

You can:
| Name | Sphingosine 1-phosphate receptor 3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | S1PR3 |
| Synonym | Sphingosine 1-phosphate receptor Edg-3 S1P3 receptor S1P3 S1P receptor Edg-3 S1P receptor 3 [ Show all ] |
| Disease | Breast cancer |
| Length | 378 |
| Amino acid sequence | MATALPPRLQPVRGNETLREHYQYVGKLAGRLKEASEGSTLTTVLFLVICSFIVLENLMVLIAIWKNNKFHNRMYFFIGNLALCDLLAGIAYKVNILMSGKKTFSLSPTVWFLREGSMFVALGASTCSLLAIAIERHLTMIKMRPYDANKRHRVFLLIGMCWLIAFTLGALPILGWNCLHNLPDCSTILPLYSKKYIAFCISIFTAILVTIVILYARIYFLVKSSSRKVANHNNSERSMALLRTVVIVVSVFIACWSPLFILFLIDVACRVQACPILFKAQWFIVLAVLNSAMNPVIYTLASKEMRRAFFRLVCNCLVRGRGARASPIQPALDPSRSKSSSSNNSSHSPKVKEDLPHTAPSSCIMDKNAALQNGIFCN |
| UniProt | Q99500 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q99500 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q99500. |
| BioLiP | N/A |
| Therapeutic Target Database | T11241 |
| ChEMBL | CHEMBL3892 |
| IUPHAR | 277 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 34 | 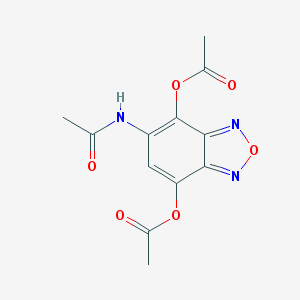 MLS000077136 MLS000077136 | C12H11N3O6 | 293.235 | 8 / 1 | -0.1 | Yes |
| 149 | 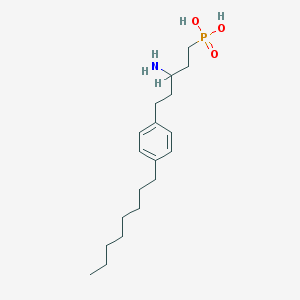 CHEMBL325198 CHEMBL325198 | C19H34NO3P | 355.459 | 4 / 3 | 2.0 | Yes |
| 165 | 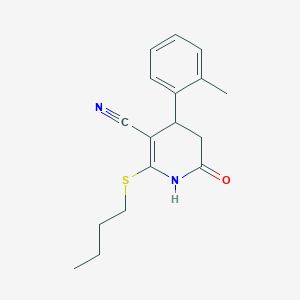 MLS000103088 MLS000103088 | C17H20N2OS | 300.42 | 3 / 1 | 3.2 | Yes |
| 302 | 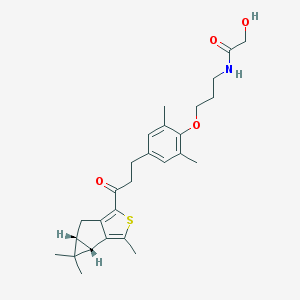 SCHEMBL2458105 SCHEMBL2458105 | C27H35NO4S | 469.64 | 5 / 2 | 4.9 | Yes |
| 331 | 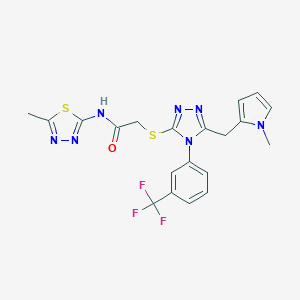 SMR000018340 SMR000018340 | C20H18F3N7OS2 | 493.527 | 10 / 1 | 3.6 | Yes |
