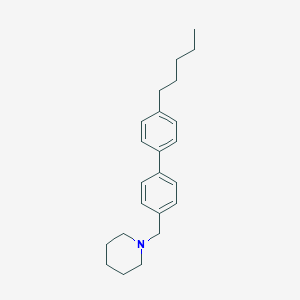 CHEMBL1934066 CHEMBL1934066 | C23H31N | 321.508 | 1 / 0 | 6.7 | No |
| 8502 | 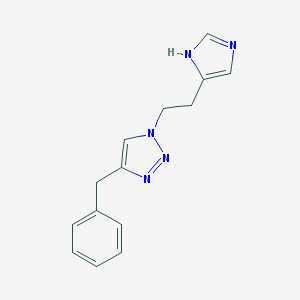 CHEMBL1688939 CHEMBL1688939 | C14H15N5 | 253.309 | 3 / 1 | 1.7 | Yes |
| 10511 | 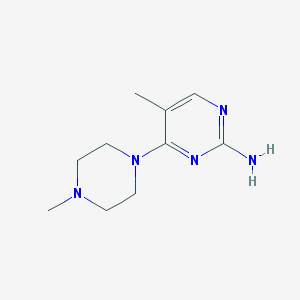 CHEMBL446896 CHEMBL446896 | C10H17N5 | 207.281 | 5 / 1 | 0.5 | Yes |
| 10587 | 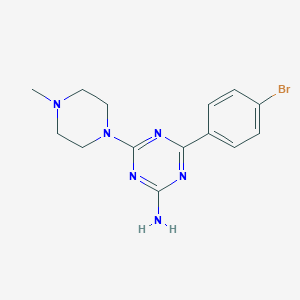 CHEMBL3290578 CHEMBL3290578 | C14H17BrN6 | 349.236 | 6 / 1 | 2.2 | Yes |
| 10745 | 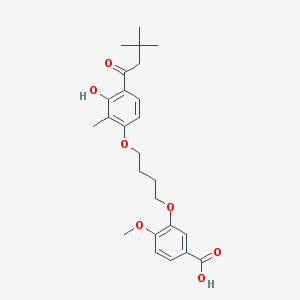 CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 11247 | 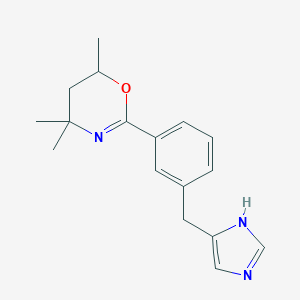 CHEMBL494982 CHEMBL494982 | C17H21N3O | 283.375 | 3 / 1 | 2.9 | Yes |
| 11513 | 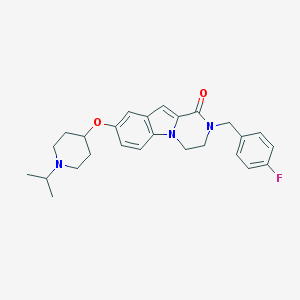 CHEMBL1257376 CHEMBL1257376 | C26H30FN3O2 | 435.543 | 4 / 0 | 4.5 | Yes |
| 11794 | 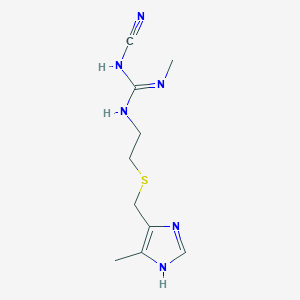 cimetidine cimetidine | C10H16N6S | 252.34 | 4 / 3 | 0.4 | Yes |
| 12692 | 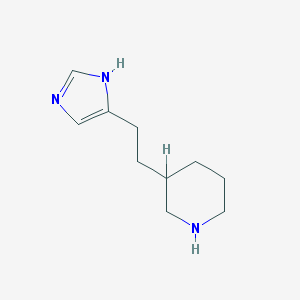 CHEMBL153207 CHEMBL153207 | C10H17N3 | 179.267 | 2 / 2 | 1.0 | Yes |
