

You can:
| Name | B2 bradykinin receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | BDKRB2 |
| Synonym | B2R B2BRA BK-2 receptor B2BKR B2 receptor [ Show all ] |
| Disease | Unspecified Cancer Hereditary angioedema Inflammatory disease Osteoarthritis [ Show all ] |
| Length | 391 |
| Amino acid sequence | MFSPWKISMFLSVREDSVPTTASFSADMLNVTLQGPTLNGTFAQSKCPQVEWLGWLNTIQPPFLWVLFVLATLENIFVLSVFCLHKSSCTVAEIYLGNLAAADLILACGLPFWAITISNNFDWLFGETLCRVVNAIISMNLYSSICFLMLVSIDRYLALVKTMSMGRMRGVRWAKLYSLVIWGCTLLLSSPMLVFRTMKEYSDEGHNVTACVISYPSLIWEVFTNMLLNVVGFLLPLSVITFCTMQIMQVLRNNEMQKFKEIQTERRATVLVLVVLLLFIICWLPFQISTFLDTLHRLGILSSCQDERIIDVITQIASFMAYSNSCLNPLVYVIVGKRFRKKSWEVYQGVCQKGGCRSEPIQMENSMGTLRTSISVERQIHKLQDWAGSRQ |
| UniProt | P30411 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P30411 |
| 3D structure model | This predicted structure model is from GPCR-EXP P30411. |
| BioLiP | N/A |
| Therapeutic Target Database | T23714 |
| ChEMBL | CHEMBL3157 |
| IUPHAR | 42 |
| DrugBank | BE0003513 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 279 | 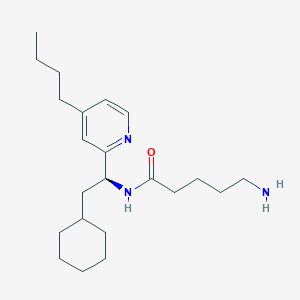 CHEMBL78487 CHEMBL78487 | C22H37N3O | 359.558 | 3 / 2 | 4.7 | Yes |
| 668 | 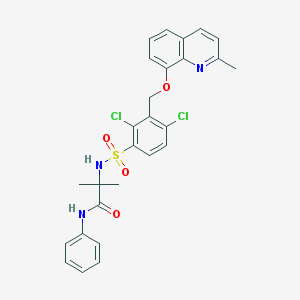 CHEMBL210427 CHEMBL210427 | C27H25Cl2N3O4S | 558.474 | 6 / 2 | 5.8 | No |
| 1065 | 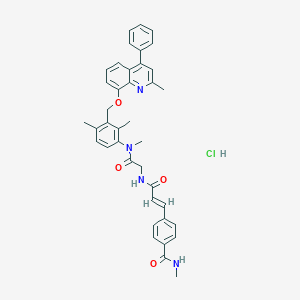 CHEMBL544720 CHEMBL544720 | C39H39ClN4O4 | 663.215 | 5 / 3 | N/A | No |
| 1883 | 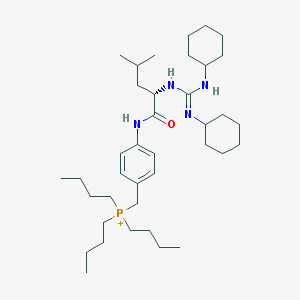 CHEMBL540897 CHEMBL540897 | C38H68N4OP+ | 627.963 | 2 / 3 | 9.0 | No |
| 1884 | 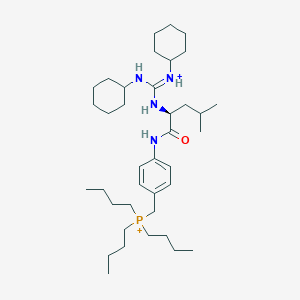 BDBM50045221 BDBM50045221 | C38H69N4OP+2 | 628.971 | 1 / 4 | 9.0 | No |