| 557317 | 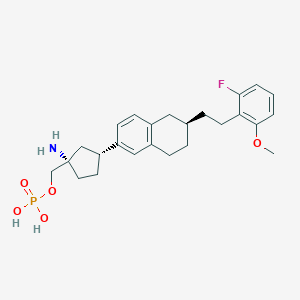 BDBM65291 BDBM65291 | C25H33FNO5P | 477.513 | 7 / 3 | 1.4 | Yes |
| 461 | 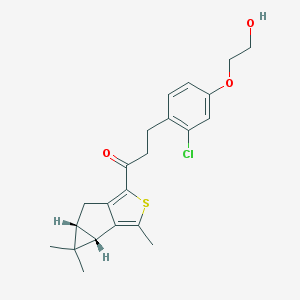 SCHEMBL2948159 SCHEMBL2948159 | C22H25ClO3S | 404.949 | 4 / 1 | 5.1 | No |
| 557324 | 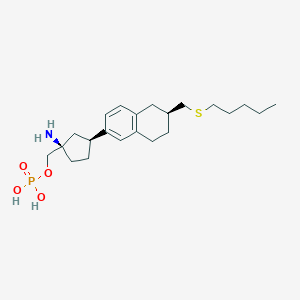 BDBM65164 BDBM65164 | C22H36NO4PS | 441.567 | 6 / 3 | 1.1 | Yes |
| 557326 | 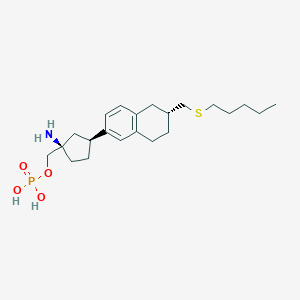 BDBM65165 BDBM65165 | C22H36NO4PS | 441.567 | 6 / 3 | 1.1 | Yes |
| 1114 | 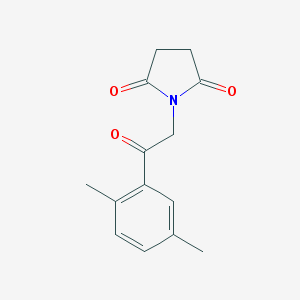 MLS000054921 MLS000054921 | C14H15NO3 | 245.278 | 3 / 0 | 1.3 | Yes |
| 1372 | 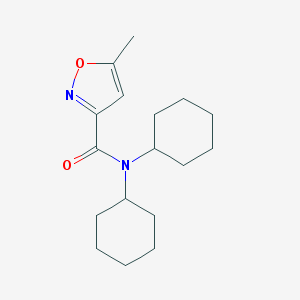 SMR000023524 SMR000023524 | C17H26N2O2 | 290.407 | 3 / 0 | 4.2 | Yes |
| 1485 | 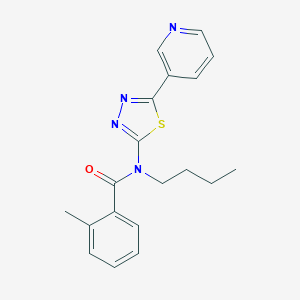 CHEMBL2011712 CHEMBL2011712 | C19H20N4OS | 352.456 | 5 / 0 | 3.9 | Yes |
| 1620 | 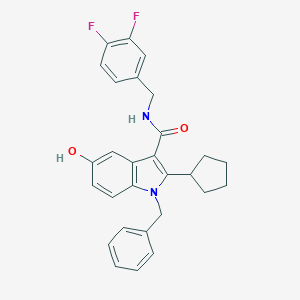 SCHEMBL786387 SCHEMBL786387 | C28H26F2N2O2 | 460.525 | 4 / 2 | 5.9 | No |
| 1955 | 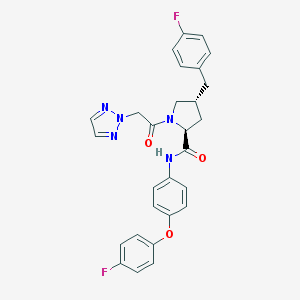 CHEMBL2018573 CHEMBL2018573 | C28H25F2N5O3 | 517.537 | 7 / 1 | 4.9 | No |
| 2072 | 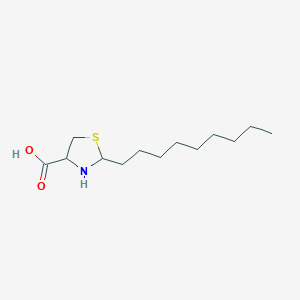 CHEMBL344354 CHEMBL344354 | C13H25NO2S | 259.408 | 4 / 2 | 2.2 | Yes |
| 2168 | 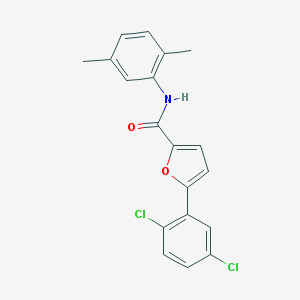 SR-02000000242 SR-02000000242 | C19H15Cl2NO2 | 360.234 | 2 / 1 | 5.8 | No |
| 2340 |  CHEMBL3126597 CHEMBL3126597 | C26H35N5O5 | 497.596 | 9 / 3 | 3.2 | Yes |
| 2963 |  CHEMBL1950564 CHEMBL1950564 | C26H24FN3O2S | 461.555 | 7 / 2 | 2.6 | Yes |
| 2970 |  CHEMBL1938927 CHEMBL1938927 | C26H24FN3O2S | 461.555 | 7 / 1 | 2.6 | Yes |