| 521841 | 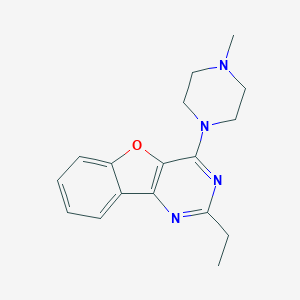 F3277-0121 F3277-0121 | C17H20N4O | 296.374 | 5 / 0 | 3.1 | Yes |
| 14125 | 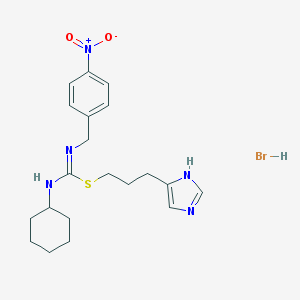 CHEMBL1923034 CHEMBL1923034 | C20H28BrN5O2S | 482.441 | 5 / 3 | N/A | N/A |
| 14525 | 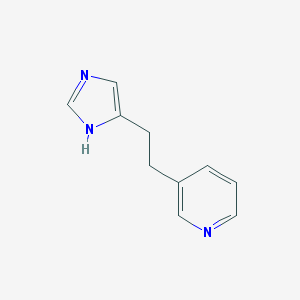 3-[2-(3H-Imidazol-4-yl)-ethyl]-pyridine 3-[2-(3H-Imidazol-4-yl)-ethyl]-pyridine | C10H11N3 | 173.219 | 2 / 1 | 1.2 | Yes |
| 14647 | 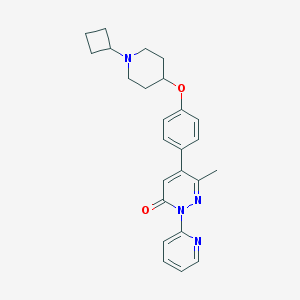 CHEMBL1945848 CHEMBL1945848 | C25H28N4O2 | 416.525 | 5 / 0 | 3.7 | Yes |
| 536376 | 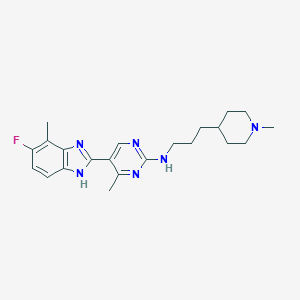 SCHEMBL603807 SCHEMBL603807 | C22H29FN6 | 396.514 | 6 / 2 | 4.1 | Yes |
| 15065 | 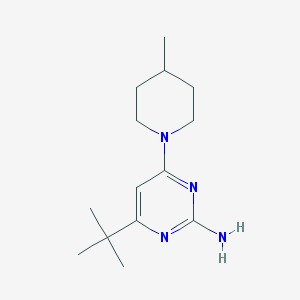 CHEMBL494687 CHEMBL494687 | C14H24N4 | 248.374 | 4 / 1 | 3.3 | Yes |
| 15468 | 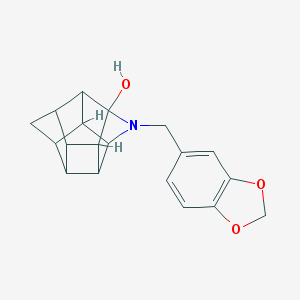 CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 15775 | 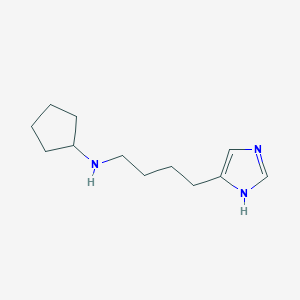 CHEMBL379447 CHEMBL379447 | C12H21N3 | 207.321 | 2 / 2 | 1.9 | Yes |
| 16032 | 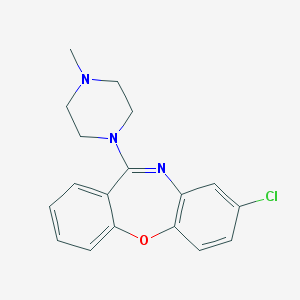 ISOLOXAPINE ISOLOXAPINE | C18H18ClN3O | 327.812 | 3 / 0 | 3.1 | Yes |