| 2125 | 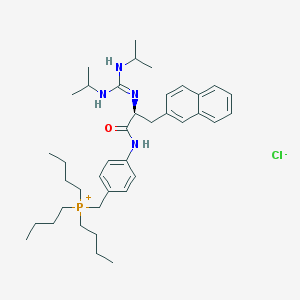 CHEMBL540895 CHEMBL540895 | C39H60ClN4OP | 667.36 | 3 / 3 | N/A | No |
| 2305 | 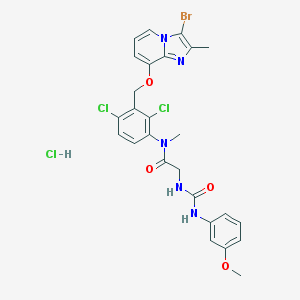 CHEMBL542473 CHEMBL542473 | C26H25BrCl3N5O4 | 657.771 | 5 / 3 | N/A | No |
| 3090 | 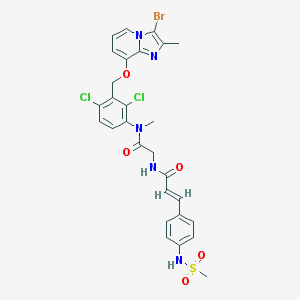 CHEMBL538281 CHEMBL538281 | C28H26BrCl2N5O5S | 695.41 | 7 / 2 | 5.7 | No |
| 4253 | 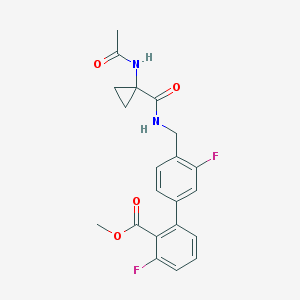 CHEMBL387331 CHEMBL387331 | C21H20F2N2O4 | 402.398 | 6 / 2 | 2.4 | Yes |
| 4300 | 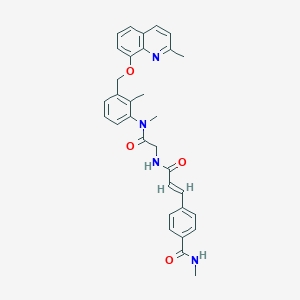 CHEMBL342195 CHEMBL342195 | C32H32N4O4 | 536.632 | 5 / 2 | 4.5 | No |
| 4435 | 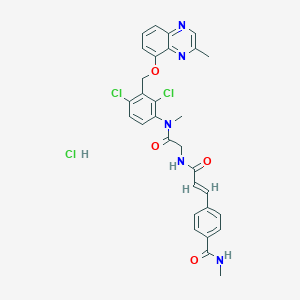 CHEMBL554206 CHEMBL554206 | C30H28Cl3N5O4 | 628.935 | 6 / 3 | N/A | No |
| 4522 | 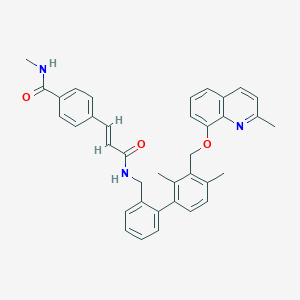 CHEMBL140383 CHEMBL140383 | C37H35N3O3 | 569.705 | 4 / 2 | 6.9 | No |
| 5022 | 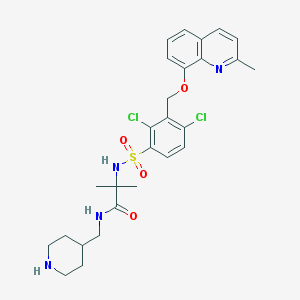 CHEMBL380260 CHEMBL380260 | C27H32Cl2N4O4S | 579.537 | 7 / 3 | 4.5 | No |
| 5197 | 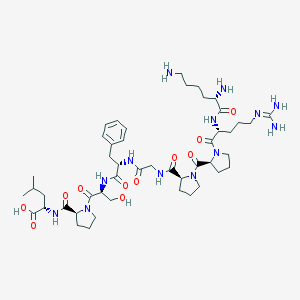 CHEMBL2369827 CHEMBL2369827 | C47H75N13O11 | 998.197 | 14 / 11 | -4.1 | No |
| 5199 | 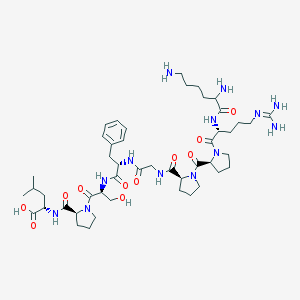 CHEMBL413382 CHEMBL413382 | C47H75N13O11 | 998.197 | 14 / 11 | -4.1 | No |
| 7479 | 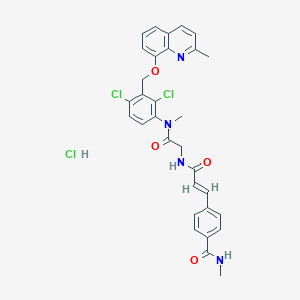 CHEMBL542096 CHEMBL542096 | C31H29Cl3N4O4 | 627.947 | 5 / 3 | N/A | No |
| 7792 |  CHEMBL563068 CHEMBL563068 | C24H24ClN5O3S | 497.998 | 7 / 0 | 2.3 | Yes |
| 8627 |  CHEMBL542175 CHEMBL542175 | C37H39Cl2N5O4 | 688.65 | 6 / 2 | 6.7 | No |
