

You can:
| Name | Smoothened homolog |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | SMO |
| Synonym | smoothened, frizzled class receptor smoothened SMOH SMO Protein Gx [ Show all ] |
| Disease | N/A |
| Length | 787 |
| Amino acid sequence | MAAARPARGPELPLLGLLLLLLLGDPGRGAASSGNATGPGPRSAGGSARRSAAVTGPPPPLSHCGRAAPCEPLRYNVCLGSVLPYGATSTLLAGDSDSQEEAHGKLVLWSGLRNAPRCWAVIQPLLCAVYMPKCENDRVELPSRTLCQATRGPCAIVERERGWPDFLRCTPDRFPEGCTNEVQNIKFNSSGQCEVPLVRTDNPKSWYEDVEGCGIQCQNPLFTEAEHQDMHSYIAAFGAVTGLCTLFTLATFVADWRNSNRYPAVILFYVNACFFVGSIGWLAQFMDGARREIVCRADGTMRLGEPTSNETLSCVIIFVIVYYALMAGVVWFVVLTYAWHTSFKALGTTYQPLSGKTSYFHLLTWSLPFVLTVAILAVAQVDGDSVSGICFVGYKNYRYRAGFVLAPIGLVLIVGGYFLIRGVMTLFSIKSNHPGLLSEKAASKINETMLRLGIFGFLAFGFVLITFSCHFYDFFNQAEWERSFRDYVLCQANVTIGLPTKQPIPDCEIKNRPSLLVEKINLFAMFGTGIAMSTWVWTKATLLIWRRTWCRLTGQSDDEPKRIKKSKMIAKAFSKRHELLQNPGQELSFSMHTVSHDGPVAGLAFDLNEPSADVSSAWAQHVTKMVARRGAILPQDISVTPVATPVPPEEQANLWLVEAEISPELQKRLGRKKKRRKRKKEVCPLAPPPELHPPAPAPSTIPRLPQLPRQKCLVAAGAWGAGDSCRQGAWTLVSNPFCPEPSPPQDPFLPSAPAPVAWAHGRRQGLGPIHSRTNLMDTELMDADSDF |
| UniProt | Q99835 |
| Protein Data Bank | 4jkv, 4n4w, 4o9r, 4qim, 4qin, 5l7d, 5l7i, 5v56, 5v57 |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4jkv. |
| BioLiP | BL0352216, BL0270837, BL0267121, BL0267120, BL0243904,BL0243905, BL0283868, BL0283867, BL0352217,BL0352218, BL0352219,BL0352220, BL0379427,BL0379429, BL0379428,BL0379430, BL0379431,BL0379433, BL0379432,BL0379434 |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL5971 |