| 557379 |  BDBM65105 BDBM65105 | C24H32NO5P | 445.496 | 6 / 3 | 0.6 | Yes |
| 557382 | 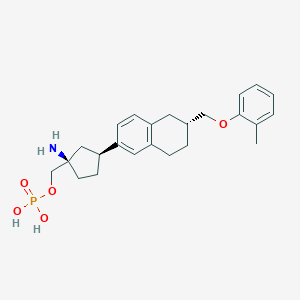 BDBM65106 BDBM65106 | C24H32NO5P | 445.496 | 6 / 3 | 0.6 | Yes |
| 3499 | 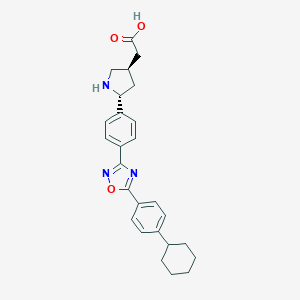 CHEMBL377828 CHEMBL377828 | C26H29N3O3 | 431.536 | 6 / 2 | 3.0 | Yes |
| 3567 | 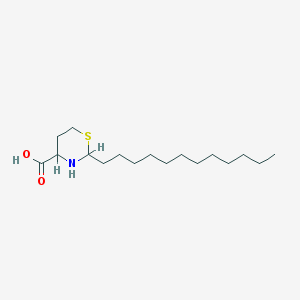 CHEMBL143172 CHEMBL143172 | C17H33NO2S | 315.516 | 4 / 2 | 4.2 | Yes |
| 521553 | 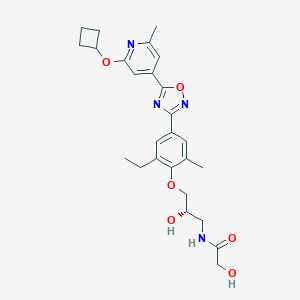 CHEMBL3800120 CHEMBL3800120 | C26H32N4O6 | 496.564 | 9 / 3 | 3.2 | Yes |
| 4069 |  SCHEMBL289923 SCHEMBL289923 | C29H30F2N2O2 | 476.568 | 4 / 1 | 6.6 | No |
| 521570 |  CHEMBL3797365 CHEMBL3797365 | C26H34N4O6 | 498.58 | 9 / 3 | 3.2 | Yes |
| 4388 |  CHEMBL117973 CHEMBL117973 | C18H40NO3P | 349.496 | 4 / 3 | 3.3 | Yes |
| 4681 |  SCHEMBL4888322 SCHEMBL4888322 | C28H28F2N4O3 | 506.554 | 7 / 1 | 4.9 | No |
| 4738 |  SCHEMBL805697 SCHEMBL805697 | C27H24F2N2O3 | 462.497 | 5 / 2 | 5.3 | No |
| 4773 |  SCHEMBL805715 SCHEMBL805715 | C29H29F2N3O2 | 489.567 | 5 / 1 | 6.0 | No |
| 5086 |  CYM50199 CYM50199 | C14H12BrCl2NO2 | 377.059 | 3 / 0 | 5.2 | No |
| 441907 |  CHEMBL3359840 CHEMBL3359840 | C24H24N4O5 | 448.479 | 9 / 1 | 0.8 | Yes |
| 5484 | 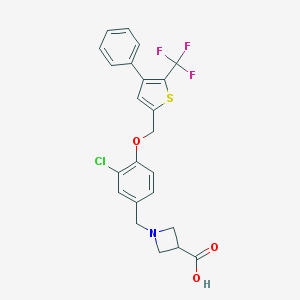 CHEMBL224571 CHEMBL224571 | C23H19ClF3NO3S | 481.914 | 8 / 1 | 3.1 | Yes |
| 5664 | 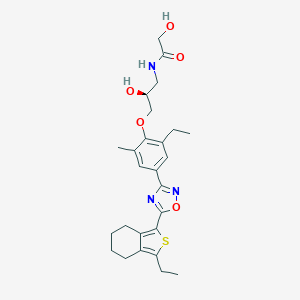 CHEMBL3121985 CHEMBL3121985 | C26H33N3O5S | 499.626 | 8 / 3 | 4.6 | Yes |
| 441920 | 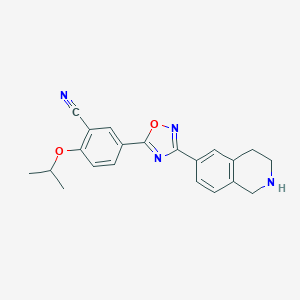 CHEMBL3360364 CHEMBL3360364 | C21H20N4O2 | 360.417 | 6 / 1 | 3.4 | Yes |