| 9344 |  CHEMBL552296 CHEMBL552296 | C22H22ClN5O3S | 471.96 | 7 / 1 | 2.5 | Yes |
| 9909 |  CHEMBL539792 CHEMBL539792 | C40H38Cl2N6O4 | 737.682 | 7 / 2 | 6.8 | No |
| 9931 | 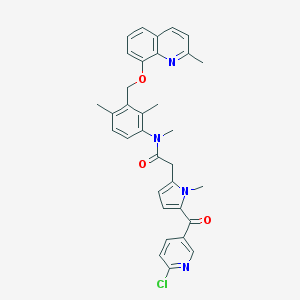 CHEMBL286710 CHEMBL286710 | C33H31ClN4O3 | 567.086 | 5 / 0 | 6.1 | No |
| 10790 | 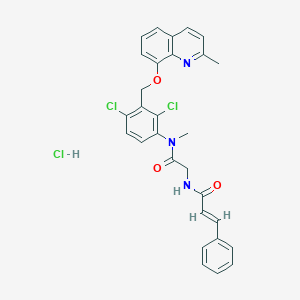 CHEMBL544601 CHEMBL544601 | C29H26Cl3N3O3 | 570.895 | 4 / 2 | N/A | No |
| 15172 | 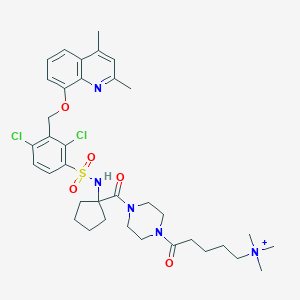 CHEMBL386794 CHEMBL386794 | C36H48Cl2N5O5S+ | 733.77 | 7 / 1 | 5.2 | No |
| 15391 | 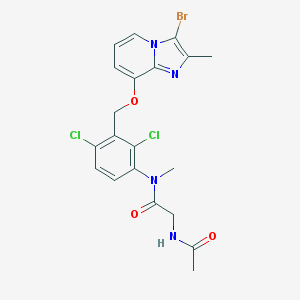 CHEMBL154188 CHEMBL154188 | C20H19BrCl2N4O3 | 514.201 | 4 / 1 | 4.6 | No |
| 15975 | 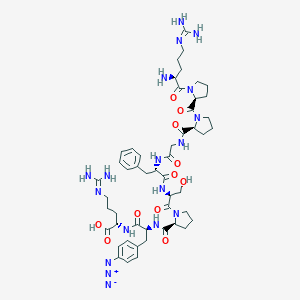 CHEMBL2372168 CHEMBL2372168 | C50H72N18O11 | 1101.24 | 16 / 12 | -3.8 | No |
| 16686 | 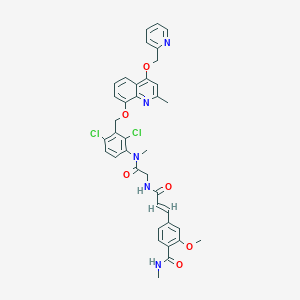 CHEMBL433214 CHEMBL433214 | C38H35Cl2N5O6 | 728.627 | 8 / 2 | 5.8 | No |
| 17026 | 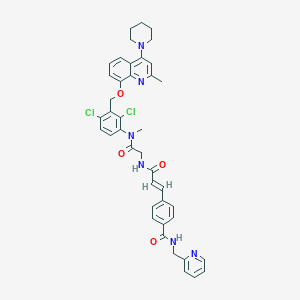 CHEMBL553897 CHEMBL553897 | C41H40Cl2N6O4 | 751.709 | 7 / 2 | 6.8 | No |
| 17155 | 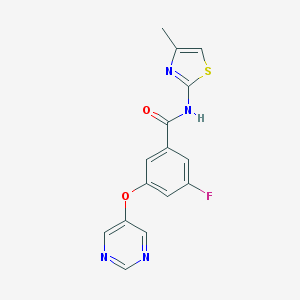 CHEMBL2440659 CHEMBL2440659 | C15H11FN4O2S | 330.337 | 7 / 1 | 2.4 | Yes |
| 17161 |  CHEMBL280869 CHEMBL280869 | C33H29Cl2N3O5S | 650.571 | 6 / 0 | 6.0 | No |
| 17173 |  CHEMBL2372142 CHEMBL2372142 | C50H79N15O11 | 1066.28 | 14 / 12 | -3.6 | No |
| 17792 |  CHEMBL379259 CHEMBL379259 | C25H30Cl2N4O4S | 553.499 | 7 / 3 | 4.0 | No |
| 18145 |  CHEMBL132510 CHEMBL132510 | C31H31Cl2N5O5 | 624.519 | 6 / 1 | 4.7 | No |
| 18680 | 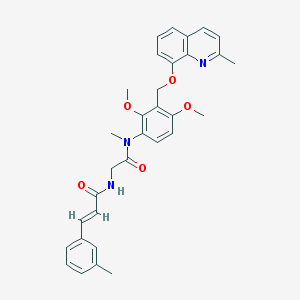 CHEMBL112360 CHEMBL112360 | C32H33N3O5 | 539.632 | 6 / 1 | 5.2 | No |
