

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MPDTNSTINLSLSTRVTLAFFMSLVAFAIMLGNALVILAFVVDKNLRHRSSYFFLNLAISDFFVGVISIPLYIPHTLFEWDFGKEICVFWLTTDYLLCTASVYNIVLISYDRYLSVSNAVSYRTQHTGVLKIVTLMVAVWVLAFLVNGPMILVSESWKDEGSECEPGFFSEWYILAITSFLEFVIPVILVAYFNMNIYWSLWKRDHLSRCQSHPGLTAVSSNICGHSFRGRLSSRRSLSASTEVPASFHSERQRRKSSLMFSSRTKMNSNTIASKMGSFSQSDSVALHQREHVELLRARRLAKSLAILLGVFAVCWAPYSLFTIVLSFYSSATGPKSVWYRIAFWLQWFNSFVNPLLYPLCHKRFQKAFLKIFCIKKQPLPSQHSRSVSS | |
| CCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCCCCHHHHHHHHHHHHHHSSSSCCHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHSSHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHSSSSSHHHHHHHHHCHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCCCCCCCCCCCCC | |
| 989998988877899999999999999999956860775773145577504677898888885216852258996608406866899999999999999999999999986276242720064120068888979999999999998556400344357998314754577499999999999999999989867876248766656512332333344443334332223343233333433223333334445544332100123432333332322222222210012234542200113555356678898253889999998637877876799999999998884020999998079999999999587788988888888899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MPDTNSTINLSLSTRVTLAFFMSLVAFAIMLGNALVILAFVVDKNLRHRSSYFFLNLAISDFFVGVISIPLYIPHTLFEWDFGKEICVFWLTTDYLLCTASVYNIVLISYDRYLSVSNAVSYRTQHTGVLKIVTLMVAVWVLAFLVNGPMILVSESWKDEGSECEPGFFSEWYILAITSFLEFVIPVILVAYFNMNIYWSLWKRDHLSRCQSHPGLTAVSSNICGHSFRGRLSSRRSLSASTEVPASFHSERQRRKSSLMFSSRTKMNSNTIASKMGSFSQSDSVALHQREHVELLRARRLAKSLAILLGVFAVCWAPYSLFTIVLSFYSSATGPKSVWYRIAFWLQWFNSFVNPLLYPLCHKRFQKAFLKIFCIKKQPLPSQHSRSVSS | |
| 424443325142011000002113102303311200000001233032100000000010011000000010000002432013100010000001000000200000000001000200302333242000000000012002010100000021254564412030243200000002101320110012001000000132122233434232433434433433423233233333322322223344333332122322233332222322232232222222234333234333102100001000100020000000020003321204300000001201300120000001004301400230000222524533443348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCCCCHHHHHHHHHHHHHHSSSSCCHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHSSHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHSSSSSHHHHHHHHHCHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCCCCCCCCCCCCC MPDTNSTINLSLSTRVTLAFFMSLVAFAIMLGNALVILAFVVDKNLRHRSSYFFLNLAISDFFVGVISIPLYIPHTLFEWDFGKEICVFWLTTDYLLCTASVYNIVLISYDRYLSVSNAVSYRTQHTGVLKIVTLMVAVWVLAFLVNGPMILVSESWKDEGSECEPGFFSEWYILAITSFLEFVIPVILVAYFNMNIYWSLWKRDHLSRCQSHPGLTAVSSNICGHSFRGRLSSRRSLSASTEVPASFHSERQRRKSSLMFSSRTKMNSNTIASKMGSFSQSDSVALHQREHVELLRARRLAKSLAILLGVFAVCWAPYSLFTIVLSFYSSATGPKSVWYRIAFWLQWFNSFVNPLLYPLCHKRFQKAFLKIFCIKKQPLPSQHSRSVSS | |||||||||||||||||||||||||
| 1 | 4ib4A | 0.19 | 0.21 | 0.95 | 2.52 | Download | --------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNS-RATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKKHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------ | |||||||||||||||||||
| 2 | 4ib4A | 0.19 | 0.21 | 0.93 | 4.52 | Download | --------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNS-RATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------ | |||||||||||||||||||
| 3 | 5uenA | 0.20 | 0.25 | 0.95 | 3.74 | Download | ---------SISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGQTYF--HTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTP-RRAAVAIAGCWILSFVVGLTPMFGWNNLSMGEPVIKCEVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDN-VKDALTKMRAAALDAPEMKDFRHGFDILVGQIDDALKLANEG---KVKEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPS-CHKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQPLEVLF---- | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.21 | 0.94 | 1.57 | Download | --------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYN-SRATAFIKITVVWLISIGIAIPVPIGIET-NPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLIEKADNAQVKDLTKMRAAALDAQKKDFR-HGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------ | |||||||||||||||||||
| 5 | 3uon | 0.22 | 0.27 | 0.92 | 1.24 | Download | -----------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTT-KMAGMMIAAAWVLSFILWAPAILFWQFIVVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKIYKDTEGKSPAAKAIGRNTNGVITKDKVRGILNAKLDAVRAGMLQQKRWEAAVNLAKSRWYNTFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC--IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------------- | |||||||||||||||||||