| 16057 | 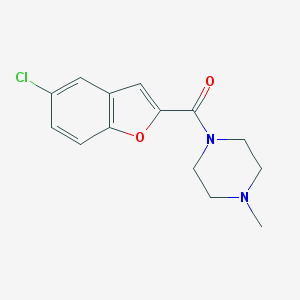 CHEMBL2047259 CHEMBL2047259 | C14H15ClN2O2 | 278.736 | 3 / 0 | 2.6 | Yes |
| 16157 | 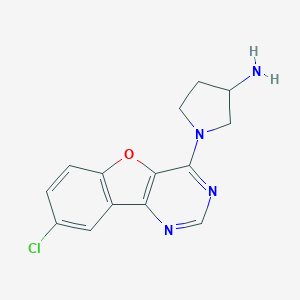 CHEMBL1090532 CHEMBL1090532 | C14H13ClN4O | 288.735 | 5 / 1 | 2.4 | Yes |
| 16339 | 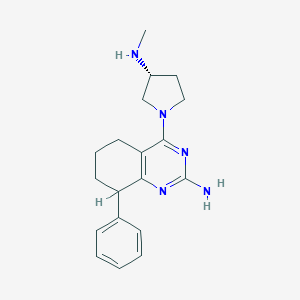 SCHEMBL605165 SCHEMBL605165 | C19H25N5 | 323.444 | 5 / 2 | 2.8 | Yes |
| 16930 | 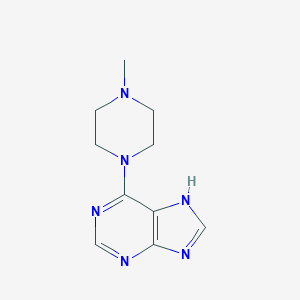 6-(4-methylpiperazin-1-yl)-9H-purine 6-(4-methylpiperazin-1-yl)-9H-purine | C10H14N6 | 218.264 | 5 / 1 | 0.5 | Yes |
| 17462 | 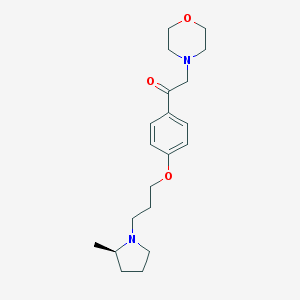 CHEMBL1951055 CHEMBL1951055 | C20H30N2O3 | 346.471 | 5 / 0 | 2.6 | Yes |
| 17762 | 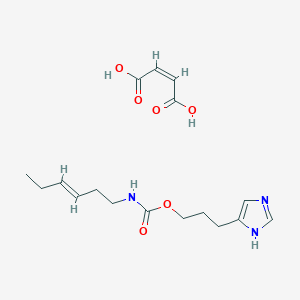 CID 54584377 CID 54584377 | C17H25N3O6 | 367.402 | 7 / 4 | N/A | N/A |
| 17969 | 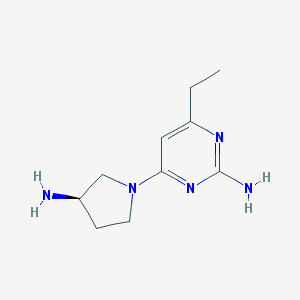 CHEMBL3236569 CHEMBL3236569 | C10H17N5 | 207.281 | 5 / 2 | 0.4 | Yes |
| 17989 | 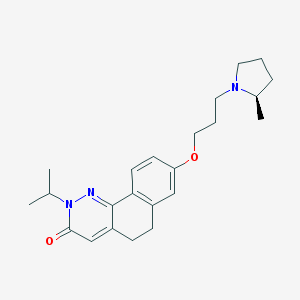 CHEMBL2031471 CHEMBL2031471 | C23H31N3O2 | 381.52 | 4 / 0 | 3.8 | Yes |
| 18670 | 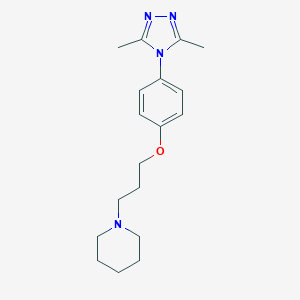 CHEMBL3238444 CHEMBL3238444 | C18H26N4O | 314.433 | 4 / 0 | 3.1 | Yes |