| 5969 | 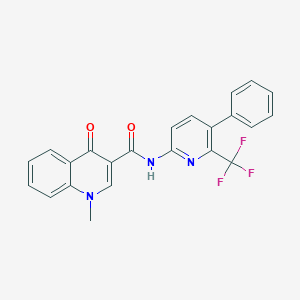 CHEMBL1938943 CHEMBL1938943 | C23H16F3N3O2 | 423.395 | 7 / 1 | 4.6 | Yes |
| 6038 | 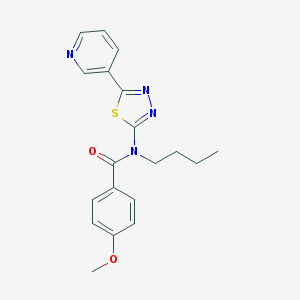 CHEMBL2011719 CHEMBL2011719 | C19H20N4O2S | 368.455 | 6 / 0 | 3.5 | Yes |
| 6323 | 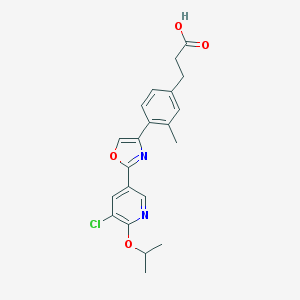 CHEMBL211407 CHEMBL211407 | C21H21ClN2O4 | 400.859 | 6 / 1 | 4.4 | Yes |
| 6598 | 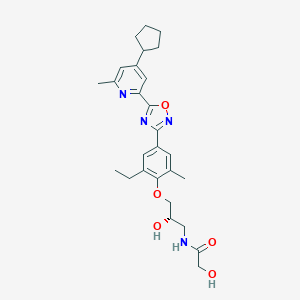 CHEMBL3126596 CHEMBL3126596 | C27H34N4O5 | 494.592 | 8 / 3 | 4.0 | Yes |
| 521640 | 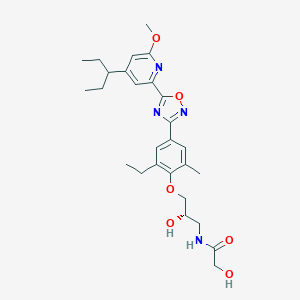 CHEMBL3797486 CHEMBL3797486 | C27H36N4O6 | 512.607 | 9 / 3 | 4.1 | No |
| 521651 | 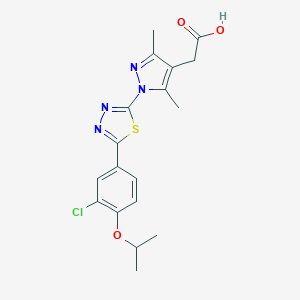 CHEMBL3770278 CHEMBL3770278 | C18H19ClN4O3S | 406.885 | 7 / 1 | 4.0 | Yes |
| 7075 | 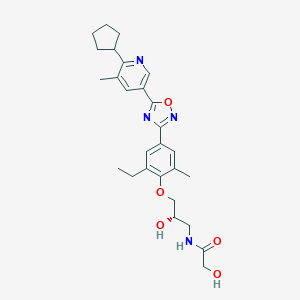 CHEMBL3126622 CHEMBL3126622 | C27H34N4O5 | 494.592 | 8 / 3 | 3.6 | Yes |
| 521656 | 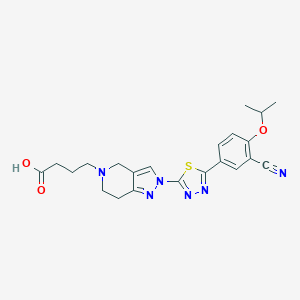 CHEMBL3771073 CHEMBL3771073 | C22H24N6O3S | 452.533 | 9 / 1 | 0.3 | Yes |
| 441997 | 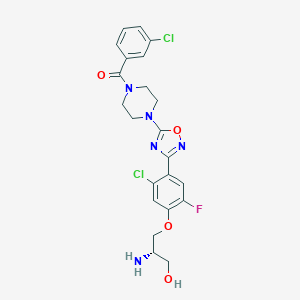 CHEMBL3344423 CHEMBL3344423 | C22H22Cl2FN5O4 | 510.347 | 9 / 2 | 3.0 | No |
| 7336 | 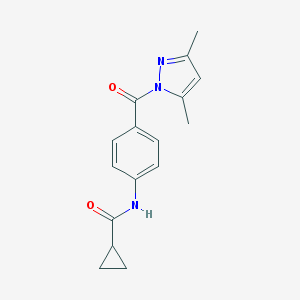 SMR000015635 SMR000015635 | C16H17N3O2 | 283.331 | 3 / 1 | 2.2 | Yes |
| 7679 | 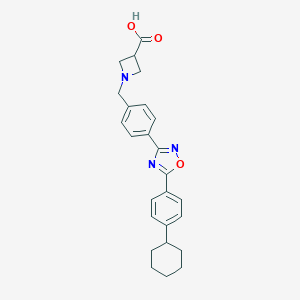 CHEMBL196534 CHEMBL196534 | C25H27N3O3 | 417.509 | 6 / 1 | 2.9 | Yes |
| 8060 | 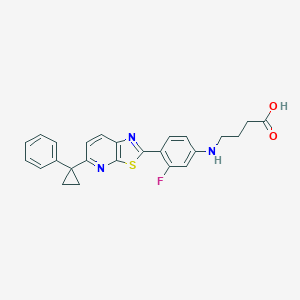 CHEMBL1950562 CHEMBL1950562 | C25H22FN3O2S | 447.528 | 7 / 2 | 5.5 | No |
| 442039 | 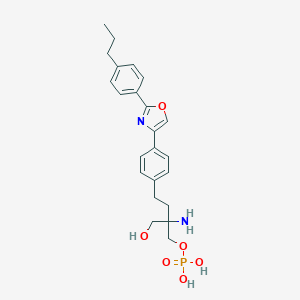 CHEMBL3341925 CHEMBL3341925 | C23H29N2O6P | 460.467 | 8 / 4 | -0.2 | Yes |