| IUPHAR | 239 |
| DrugBank | BE0004659 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 459239 |  CHEMBL3692739 CHEMBL3692739 | C16H20ClN5O2S | 381.879 | 7 / 1 | 1.9 | Yes |
| 972 |  BDBM50349278 BDBM50349278 | C22H20Cl2N4O3 | 459.327 | 3 / 2 | 3.0 | Yes |
| 1151 |  CID 56666585 CID 56666585 | C17H19N3O4 | 329.356 | 4 / 0 | 1.0 | Yes |
| 535953 |  CHEMBL3955601 CHEMBL3955601 | C25H28N6 | 412.541 | 5 / 1 | 4.6 | Yes |
| 1958 |  CHEMBL2031277 CHEMBL2031277 | C31H29N3O6 | 539.588 | 6 / 3 | 5.7 | No |
| 1998 |  CHEMBL2031274 CHEMBL2031274 | C27H29N3O6 | 491.544 | 6 / 3 | 4.8 | Yes |
| 2339 |  BDBM50349294 BDBM50349294 | C22H19Cl2FN4O3 | 477.317 | 4 / 2 | 3.1 | Yes |
| 459249 |  3-chloro-6-(4-((2-methoxyethyl)sulfonyl)piperazin-1-yl)-2,2'-bipyridine 3-chloro-6-(4-((2-methoxyethyl)sulfonyl)piperazin-1-yl)-2,2'-bipyridine | C17H21ClN4O3S | 396.89 | 7 / 0 | 1.5 | Yes |
| 3320 |  CHEMBL3126705 CHEMBL3126705 | C23H20Cl2N2O4S | 491.383 | 4 / 2 | 4.8 | Yes |
| 3926 |  CHEMBL1823844 CHEMBL1823844 | C24H22ClN5O4S | 511.981 | 7 / 1 | 3.4 | No |
| 4587 |  CHEMBL3262652 CHEMBL3262652 | C20H18ClF3N6O | 450.85 | 8 / 1 | 3.2 | Yes |
| 536057 |  CHEMBL568916 CHEMBL568916 | C19H20FN3O | 325.387 | 3 / 1 | 3.8 | Yes |
| 5130 | 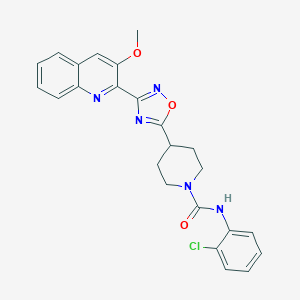 CHEMBL1823847 CHEMBL1823847 | C24H22ClN5O3 | 463.922 | 6 / 1 | 4.2 | Yes |
| 536087 | 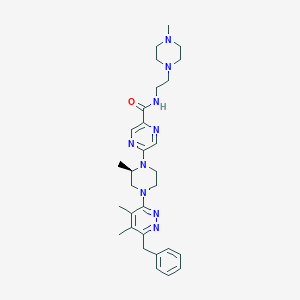 CHEMBL3938334 CHEMBL3938334 | C30H41N9O | 543.72 | 9 / 1 | 2.7 | No |