| 18809 | 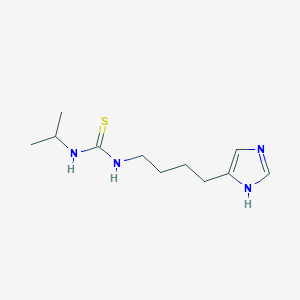 CHEMBL306077 CHEMBL306077 | C11H20N4S | 240.369 | 2 / 3 | 1.2 | Yes |
| 19308 | 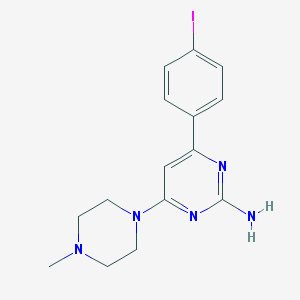 CHEMBL492288 CHEMBL492288 | C15H18IN5 | 395.248 | 5 / 1 | 2.4 | Yes |
| 19467 | 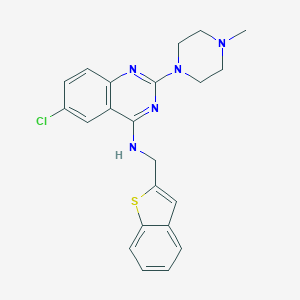 CHEMBL509961 CHEMBL509961 | C22H22ClN5S | 423.963 | 6 / 1 | 5.3 | No |
| 19823 | 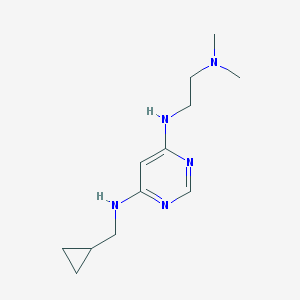 CHEMBL1916505 CHEMBL1916505 | C12H21N5 | 235.335 | 5 / 2 | 1.6 | Yes |
| 20234 | 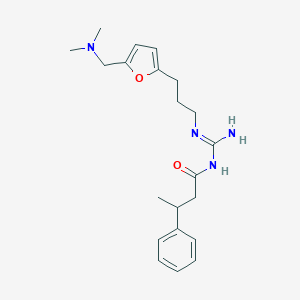 CHEMBL3220888 CHEMBL3220888 | C21H30N4O2 | 370.497 | 4 / 2 | 2.9 | Yes |
| 20696 | 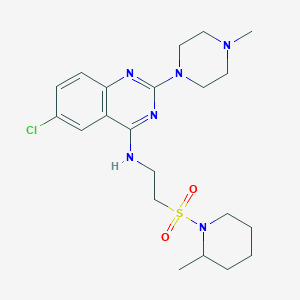 CHEMBL591249 CHEMBL591249 | C21H31ClN6O2S | 467.029 | 8 / 1 | 3.9 | Yes |
| 20777 | 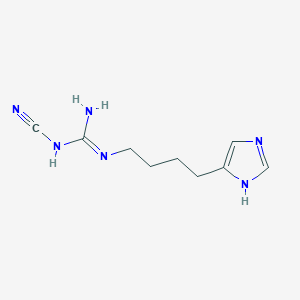 CHEMBL1077861 CHEMBL1077861 | C9H14N6 | 206.253 | 3 / 3 | 0.0 | Yes |
| 465456 | 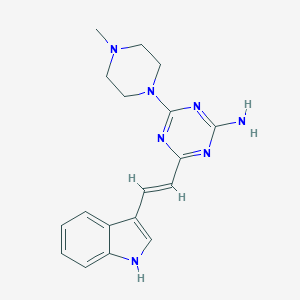 CHEMBL3628052 CHEMBL3628052 | C18H21N7 | 335.415 | 6 / 2 | 2.3 | Yes |
| 21747 |  CHEMBL1914767 CHEMBL1914767 | C14H15N5O | 269.308 | 6 / 2 | 1.5 | Yes |