| 8309 | 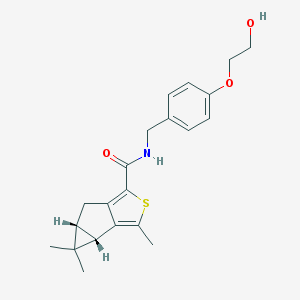 SCHEMBL2945721 SCHEMBL2945721 | C21H25NO3S | 371.495 | 4 / 2 | 3.7 | Yes |
| 8342 | 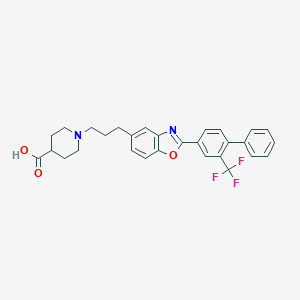 CHEMBL2032437 CHEMBL2032437 | C29H27F3N2O3 | 508.541 | 8 / 1 | 4.3 | No |
| 8589 | 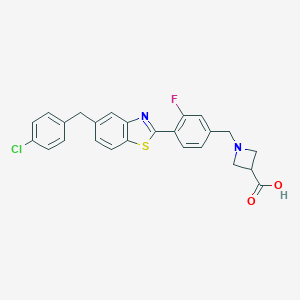 CHEMBL1672569 CHEMBL1672569 | C25H20ClFN2O2S | 466.955 | 6 / 1 | 3.6 | Yes |
| 557519 | 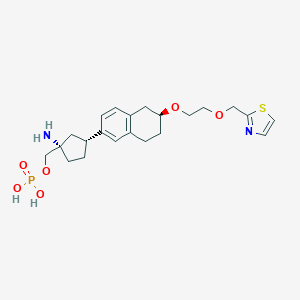 BDBM65262 BDBM65262 | C22H31N2O6PS | 482.532 | 9 / 3 | -1.3 | Yes |
| 8957 | 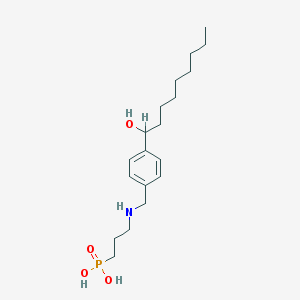 CHEMBL117007 CHEMBL117007 | C19H34NO4P | 371.458 | 5 / 4 | 0.6 | Yes |
| 9120 | 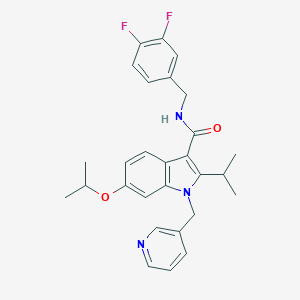 SCHEMBL4886369 SCHEMBL4886369 | C28H29F2N3O2 | 477.556 | 5 / 1 | 5.4 | No |
| 9256 | 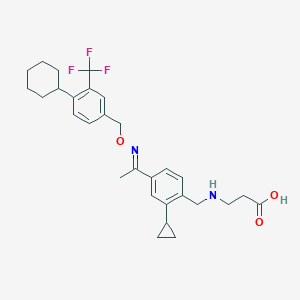 CHEMBL2336069 CHEMBL2336069 | C29H35F3N2O3 | 516.605 | 8 / 2 | 4.6 | No |
| 9787 | 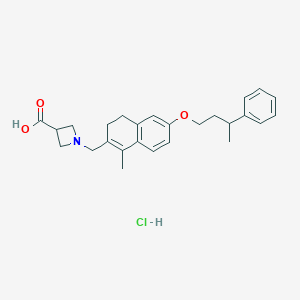 CHEMBL1935579 CHEMBL1935579 | C26H32ClNO3 | 441.996 | 4 / 2 | N/A | N/A |
| 9976 | 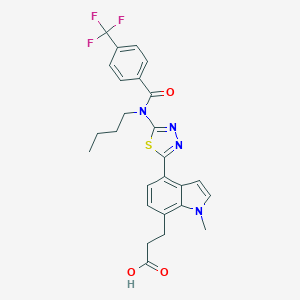 CHEMBL2011747 CHEMBL2011747 | C26H25F3N4O3S | 530.566 | 9 / 1 | 5.4 | No |
| 10099 | 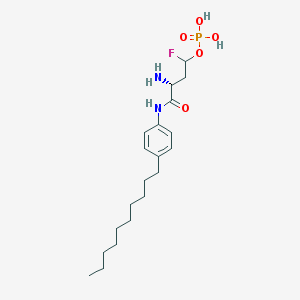 CHEMBL226612 CHEMBL226612 | C20H34FN2O5P | 432.473 | 7 / 4 | 2.2 | Yes |
| 557573 | 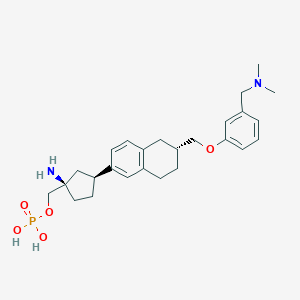 BDBM65133 BDBM65133 | C26H37N2O5P | 488.565 | 7 / 3 | 0.1 | Yes |
| 521730 | 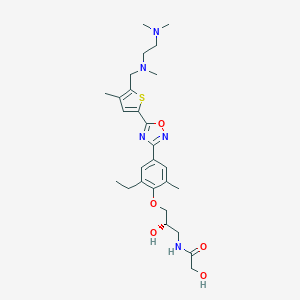 CHEMBL3799923 CHEMBL3799923 | C27H39N5O5S | 545.699 | 10 / 3 | 2.6 | No |
| 10258 |  CHEMBL3133594 CHEMBL3133594 | C21H25N2O7P | 448.412 | 9 / 4 | -1.3 | Yes |
