

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MFSPWKISMFLSVREDSVPTTASFSADMLNVTLQGPTLNGTFAQSKCPQVEWLGWLNTIQPPFLWVLFVLATLENIFVLSVFCLHKSSCTVAEIYLGNLAAADLILACGLPFWAITISNNFDWLFGETLCRVVNAIISMNLYSSICFLMLVSIDRYLALVKTMSMGRMRGVRWAKLYSLVIWGCTLLLSSPMLVFRTMKEYSDEGHNVTACVISYPSLIWEVFTNMLLNVVGFLLPLSVITFCTMQIMQVLRNNEMQKFKEIQTERRATVLVLVVLLLFIICWLPFQISTFLDTLHRLGILSSCQDERIIDVITQIASFMAYSNSCLNPLVYVIVGKRFRKKSWEVYQGVCQKGGCRSEPIQMENSMGTLRTSISVERQIHKLQDWAGSRQ | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9987655446567789998878875455666788888788877777864103757898699999999999998899978976505776858899999999999999998899999995789986987479899999999999999999999997036322026555464503476667999999999998999983023324689856886798980789999999999999999999999999999999946776643323446324541258899999986168899999999984578884588999999999999999999899999993089999999999988612356768887677778886788877788765467777899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MFSPWKISMFLSVREDSVPTTASFSADMLNVTLQGPTLNGTFAQSKCPQVEWLGWLNTIQPPFLWVLFVLATLENIFVLSVFCLHKSSCTVAEIYLGNLAAADLILACGLPFWAITISNNFDWLFGETLCRVVNAIISMNLYSSICFLMLVSIDRYLALVKTMSMGRMRGVRWAKLYSLVIWGCTLLLSSPMLVFRTMKEYSDEGHNVTACVISYPSLIWEVFTNMLLNVVGFLLPLSVITFCTMQIMQVLRNNEMQKFKEIQTERRATVLVLVVLLLFIICWLPFQISTFLDTLHRLGILSSCQDERIIDVITQIASFMAYSNSCLNPLVYVIVGKRFRKKSWEVYQGVCQKGGCRSEPIQMENSMGTLRTSISVERQIHKLQDWAGSRQ | |
| 6345242322221354423333323332322333333333323433043430430010000220120022033232200100011233210000000000100000000000000000353301001000100000122111000000000031020000000202321321100000000010000001000000202324566321000103134410210010111110122111202230000001013143464554443200000000000001003110100000100230312440423300100000000000200031000000004401410140024004434344443435444434444434544434345455658 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MFSPWKISMFLSVREDSVPTTASFSADMLNVTLQGPTLNGTFAQSKCPQVEWLGWLNTIQPPFLWVLFVLATLENIFVLSVFCLHKSSCTVAEIYLGNLAAADLILACGLPFWAITISNNFDWLFGETLCRVVNAIISMNLYSSICFLMLVSIDRYLALVKTMSMGRMRGVRWAKLYSLVIWGCTLLLSSPMLVFRTMKEYSDEGHNVTACVISYPSLIWEVFTNMLLNVVGFLLPLSVITFCTMQIMQVLRNNEMQKFKEIQTERRATVLVLVVLLLFIICWLPFQISTFLDTLHRLGILSSCQDERIIDVITQIASFMAYSNSCLNPLVYVIVGKRFRKKSWEVYQGVCQKGGCRSEPIQMENSMGTLRTSISVERQIHKLQDWAGSRQ | |||||||||||||||||||||||||
| 1 | 5unfA | 0.29 | 0.29 | 0.86 | 2.77 | Download | TPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFPWQ-------ASYIVPLVWCMACLSSLPTFYFRDVRTIEYLG--VNACIMAFPPAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNS-YGKNRITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVINSCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR------------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.25 | 0.21 | 0.76 | 4.03 | Download | ---------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK----EGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKEEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.21 | 0.76 | 3.80 | Download | --------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG----AVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKERSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-----DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.26 | 0.21 | 0.72 | 1.53 | Download | ----------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKLD-FRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR--EDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------ | |||||||||||||||||||
| 5 | 5t1a | 0.26 | 0.22 | 0.74 | 1.17 | Download | ---------------------------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE-----DSVYVCGPYFPRG-WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIFYPPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF-------------------------------------------- | |||||||||||||||||||
| 6 | 5unfA | 0.29 | 0.29 | 0.86 | 3.21 | Download | PKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFP-------WQASYIVPLVWCMACLSSLPTFYFRDVRTIEYL--GVNACIMAFPYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNS-YGKNRITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVINSCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR------------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.26 | 0.21 | 0.71 | 1.70 | Download | -------------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVY-LMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDV--DVIECSLQFPDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-----AA------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC-------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.25 | 0.20 | 0.74 | 4.50 | Download | ---------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL----HYTCFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.26 | 0.21 | 0.75 | 3.29 | Download | ---------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK----EGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK----EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.26 | 0.21 | 0.75 | 3.45 | Download | -----------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKY--RQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALI------TIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
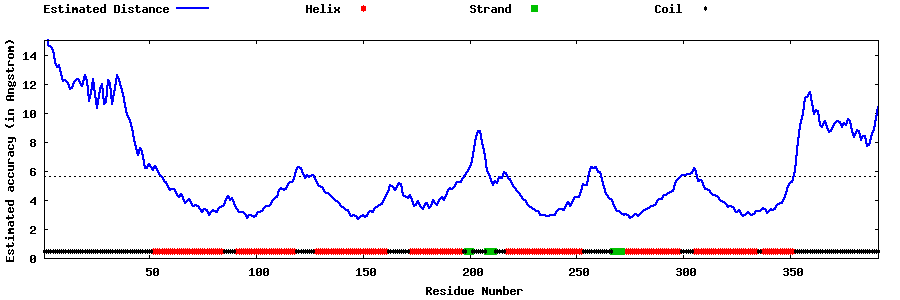 | |||
|
|
|||
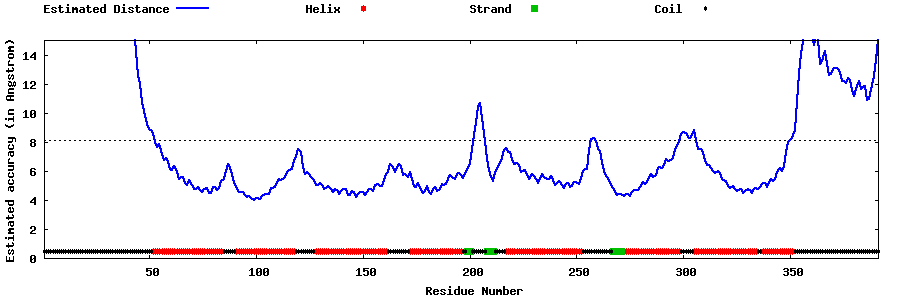 | |||
|
| |||
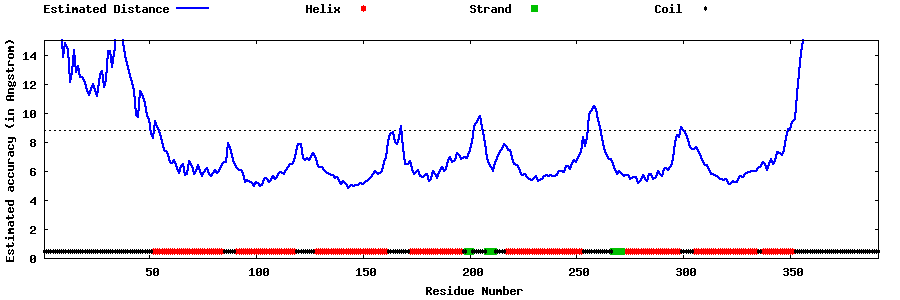 | |||
|
| |||
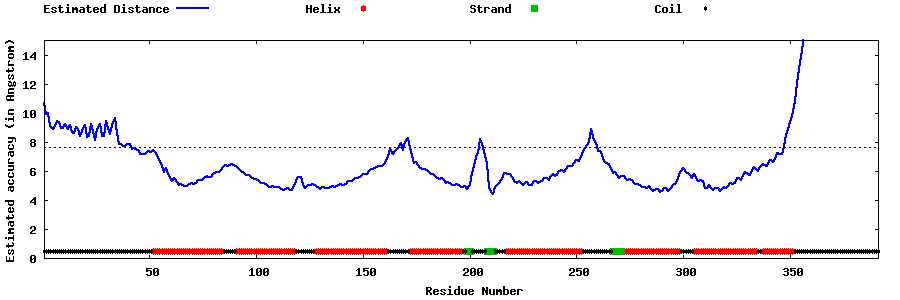 | |||
|
| |||
 | |||
|
| |||