| 22036 |  CHEMBL1770975 CHEMBL1770975 | C15H27N5 | 277.416 | 5 / 3 | 2.4 | Yes |
| 22404 |  CHEMBL2013045 CHEMBL2013045 | C23H34N2O4S | 434.595 | 6 / 0 | 3.4 | Yes |
| 22735 |  CHEMBL1914794 CHEMBL1914794 | C16H19N5OS | 329.422 | 7 / 2 | 2.5 | Yes |
| 22736 | 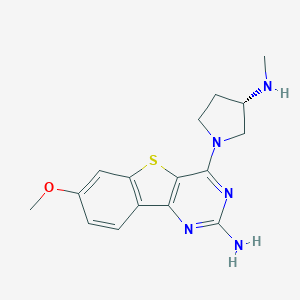 CHEMBL1915022 CHEMBL1915022 | C16H19N5OS | 329.422 | 7 / 2 | 2.5 | Yes |
| 24679 | 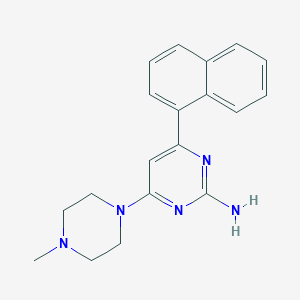 SCHEMBL607510 SCHEMBL607510 | C19H21N5 | 319.412 | 5 / 1 | 3.0 | Yes |
| 24691 | 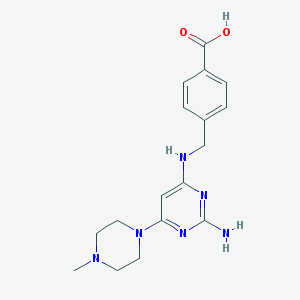 CHEMBL594266 CHEMBL594266 | C17H22N6O2 | 342.403 | 8 / 3 | -0.8 | Yes |
| 24729 | 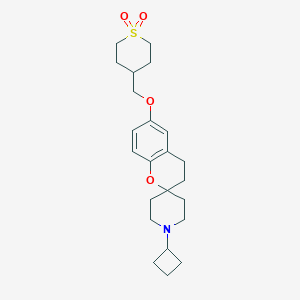 CHEMBL1935426 CHEMBL1935426 | C23H33NO4S | 419.58 | 5 / 0 | 3.7 | Yes |
| 24733 | 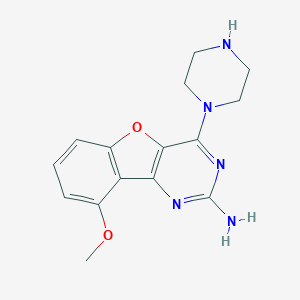 CHEMBL1914771 CHEMBL1914771 | C15H17N5O2 | 299.334 | 7 / 2 | 1.5 | Yes |
| 24767 | 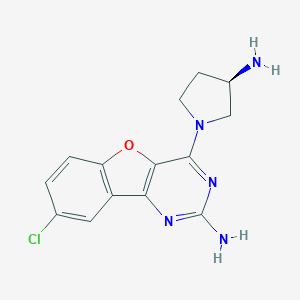 CHEMBL1914543 CHEMBL1914543 | C14H14ClN5O | 303.75 | 6 / 2 | 2.0 | Yes |
| 24829 | 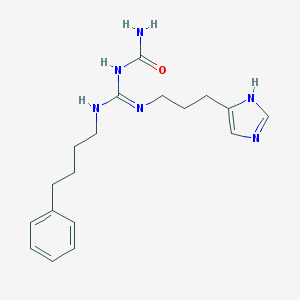 CHEMBL1078746 CHEMBL1078746 | C18H26N6O | 342.447 | 3 / 4 | 2.4 | Yes |