| 10338 |  MLS000043821 MLS000043821 | C16H11ClFN5O | 343.746 | 6 / 1 | 2.9 | Yes |
| 10695 |  CHEMBL206940 CHEMBL206940 | C21H21ClN2O4 | 400.859 | 6 / 1 | 5.0 | Yes |
| 11001 |  CHEMBL334038 CHEMBL334038 | C19H33N2O5P | 400.456 | 6 / 4 | 1.3 | Yes |
| 11619 | 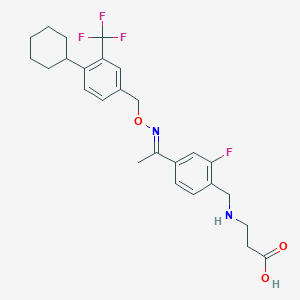 CHEMBL2336064 CHEMBL2336064 | C26H30F4N2O3 | 494.531 | 9 / 2 | 3.8 | Yes |
| 11895 | 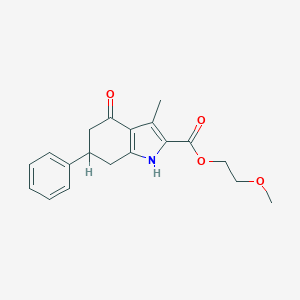 MLS000090902 MLS000090902 | C19H21NO4 | 327.38 | 4 / 1 | 2.6 | Yes |
| 521801 | 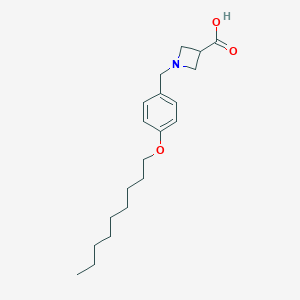 CHEMBL3739440 CHEMBL3739440 | C20H31NO3 | 333.472 | 4 / 1 | 2.7 | Yes |
| 12295 | 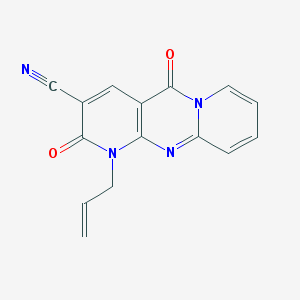 MLS000094258 MLS000094258 | C15H10N4O2 | 278.271 | 4 / 0 | 0.5 | Yes |
| 12308 | 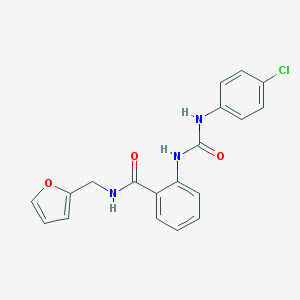 MLS000051886 MLS000051886 | C19H16ClN3O3 | 369.805 | 3 / 3 | 3.5 | Yes |
| 12448 | 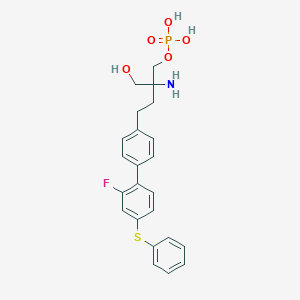 CHEMBL1092286 CHEMBL1092286 | C23H25FNO5PS | 477.487 | 8 / 4 | 0.5 | Yes |
| 517383 | 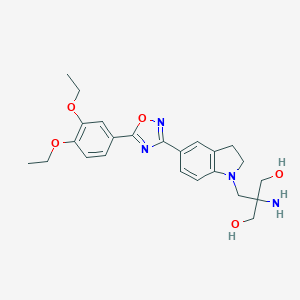 SCHEMBL566013 SCHEMBL566013 | C24H30N4O5 | 454.527 | 9 / 3 | 1.9 | Yes |
| 13018 | 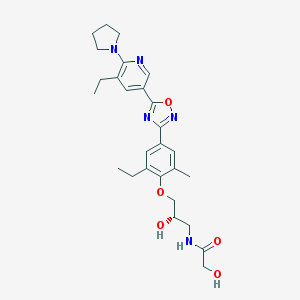 SCHEMBL1187087 SCHEMBL1187087 | C27H35N5O5 | 509.607 | 9 / 3 | 3.3 | No |
| 13107 | 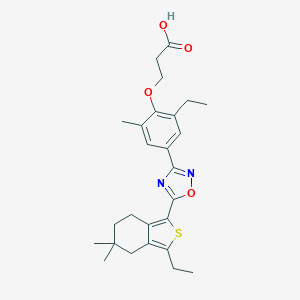 CHEMBL3121960 CHEMBL3121960 | C26H32N2O4S | 468.612 | 7 / 1 | 6.7 | No |
| 13212 | 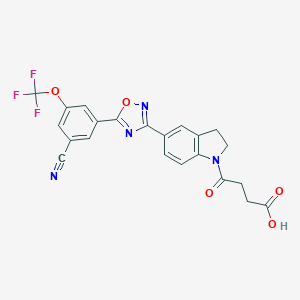 CHEMBL1916399 CHEMBL1916399 | C22H15F3N4O5 | 472.38 | 11 / 1 | 3.2 | No |
| 13506 | 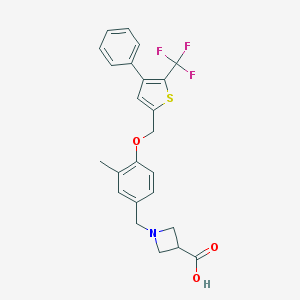 CHEMBL224005 CHEMBL224005 | C24H22F3NO3S | 461.499 | 8 / 1 | 2.9 | Yes |