| 19574 | 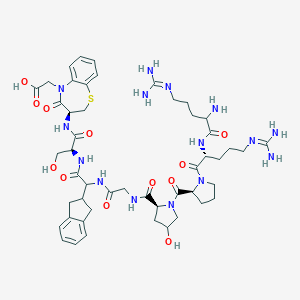 CHEMBL216618 CHEMBL216618 | C49H69N15O12S | 1092.24 | 16 / 13 | -5.6 | No |
| 19828 | 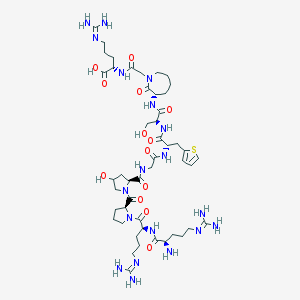 CHEMBL415032 CHEMBL415032 | C48H79N19O13S | 1162.34 | 18 / 16 | -8.6 | No |
| 19829 | 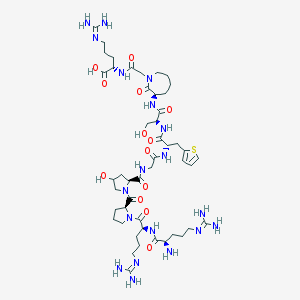 CHEMBL427805 CHEMBL427805 | C48H79N19O13S | 1162.34 | 18 / 16 | -8.6 | No |
| 20030 | 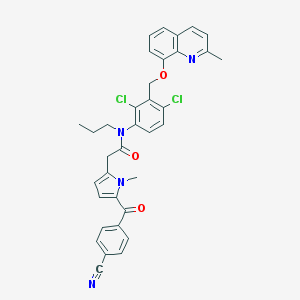 CHEMBL284865 CHEMBL284865 | C35H30Cl2N4O3 | 625.55 | 5 / 0 | 7.3 | No |
| 20925 | 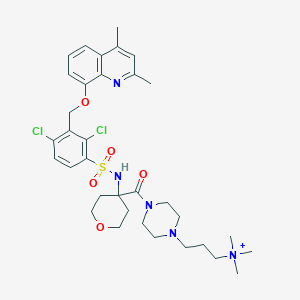 CHEMBL1956869 CHEMBL1956869 | C34H46Cl2N5O5S+ | 707.732 | 8 / 1 | 4.6 | No |
| 522148 | 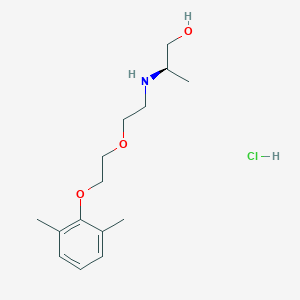 CHEMBL3775436 CHEMBL3775436 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 522161 | 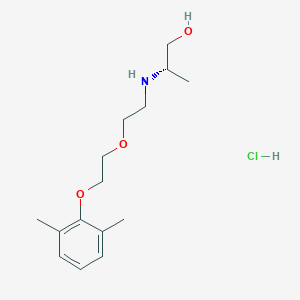 CHEMBL3775769 CHEMBL3775769 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 22648 | 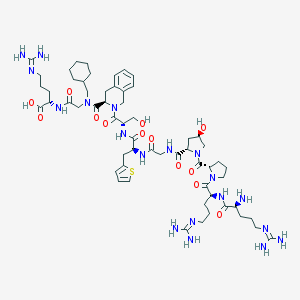 CHEMBL3038098 CHEMBL3038098 | C59H91N19O13S | 1306.56 | 18 / 15 | -5.9 | No |
| 22795 | 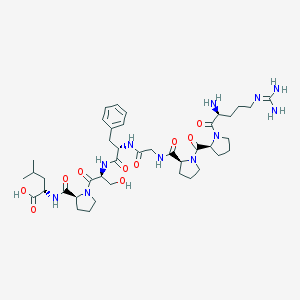 8-Leu-9-des-arg-BK 8-Leu-9-des-arg-BK | C41H63N11O10 | 870.022 | 12 / 9 | -3.6 | No |
| 22909 | 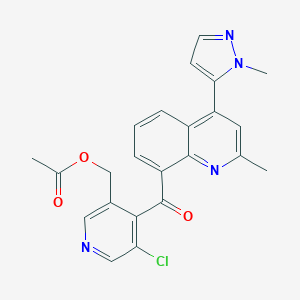 CHEMBL562296 CHEMBL562296 | C23H19ClN4O3 | 434.88 | 6 / 0 | 3.2 | Yes |
| 23063 | 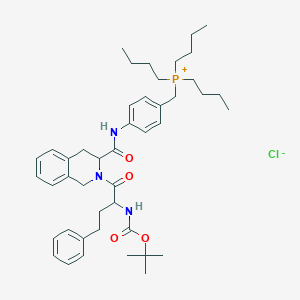 CHEMBL123124 CHEMBL123124 | C44H63ClN3O4P | 764.429 | 5 / 2 | N/A | No |
| 23386 | 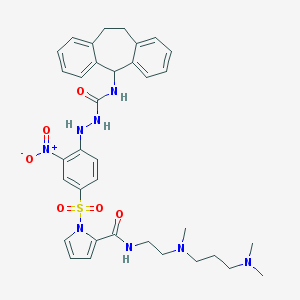 CHEMBL2112220 CHEMBL2112220 | C35H42N8O6S | 702.831 | 9 / 4 | 5.1 | No |