| 24955 | 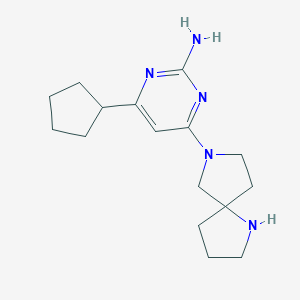 CHEMBL3236564 CHEMBL3236564 | C16H25N5 | 287.411 | 5 / 2 | 2.0 | Yes |
| 25580 | 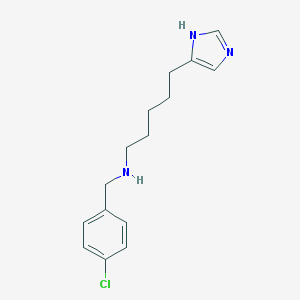 CHEMBL203263 CHEMBL203263 | C15H20ClN3 | 277.796 | 2 / 2 | 3.1 | Yes |
| 25760 | 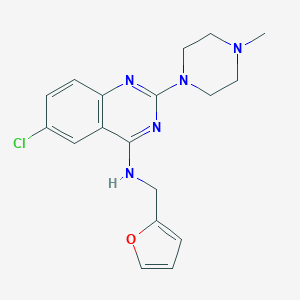 CHEMBL452925 CHEMBL452925 | C18H20ClN5O | 357.842 | 6 / 1 | 3.3 | Yes |
| 26063 | 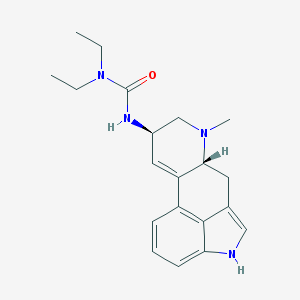 S-(-)-Lisuride S-(-)-Lisuride | C20H26N4O | 338.455 | 2 / 2 | 2.7 | Yes |
| 26294 | 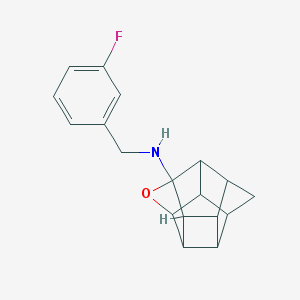 CHEMBL600053 CHEMBL600053 | C18H18FNO | 283.346 | 3 / 1 | 2.5 | Yes |
| 522379 | 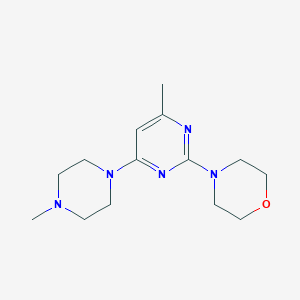 4-[4-methyl-6-(4-methylpiperazin-1-yl)pyrimidin-2-yl]morpholine 4-[4-methyl-6-(4-methylpiperazin-1-yl)pyrimidin-2-yl]morpholine | C14H23N5O | 277.372 | 6 / 0 | 1.0 | Yes |
| 27316 | 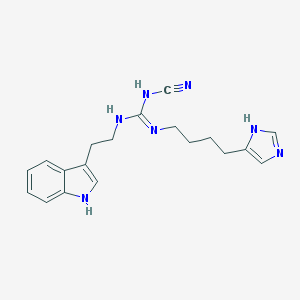 CHEMBL1078735 CHEMBL1078735 | C19H23N7 | 349.442 | 3 / 4 | 2.5 | Yes |
| 27507 | 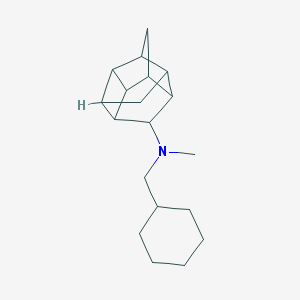 CHEMBL2432063 CHEMBL2432063 | C19H29N | 271.448 | 1 / 0 | 4.7 | Yes |