| 463642 | 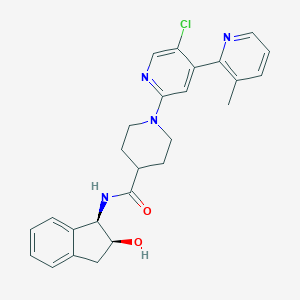 CHEMBL3692763 CHEMBL3692763 | C26H27ClN4O2 | 462.978 | 5 / 2 | 3.6 | Yes |
| 459281 | 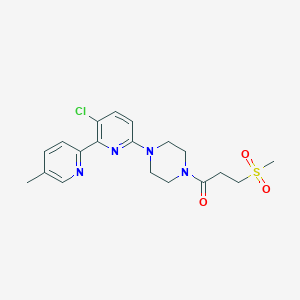 CHEMBL3696804 CHEMBL3696804 | C19H23ClN4O3S | 422.928 | 6 / 0 | 1.5 | Yes |
| 7117 |  CHEMBL2063140 CHEMBL2063140 | C27H50O | 390.696 | 1 / 1 | 10.0 | No |
| 7737 |  CHEMBL1209249 CHEMBL1209249 | C25H23N5O | 409.493 | 5 / 0 | 3.5 | Yes |
| 521673 |  CHEMBL3817945 CHEMBL3817945 | C27H29N5OS | 471.623 | 5 / 0 | 4.1 | Yes |
| 536184 |  CHEMBL3983372 CHEMBL3983372 | C25H28FN5O2 | 449.53 | 8 / 0 | 4.2 | Yes |
| 8898 | 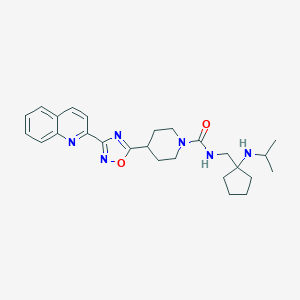 CHEMBL1822447 CHEMBL1822447 | C26H34N6O2 | 462.598 | 6 / 2 | 3.5 | Yes |
| 463975 | 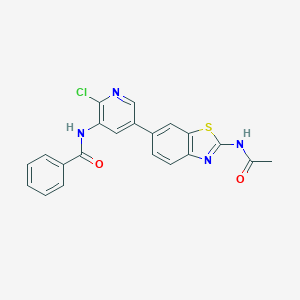 CHEMBL3604610 CHEMBL3604610 | C21H15ClN4O2S | 422.887 | 5 / 2 | 4.2 | Yes |
| 9165 | 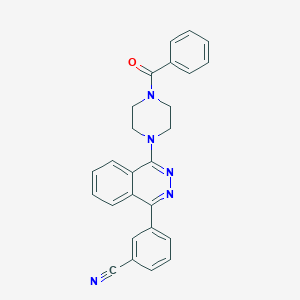 CHEMBL1082386 CHEMBL1082386 | C26H21N5O | 419.488 | 5 / 0 | 3.9 | Yes |
| 459298 | 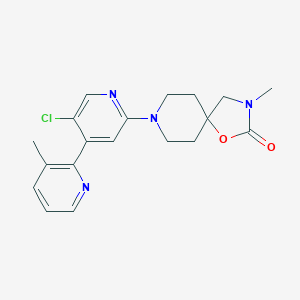 SCHEMBL2712319 SCHEMBL2712319 | C19H21ClN4O2 | 372.853 | 5 / 0 | 2.9 | Yes |
| 10213 | 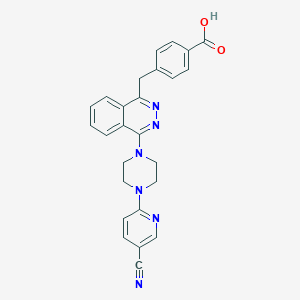 CHEMBL497218 CHEMBL497218 | C26H22N6O2 | 450.502 | 8 / 1 | 3.6 | Yes |
| 10467 | 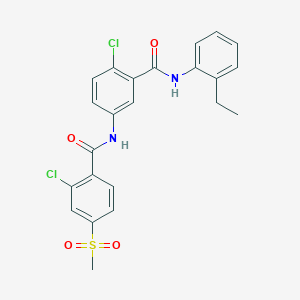 CHEMBL3126704 CHEMBL3126704 | C23H20Cl2N2O4S | 491.383 | 4 / 2 | 4.9 | Yes |
| 10757 | 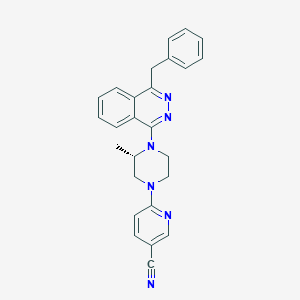 CHEMBL497238 CHEMBL497238 | C26H24N6 | 420.52 | 6 / 0 | 4.5 | Yes |
| 10760 | 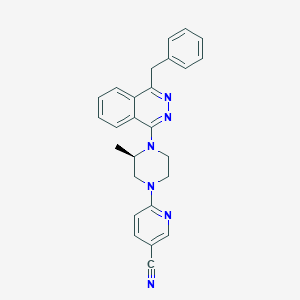 CHEMBL497639 CHEMBL497639 | C26H24N6 | 420.52 | 6 / 0 | 4.5 | Yes |
