

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MATALPPRLQPVRGNETLREHYQYVGKLAGRLKEASEGSTLTTVLFLVICSFIVLENLMVLIAIWKNNKFHNRMYFFIGNLALCDLLAGIAYKVNILMSGKKTFSLSPTVWFLREGSMFVALGASTCSLLAIAIERHLTMIKMRPYDANKRHRVFLLIGMCWLIAFTLGALPILGWNCLHNLPDCSTILPLYSKKYIAFCISIFTAILVTIVILYARIYFLVKSSSRKVANHNNSERSMALLRTVVIVVSVFIACWSPLFILFLIDVACRVQACPILFKAQWFIVLAVLNSAMNPVIYTLASKEMRRAFFRLVCNCLVRGRGARASPIQPALDPSRSKSSSSNNSSHSPKVKEDLPHTAPSSCIMDKNAALQNGIFCN | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 998999988888897621133435576456778980689999999999999999988861543660699997799999999999999999999999998378346051160399999999999999999999997598811788798560888888456999999999999998625322568877432314541689865668999999999999999999998745316663156789999999888999998899999999999779887718999999999999999885999994899999999999800578898776777777778777778888986788775677898899877758875232466669 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MATALPPRLQPVRGNETLREHYQYVGKLAGRLKEASEGSTLTTVLFLVICSFIVLENLMVLIAIWKNNKFHNRMYFFIGNLALCDLLAGIAYKVNILMSGKKTFSLSPTVWFLREGSMFVALGASTCSLLAIAIERHLTMIKMRPYDANKRHRVFLLIGMCWLIAFTLGALPILGWNCLHNLPDCSTILPLYSKKYIAFCISIFTAILVTIVILYARIYFLVKSSSRKVANHNNSERSMALLRTVVIVVSVFIACWSPLFILFLIDVACRVQACPILFKAQWFIVLAVLNSAMNPVIYTLASKEMRRAFFRLVCNCLVRGRGARASPIQPALDPSRSKSSSSNNSSHSPKVKEDLPHTAPSSCIMDKNAALQNGIFCN | |
| 752432331432331312332222224333434423111000000111002102313200000001033000000000000010000001110100010023101002000101000100000001300000000000000100314322311000000101120222021000000122424300000013332100000231332121002001100010131044145364334310000000001000000121000000000003232111000000000100210022000100116400300120001001234445444344444444444444444444443455444434442436464413313128 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MATALPPRLQPVRGNETLREHYQYVGKLAGRLKEASEGSTLTTVLFLVICSFIVLENLMVLIAIWKNNKFHNRMYFFIGNLALCDLLAGIAYKVNILMSGKKTFSLSPTVWFLREGSMFVALGASTCSLLAIAIERHLTMIKMRPYDANKRHRVFLLIGMCWLIAFTLGALPILGWNCLHNLPDCSTILPLYSKKYIAFCISIFTAILVTIVILYARIYFLVKSSSRKVANHNNSERSMALLRTVVIVVSVFIACWSPLFILFLIDVACRVQACPILFKAQWFIVLAVLNSAMNPVIYTLASKEMRRAFFRLVCNCLVRGRGARASPIQPALDPSRSKSSSSNNSSHSPKVKEDLPHTAPSSCIMDKNAALQNGIFCN | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.28 | 0.23 | 0.76 | 2.75 | Download | -------------ENFMDIECFMVL---------NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSF--SDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-KLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4z34A | 0.37 | 0.31 | 0.80 | 3.70 | Download | -------NEPQCFYNESIAFFYNRSGKHLA--TEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVLAYEKF--FLLLAEFNSAMNPIIYSYRDKEMSATFRQIL----------------------------------------------------------------- | |||||||||||||||||||
| 3 | 5tgzA | 0.28 | 0.24 | 0.76 | 3.25 | Download | ------------------GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHID--KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA---PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK-LIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------- | |||||||||||||||||||