| 13893 | 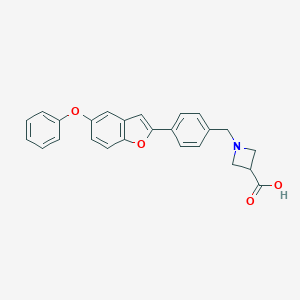 CHEMBL1651707 CHEMBL1651707 | C25H21NO4 | 399.446 | 5 / 1 | 2.4 | Yes |
| 459345 | 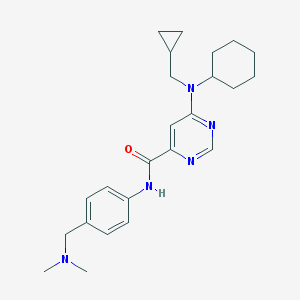 CHEMBL1087911 CHEMBL1087911 | C24H33N5O | 407.562 | 5 / 1 | 4.2 | Yes |
| 13988 | 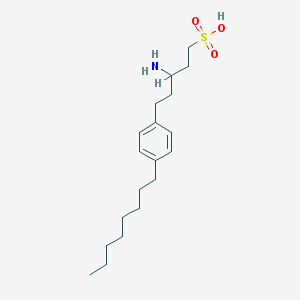 CHEMBL115131 CHEMBL115131 | C19H33NO3S | 355.537 | 4 / 2 | 2.7 | Yes |
| 14403 | 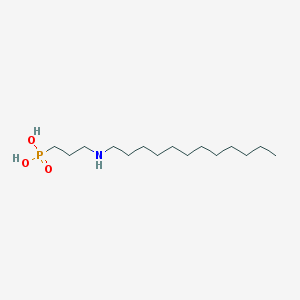 CHEMBL119562 CHEMBL119562 | C15H34NO3P | 307.415 | 4 / 3 | 1.6 | Yes |
| 14464 | 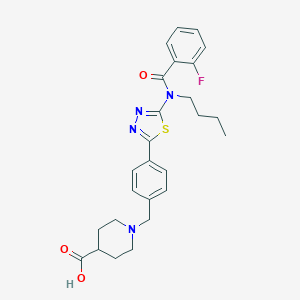 CHEMBL2207780 CHEMBL2207780 | C26H29FN4O3S | 496.601 | 8 / 1 | 2.3 | Yes |
| 14697 | 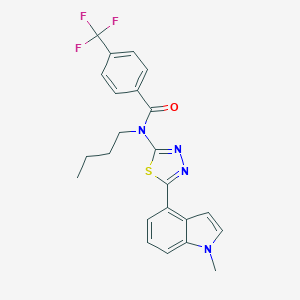 CHEMBL2011740 CHEMBL2011740 | C23H21F3N4OS | 458.503 | 7 / 0 | 5.6 | No |
| 14859 | 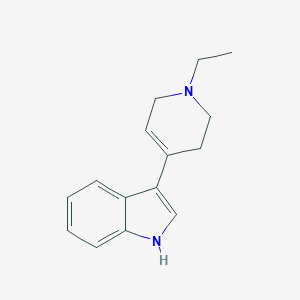 TCMDC-124447 TCMDC-124447 | C15H18N2 | 226.323 | 1 / 1 | 2.6 | Yes |
| 557714 | 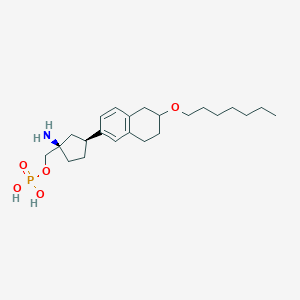 BDBM65063 BDBM65063 | C23H38NO5P | 439.533 | 6 / 3 | 1.2 | Yes |
| 15905 | 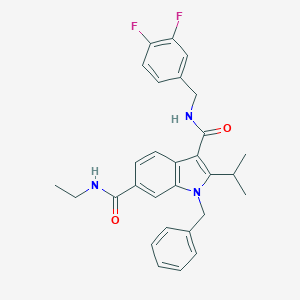 SCHEMBL805638 SCHEMBL805638 | C29H29F2N3O2 | 489.567 | 4 / 2 | 5.4 | No |
| 16069 | 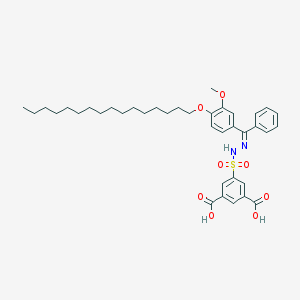 CHEMBL218446 CHEMBL218446 | C38H50N2O8S | 694.884 | 10 / 3 | 11.1 | No |
| 16070 | 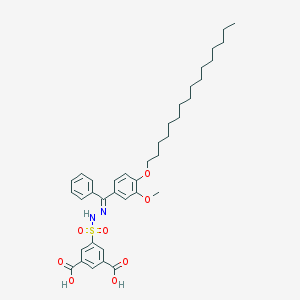 CHEMBL218445 CHEMBL218445 | C38H50N2O8S | 694.884 | 10 / 3 | 11.1 | No |
| 521961 | 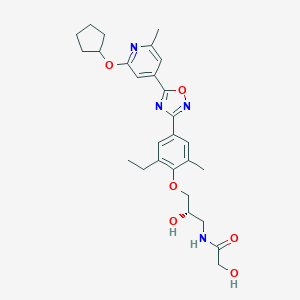 CHEMBL3800019 CHEMBL3800019 | C27H34N4O6 | 510.591 | 9 / 3 | 3.5 | No |