| 13403 |  CHEMBL1209455 CHEMBL1209455 | C26H22F3N5O | 477.491 | 8 / 0 | 4.4 | Yes |
| 459342 |  SCHEMBL2714472 SCHEMBL2714472 | C19H23ClN4O2 | 374.869 | 5 / 1 | 1.7 | Yes |
| 14130 |  Grundmann's ketone Grundmann's ketone | C18H32O | 264.453 | 1 / 0 | 5.9 | No |
| 459347 |  SCHEMBL2713794 SCHEMBL2713794 | C16H18ClN5O | 331.804 | 5 / 0 | 1.6 | Yes |
| 536368 |  CHEMBL3920191 CHEMBL3920191 | C25H28N6O2 | 444.539 | 8 / 0 | 3.5 | Yes |
| 459354 |  CHEMBL3696810 CHEMBL3696810 | C20H26ClFN4O3S | 456.961 | 8 / 2 | 3.1 | Yes |
| 459357 |  CHEMBL3692736 CHEMBL3692736 | C20H26ClN5O | 387.912 | 5 / 1 | 2.8 | Yes |
| 16385 |  CHEMBL1822453 CHEMBL1822453 | C30H34N6O2 | 510.642 | 6 / 1 | 4.4 | No |
| 536425 | 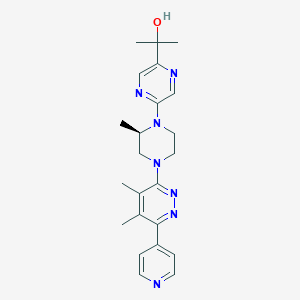 CHEMBL3922084 CHEMBL3922084 | C23H29N7O | 419.533 | 8 / 1 | 1.6 | Yes |
| 459365 | 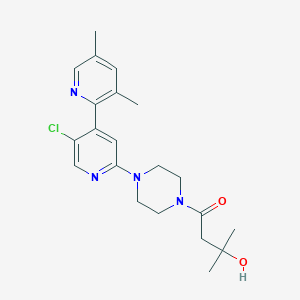 SCHEMBL2712382 SCHEMBL2712382 | C21H27ClN4O2 | 402.923 | 5 / 1 | 2.4 | Yes |
| 17960 | 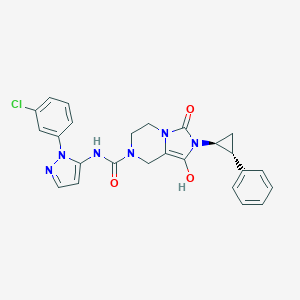 BDBM50350338 BDBM50350338 | C25H23ClN6O3 | 490.948 | 4 / 2 | 2.7 | Yes |
| 18421 | 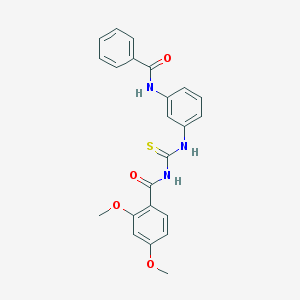 CHEMBL2031089 CHEMBL2031089 | C23H21N3O4S | 435.498 | 5 / 3 | 5.1 | No |
| 536460 |  CHEMBL3965479 CHEMBL3965479 | C27H32N8O2 | 500.607 | 8 / 1 | 2.1 | No |
| 459392 |  PF-5274857 PF-5274857 | C20H25ClN4O3S | 436.955 | 6 / 0 | 1.8 | Yes |
| 557925 |  SCHEMBL15169489 SCHEMBL15169489 | C20H23ClN4O4S | 450.938 | 7 / 1 | 0.7 | Yes |