| 24529 | 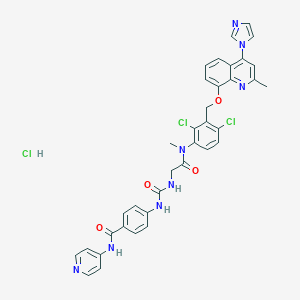 CHEMBL542598 CHEMBL542598 | C36H31Cl3N8O4 | 746.046 | 7 / 4 | N/A | No |
| 24699 | 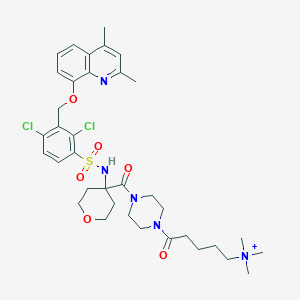 CHEMBL1956722 CHEMBL1956722 | C36H48Cl2N5O6S+ | 749.769 | 8 / 1 | 4.3 | No |
| 25746 | 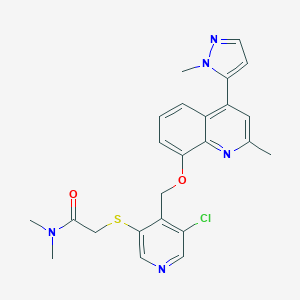 CHEMBL564841 CHEMBL564841 | C24H24ClN5O2S | 481.999 | 6 / 0 | 3.6 | Yes |
| 27102 | 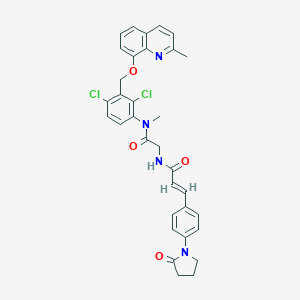 CHEMBL553293 CHEMBL553293 | C33H30Cl2N4O4 | 617.527 | 5 / 1 | 5.5 | No |
| 27181 | 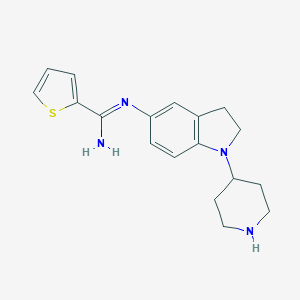 CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27304 | 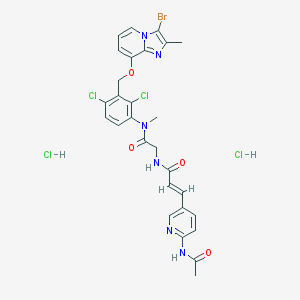 CHEMBL540840 CHEMBL540840 | C28H27BrCl4N6O4 | 733.266 | 6 / 4 | N/A | No |
| 28775 | 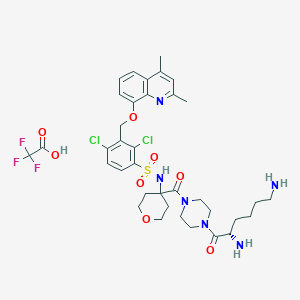 CHEMBL1956857 CHEMBL1956857 | C36H45Cl2F3N6O8S | 849.745 | 15 / 4 | N/A | No |
| 29428 |  CHEMBL2372164 CHEMBL2372164 | C50H72N17O11+ | 1087.23 | 15 / 12 | -4.1 | No |
| 30312 |  CHEMBL222156 CHEMBL222156 | C35H45Cl2N7O5S | 746.749 | 8 / 3 | 4.0 | No |
| 536782 |  CHEMBL3889627 CHEMBL3889627 | C23H27N5O3S | 453.561 | 5 / 2 | 2.6 | Yes |
| 32461 |  CHEMBL550142 CHEMBL550142 | C19H13Cl2N3OS | 402.293 | 5 / 0 | 4.9 | Yes |
| 33499 | 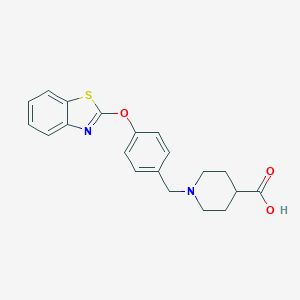 UNII-7W7511NPF8 UNII-7W7511NPF8 | C20H20N2O3S | 368.451 | 6 / 1 | 1.8 | Yes |
| 33938 | 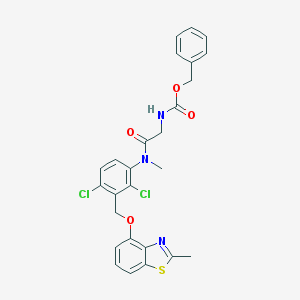 CHEMBL325742 CHEMBL325742 | C26H23Cl2N3O4S | 544.447 | 6 / 1 | 6.1 | No |
| 33986 | 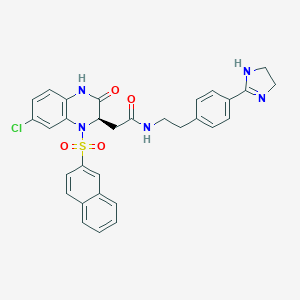 CHEMBL160547 CHEMBL160547 | C31H28ClN5O4S | 602.106 | 6 / 3 | 3.7 | No |