| 16342 | 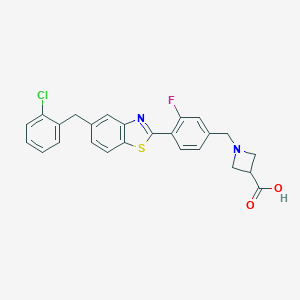 CHEMBL1672567 CHEMBL1672567 | C25H20ClFN2O2S | 466.955 | 6 / 1 | 3.6 | Yes |
| 16492 | 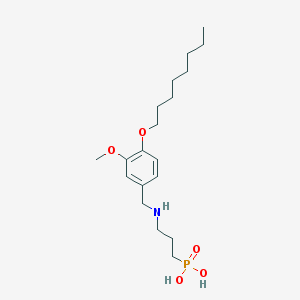 CHEMBL118508 CHEMBL118508 | C19H34NO5P | 387.457 | 6 / 3 | 0.8 | Yes |
| 16506 | 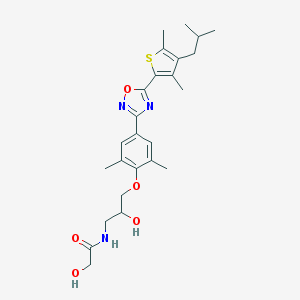 SCHEMBL2294162 SCHEMBL2294162 | C25H33N3O5S | 487.615 | 8 / 3 | 4.3 | Yes |
| 16734 | 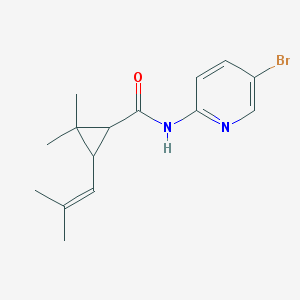 MLS000109722 MLS000109722 | C15H19BrN2O | 323.234 | 2 / 1 | 3.7 | Yes |
| 17241 | 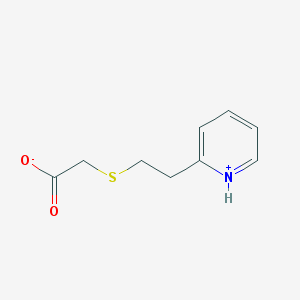 AC1M0X0O AC1M0X0O | C9H11NO2S | 197.252 | 3 / 1 | 2.2 | Yes |
| 17366 | 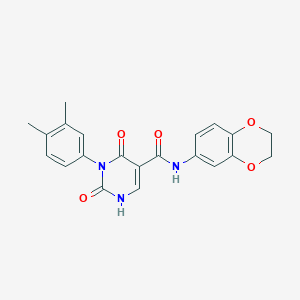 SMR000016951 SMR000016951 | C21H19N3O5 | 393.399 | 5 / 2 | 2.3 | Yes |
| 521997 | 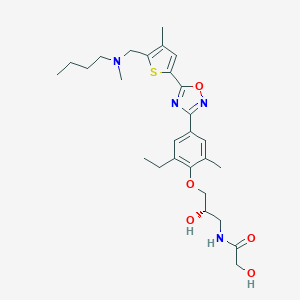 CHEMBL3800591 CHEMBL3800591 | C27H38N4O5S | 530.684 | 9 / 3 | 3.8 | No |
| 17807 | 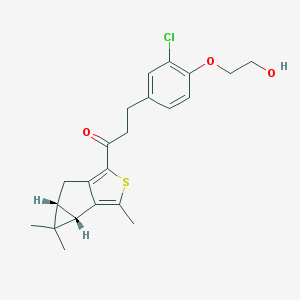 SCHEMBL4685937 SCHEMBL4685937 | C22H25ClO3S | 404.949 | 4 / 1 | 5.1 | No |
| 17997 | 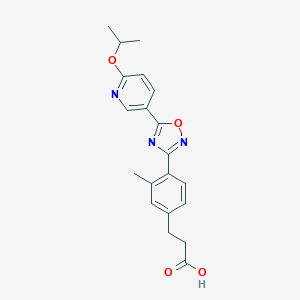 CHEMBL210301 CHEMBL210301 | C20H21N3O4 | 367.405 | 7 / 1 | 3.6 | Yes |
| 18086 | 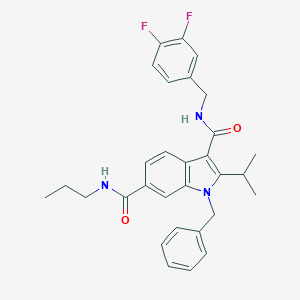 SCHEMBL805679 SCHEMBL805679 | C30H31F2N3O2 | 503.594 | 4 / 2 | 5.9 | No |
| 18175 | 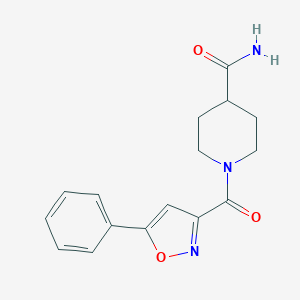 BAS 09529732 BAS 09529732 | C16H17N3O3 | 299.33 | 4 / 1 | 1.2 | Yes |
| 18190 | 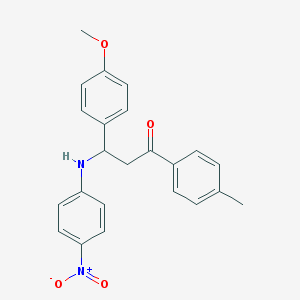 MLS000583267 MLS000583267 | C23H22N2O4 | 390.439 | 5 / 1 | 5.5 | No |
| 18467 | 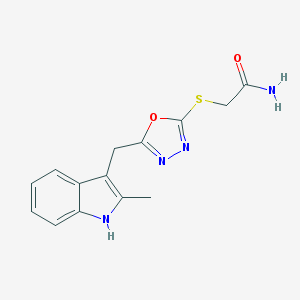 SMR000092447 SMR000092447 | C14H14N4O2S | 302.352 | 5 / 2 | 2.0 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417