| 6 | 4ib4A | 0.19 | 0.21 | 0.94 | 3.48 | Download | ----------QGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNS-RATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKKDFRHGFDILVGQIDDALKLGKVKEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------ | |||||||||||||||||||
| 7 | 4ib4 | 0.20 | 0.21 | 0.92 | 1.74 | Download | --------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSR-ATAFIKITVVWLISIGIAIPVPIGIETN-PNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADN-AAQVKD--ALTKMRAAALDAQKKDFRHGFDILVGIDLKLANEGKVKEAQAAAEQTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------ | |||||||||||||||||||
| 8 | 4dajA | 0.25 | 0.28 | 0.92 | 3.91 | Download | -----------IWQVVFIAFLTGFLALVTIIGNILVIVAFKVNKQLKTVNNYFLLSLACADLIIGVISMNLFTTYIIMNWALGNLACDLWLSIDYVASNASVMNLLVISFDRYFSITRPLTYRAKRT-TKRAGVMIGLAWVISFVLWAPAILFWQYFVGKRGECFIQFLSEPTITFGTAIAAFYMPVTIMTILYWRIYKETAKSELDKAIGRNTNGVITKDEAEKLFNQDVDAAVRGILRNAKAALINMVFQMGETGVAGFTNSLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTGTLSAILLAFIITWTPYNIMVLVNTFCDSC--IPKTYWNLGYWLCYINSTVNPVCYALCNKTFRTTFKT------------------- | |||||||||||||||||||
| 9 | 4ib4A | 0.19 | 0.21 | 0.95 | 2.82 | Download | --------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYN-SRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKKDFRHILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------ | |||||||||||||||||||
| 10 | 5dsgA | 0.22 | 0.26 | 0.95 | 2.43 | Download | --GPSSHNRYETVEMVFIATVTGSLSLVTVVGNILVMLSIKVNRQLQTVNNYFLFSLACADLIIGAFSMNLYTVYIIGYWPLGAVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPARRT-TKMAGLMIAAAWVLSFVLWAPAILFWQKRTVPDNQCFIQFLSNPAVTFGTAIAAFYLPVVIMTVLYIHISLASRSRVNIFEMLRIDEEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDFQMGETGVAGFTNSLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTRKVTRTIFAILLAFILTWTPYNVMVLVNTFCQSCI--PDTVWSIGYWLCYVNSTINPACYALCNATFKKTFRHLLLC--------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
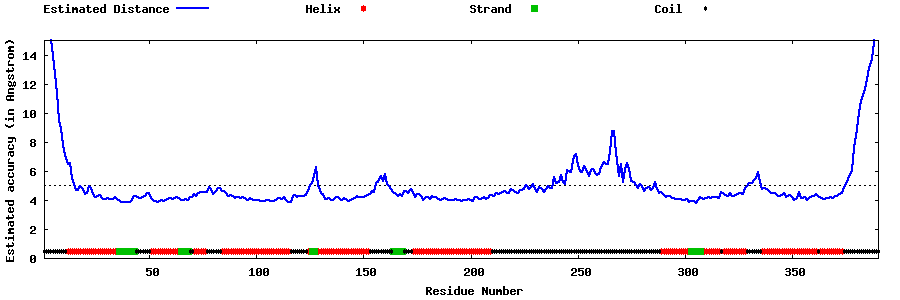
| Generated 3D models | Estimated local accuracy of models | ||
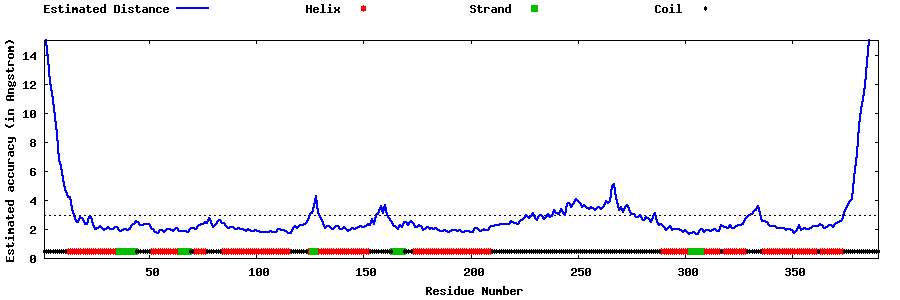 | |||
|
|
|||
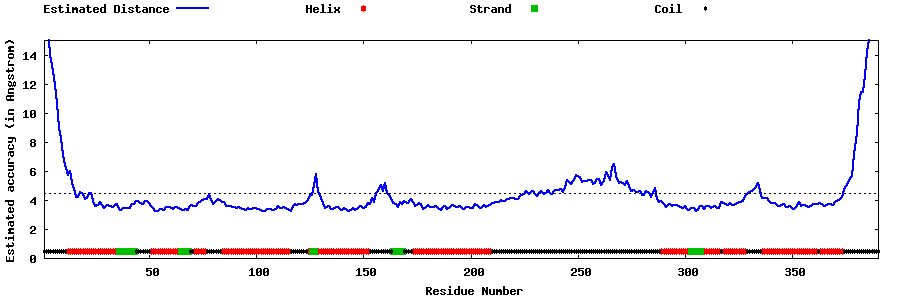 | |||
|
| |||
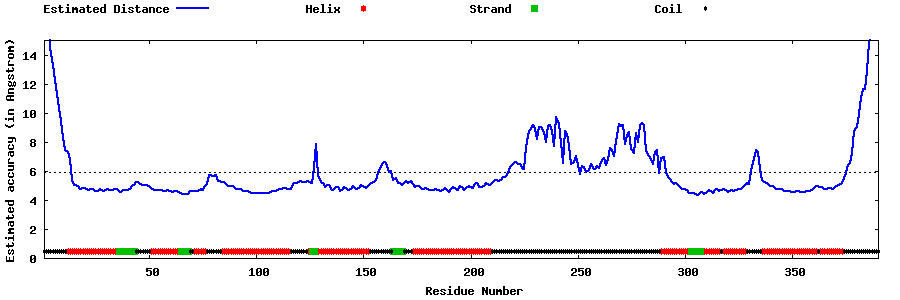 | |||
|
| |||
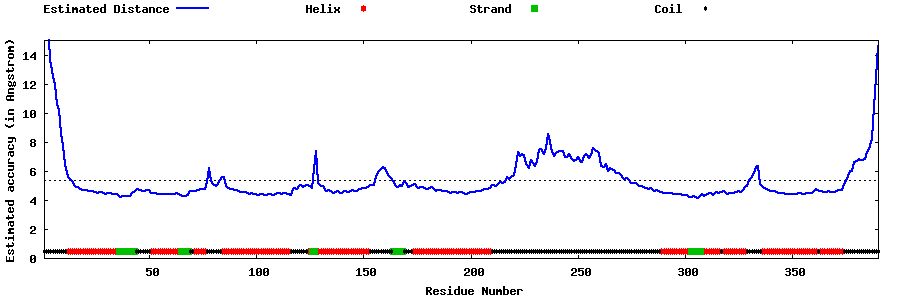 | |||
|
| |||
 | |||
|
| |||