| 34876 | 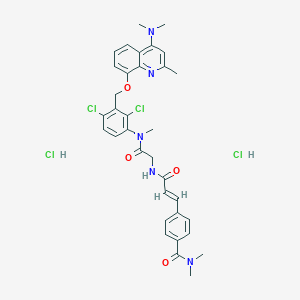 CHEMBL3143958 CHEMBL3143958 | C34H37Cl4N5O4 | 721.501 | 6 / 3 | N/A | No |
| 34898 | 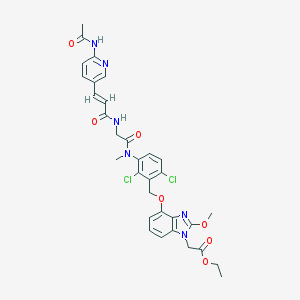 CHEMBL98292 CHEMBL98292 | C32H32Cl2N6O7 | 683.543 | 9 / 2 | 4.1 | No |
| 35702 | 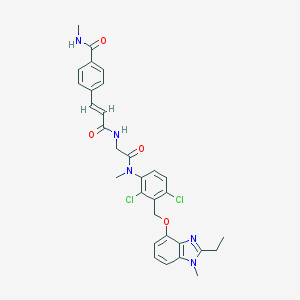 CHEMBL542836 CHEMBL542836 | C31H31Cl2N5O4 | 608.52 | 5 / 2 | 5.1 | No |
| 35709 | 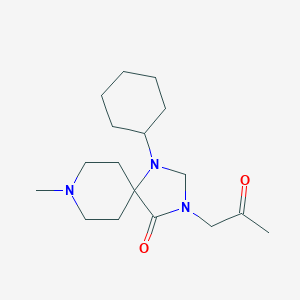 CHEMBL110188 CHEMBL110188 | C17H29N3O2 | 307.438 | 4 / 0 | 1.6 | Yes |
| 35885 | 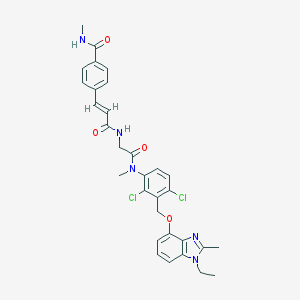 CHEMBL43746 CHEMBL43746 | C31H31Cl2N5O4 | 608.52 | 5 / 2 | 4.9 | No |
| 35948 | 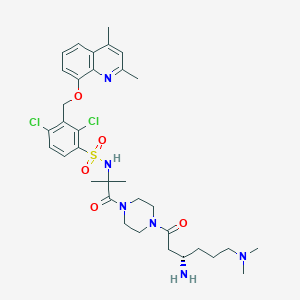 CHEMBL205093 CHEMBL205093 | C34H46Cl2N6O5S | 721.739 | 9 / 2 | 3.8 | No |
| 36157 | 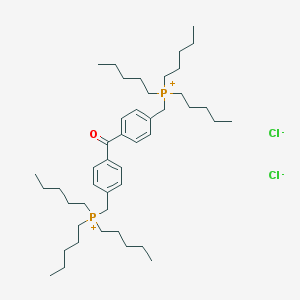 CHEMBL121565 CHEMBL121565 | C45H78Cl2OP2 | 767.966 | 3 / 0 | N/A | No |
| 36162 | 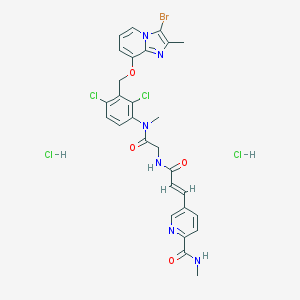 CHEMBL553844 CHEMBL553844 | C28H27BrCl4N6O4 | 733.266 | 6 / 4 | N/A | No |
| 36350 | 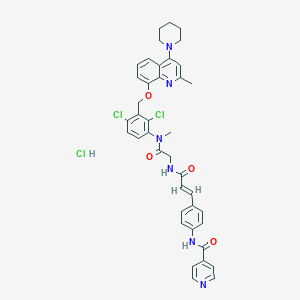 CHEMBL539792 CHEMBL539792 | C40H39Cl3N6O4 | 774.14 | 7 / 3 | N/A | No |
| 36668 | 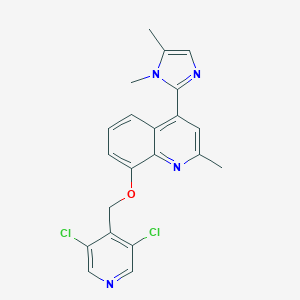 CHEMBL561839 CHEMBL561839 | C21H18Cl2N4O | 413.302 | 4 / 0 | 4.4 | Yes |
| 38102 | 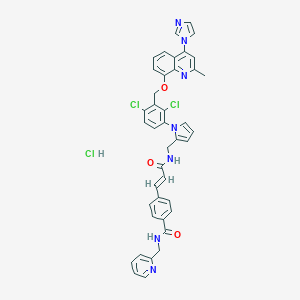 CHEMBL543068 CHEMBL543068 | C41H34Cl3N7O3 | 779.119 | 6 / 3 | N/A | No |
| 38934 | 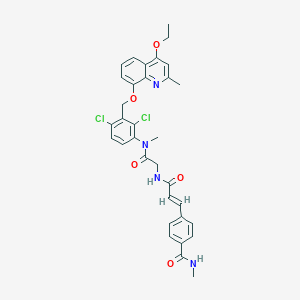 CHEMBL555403 CHEMBL555403 | C33H32Cl2N4O5 | 635.542 | 6 / 2 | 5.7 | No |
| 39070 | 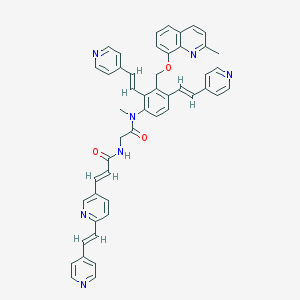 CHEMBL357075 CHEMBL357075 | C49H41N7O3 | 775.913 | 8 / 1 | 7.4 | No |