| 27919 | 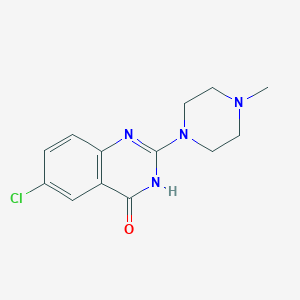 BRN 0805832 BRN 0805832 | C13H15ClN4O | 278.74 | 3 / 1 | 1.1 | Yes |
| 27929 | 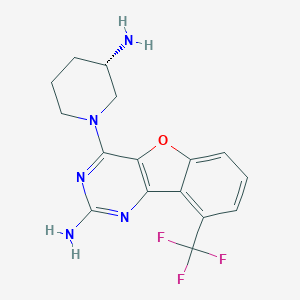 CHEMBL1915024 CHEMBL1915024 | C16H16F3N5O | 351.333 | 9 / 2 | 2.6 | Yes |
| 28129 | 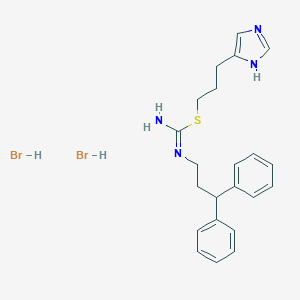 CHEMBL1202323 CHEMBL1202323 | C22H28Br2N4S | 540.362 | 3 / 4 | N/A | No |
| 28390 | 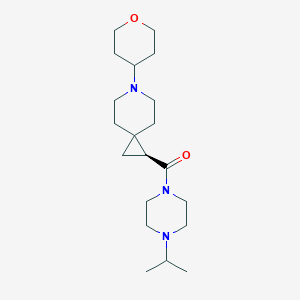 CHEMBL3124968 CHEMBL3124968 | C20H35N3O2 | 349.519 | 4 / 0 | 1.5 | Yes |
| 28589 | 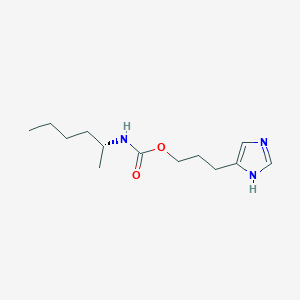 CHEMBL1079553 CHEMBL1079553 | C13H23N3O2 | 253.346 | 3 / 2 | 2.7 | Yes |
| 28592 | 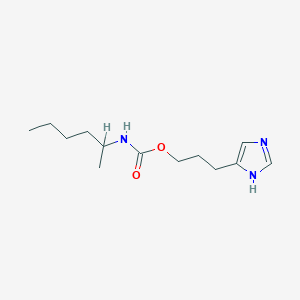 CHEMBL1079552 CHEMBL1079552 | C13H23N3O2 | 253.346 | 3 / 2 | 2.7 | Yes |
| 29044 | 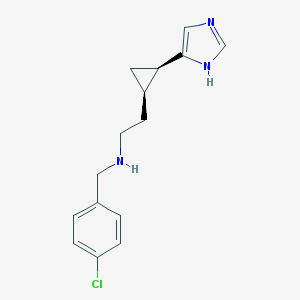 CHEMBL377572 CHEMBL377572 | C15H18ClN3 | 275.78 | 2 / 2 | 2.6 | Yes |
| 29049 | 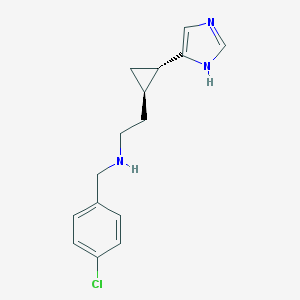 CHEMBL214311 CHEMBL214311 | C15H18ClN3 | 275.78 | 2 / 2 | 2.6 | Yes |
| 29050 | 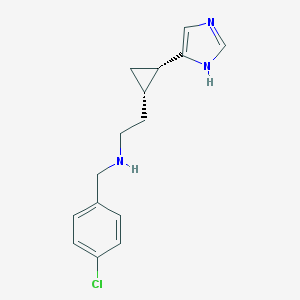 CHEMBL215628 CHEMBL215628 | C15H18ClN3 | 275.78 | 2 / 2 | 2.6 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417