| 4 | 4z34 | 0.38 | 0.31 | 0.80 | 1.63 | Download | --------NEPQCFNESIAFFYNRSGKH--LATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLERKAMYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVL--AYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG---------------------------------------------------------------- | |||||||||||||||||||
| 5 | 3uon | 0.21 | 0.20 | 0.73 | 1.17 | Download | -----------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTKMAGMMIAAAWVLSFILWAPAILFWQFIVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPN--TVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------------------------------------------------------------- | |||||||||||||||||||
| 6 | 5tgzA | 0.30 | 0.24 | 0.75 | 3.02 | Download | -------------ENFMDIECFMVLN---------PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA-----PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-KLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------- | |||||||||||||||||||
| 7 | 4z34 | 0.39 | 0.31 | 0.78 | 1.73 | Download | --------------NESIAFFYNRSGKHLA--TEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFR-MQLHRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLERKAMKLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV--LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG---------------------------------------------------------------- | |||||||||||||||||||
| 8 | 3v2yA | 0.56 | 0.47 | 0.80 | 3.03 | Download | --------VSDYVNYDIIVRHYNYTGKL-NISADKENSIKLTSVVFILICCFIILENIFVLLTIWKTKKFHRPMYYFIGNLALSDLLAGVAYTANLLLSGATTYKLTPAQWFLREGSMFVALSASVFSLLAIAIERYITMLKN-------NFRLFLLISACWVISLILGGLPIMGWNCISALSSCSTVLPLYHKHYILFCTTVFTLLLLSIVILYCRIYSLVRTRNIFEMLRIDESENVALLKTVIIVLSVFIACWAPLFILLLLDVGCKVKTCDILFRAEYFLVLAVLNSGTNPIIYTLTNKEMRRAFIRIMGRPL------------------------------------------------------------- | |||||||||||||||||||
| 9 | 5tjvA | 0.28 | 0.23 | 0.76 | 2.96 | Download | ----------------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSF--SDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKM-NKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5g53A | 0.24 | 0.21 | 0.74 | 3.05 | Download | --------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCCGEGQVACLFEVVPMNYMVNFFACVLVPLLLMLGVYLRIFLAARRQLKQM----TLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLENLY-------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
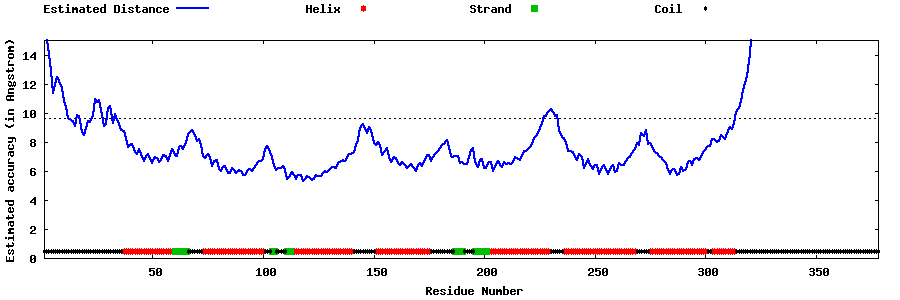
| Generated 3D models | Estimated local accuracy of models | ||
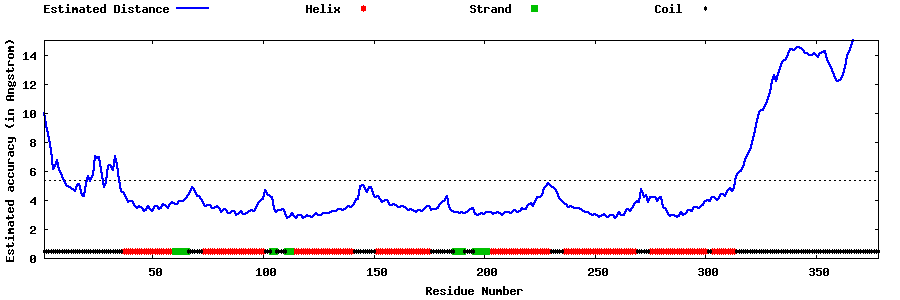 | |||
|
|
|||
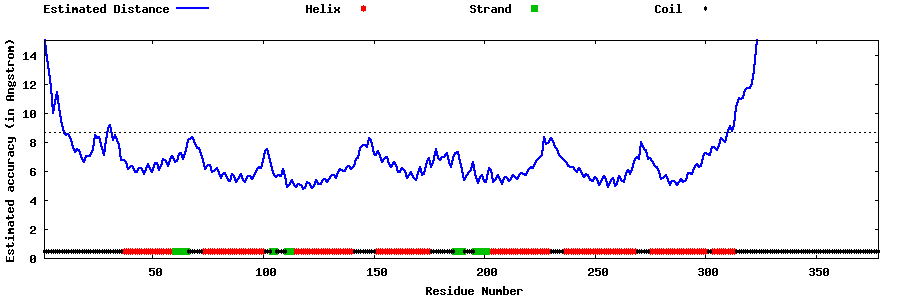 | |||
|
| |||
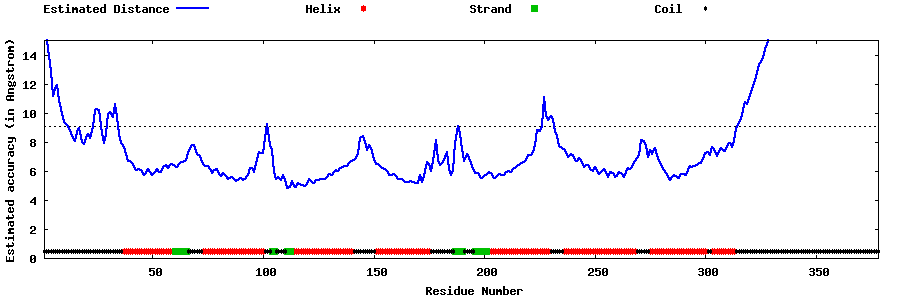 | |||
|
| |||
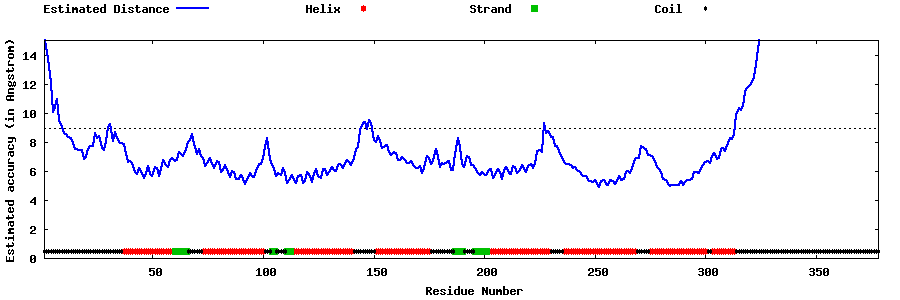 | |||
|
| |||
 | |||
|
| |||