| 41664 | 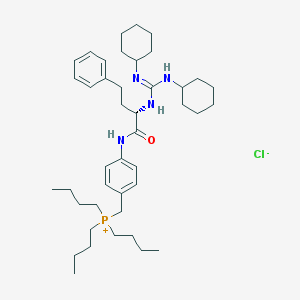 CHEMBL554255 CHEMBL554255 | C42H68ClN4OP | 711.457 | 3 / 3 | N/A | No |
| 42325 | 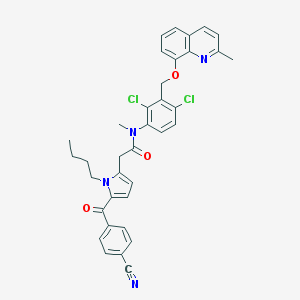 CHEMBL286520 CHEMBL286520 | C36H32Cl2N4O3 | 639.577 | 5 / 0 | 7.6 | No |
| 42910 | 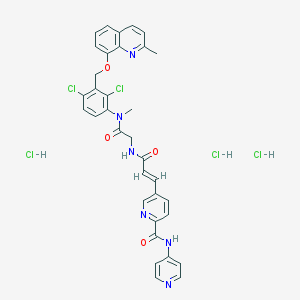 CHEMBL543659 CHEMBL543659 | C34H31Cl5N6O4 | 764.91 | 7 / 5 | N/A | No |
| 43707 | 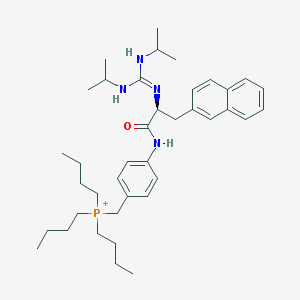 CHEMBL540895 CHEMBL540895 | C39H60N4OP+ | 631.91 | 2 / 3 | 8.7 | No |
| 43791 |  CHEMBL544083 CHEMBL544083 | C35H35Cl2N5O5 | 676.595 | 7 / 2 | 5.1 | No |
| 44126 |  CHEMBL107234 CHEMBL107234 | C14H25N3O2 | 267.373 | 4 / 0 | 0.7 | Yes |
| 44214 |  Cipargamin Cipargamin | C19H14Cl2FN3O | 390.239 | 3 / 3 | 3.9 | Yes |
| 45269 |  CHEMBL538732 CHEMBL538732 | C29H27BrCl3N5O4 | 695.82 | 5 / 3 | N/A | No |
| 46024 | 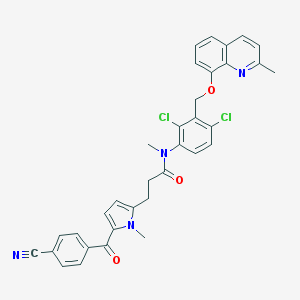 CHEMBL284727 CHEMBL284727 | C34H28Cl2N4O3 | 611.523 | 5 / 0 | 6.7 | No |
| 46599 | 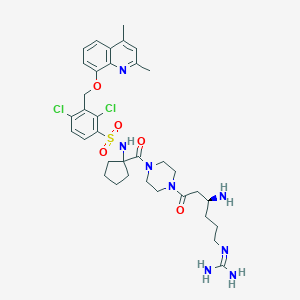 CHEMBL267493 CHEMBL267493 | C35H46Cl2N8O5S | 761.764 | 9 / 4 | 2.7 | No |
| 46890 | 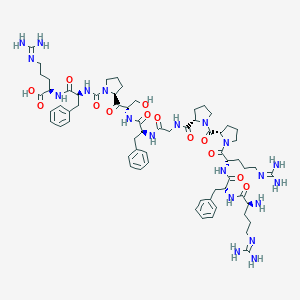 CHEMBL2372276 CHEMBL2372276 | C65H94N20O13 | 1363.59 | 17 / 16 | -4.7 | No |
| 443551 | 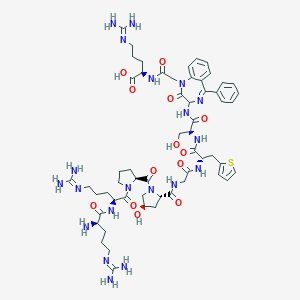 CHEMBL3350763 CHEMBL3350763 | C57H80N20O13S | 1285.45 | 19 / 16 | -6.5 | No |
| 47171 | 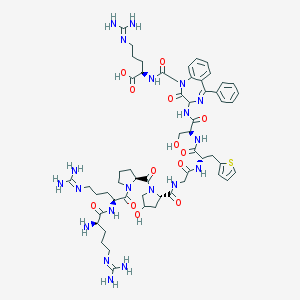 CHEMBL408645 CHEMBL408645 | C57H80N20O13S | 1285.45 | 19 / 16 | -6.5 | No |
| 47389 | 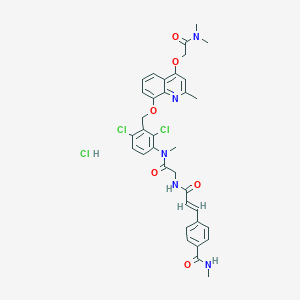 CHEMBL538020 CHEMBL538020 | C35H36Cl3N5O6 | 729.052 | 7 / 3 | N/A | No |
| 48226 | 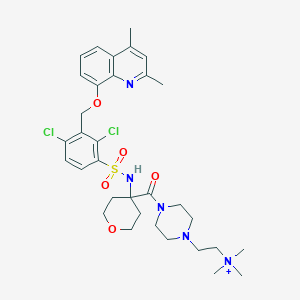 CHEMBL1956868 CHEMBL1956868 | C33H44Cl2N5O5S+ | 693.705 | 8 / 1 | 4.2 | No |
| 48431 | 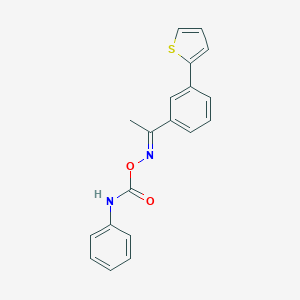 CHEMBL1210154 CHEMBL1210154 | C19H16N2O2S | 336.409 | 4 / 1 | 4.7 | Yes |
| 48472 | 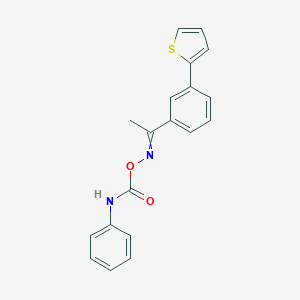 SCHEMBL4318956 SCHEMBL4318956 | C19H16N2O2S | 336.409 | 4 / 1 | 4.7 | Yes |
| 49162 | 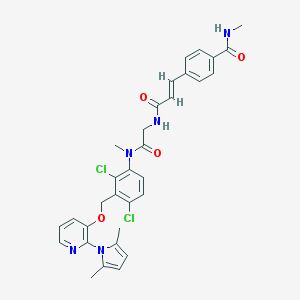 CHEMBL133186 CHEMBL133186 | C32H31Cl2N5O4 | 620.531 | 5 / 2 | 5.2 | No |
| 49576 | 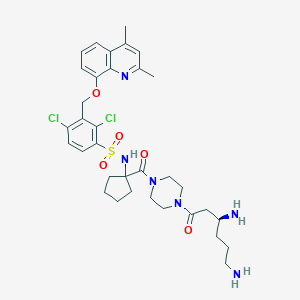 CHEMBL375576 CHEMBL375576 | C34H44Cl2N6O5S | 719.723 | 9 / 3 | 3.3 | No |
| 49714 | 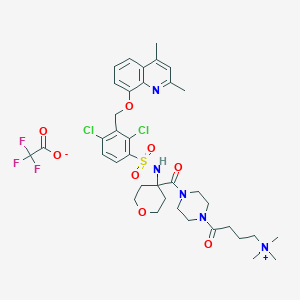 CHEMBL1956721 CHEMBL1956721 | C37H46Cl2F3N5O8S | 848.757 | 13 / 1 | N/A | No |
| 49924 |  CHEMBL3142396 CHEMBL3142396 | C59H83N19O13S | 1298.49 | 18 / 15 | -6.6 | No |
| 49926 |  CHEMBL412349 CHEMBL412349 | C59H83N19O13S | 1298.49 | 18 / 15 | -6.6 | No |
| 51258 |  CHEMBL27701 CHEMBL27701 | C33H26Cl2F3N3O3 | 640.484 | 7 / 0 | 7.6 | No |
| 51404 |  CHEMBL218636 CHEMBL218636 | C38H52Cl2N7O6S+ | 805.837 | 9 / 2 | 3.2 | No |
| 51428 | 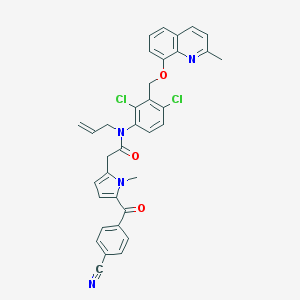 CHEMBL284726 CHEMBL284726 | C35H28Cl2N4O3 | 623.534 | 5 / 0 | 7.1 | No |
| 51679 | 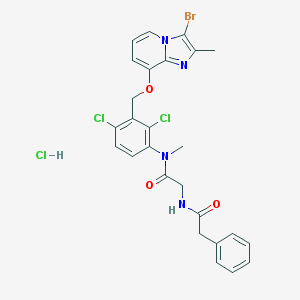 CHEMBL544600 CHEMBL544600 | C26H24BrCl3N4O3 | 626.757 | 4 / 2 | N/A | No |
| 52315 | 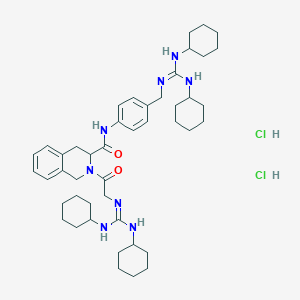 CHEMBL124221 CHEMBL124221 | C45H68Cl2N8O2 | 823.993 | 4 / 7 | N/A | No |
| 53313 | 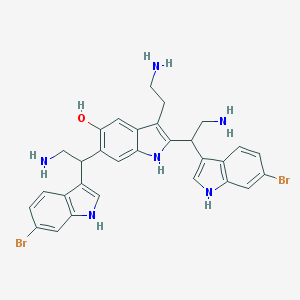 Gelliusine A Gelliusine A | C30H30Br2N6O | 650.419 | 4 / 7 | 4.2 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417