| 548124 |  CHEMBL3963716 CHEMBL3963716 | C20H23ClN4O4S | 450.938 | 7 / 1 | 0.7 | Yes |
| 459402 | 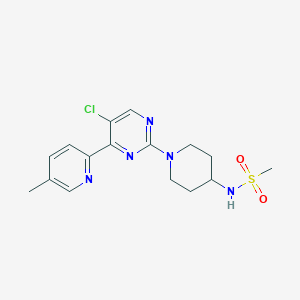 SCHEMBL4440843 SCHEMBL4440843 | C16H20ClN5O2S | 381.879 | 7 / 1 | 1.9 | Yes |
| 459403 | 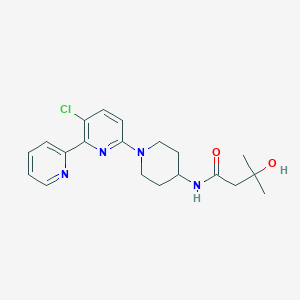 CHEMBL3696789 CHEMBL3696789 | C20H25ClN4O2 | 388.896 | 5 / 2 | 2.9 | Yes |
| 21112 | 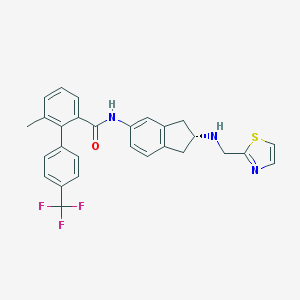 CHEMBL474892 CHEMBL474892 | C28H24F3N3OS | 507.575 | 7 / 2 | 6.1 | No |
| 21113 | 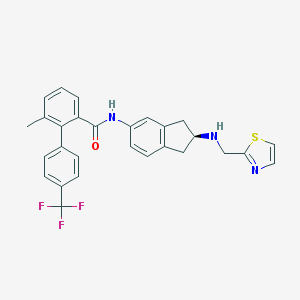 CHEMBL474893 CHEMBL474893 | C28H24F3N3OS | 507.575 | 7 / 2 | 6.1 | No |
| 22473 | 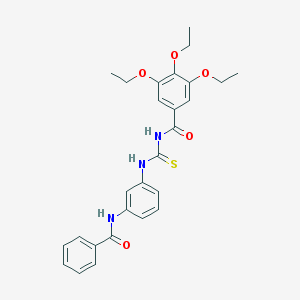 CHEMBL2031090 CHEMBL2031090 | C27H29N3O5S | 507.605 | 6 / 3 | 5.4 | No |
| 22719 | 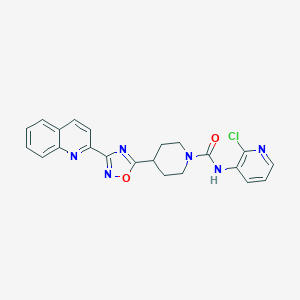 CHEMBL1823835 CHEMBL1823835 | C22H19ClN6O2 | 434.884 | 6 / 1 | 3.5 | Yes |
| 23656 | 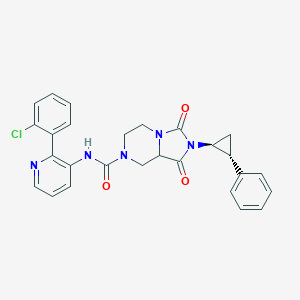 CID 56683603 CID 56683603 | C27H24ClN5O3 | 501.971 | 4 / 1 | 3.2 | No |
| 536587 | 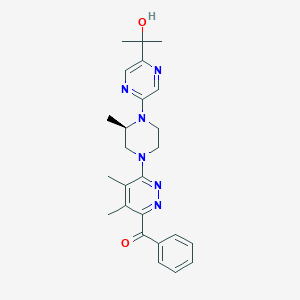 CHEMBL3953149 CHEMBL3953149 | C25H30N6O2 | 446.555 | 8 / 1 | 2.7 | Yes |
| 536606 | 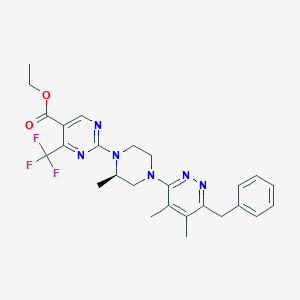 CHEMBL3921763 CHEMBL3921763 | C26H29F3N6O2 | 514.553 | 11 / 0 | 4.8 | No |
| 536609 | 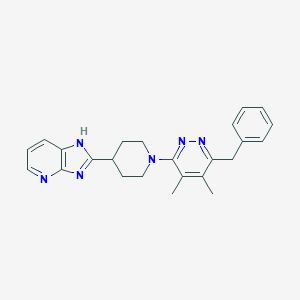 SCHEMBL1833847 SCHEMBL1833847 | C24H26N6 | 398.514 | 5 / 1 | 4.1 | Yes |
| 25253 | 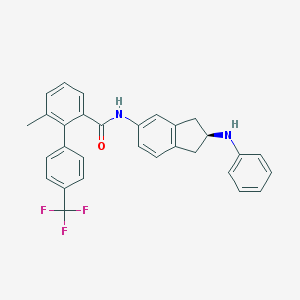 CHEMBL474302 CHEMBL474302 | C30H25F3N2O | 486.538 | 5 / 2 | 7.7 | No |
| 26148 | 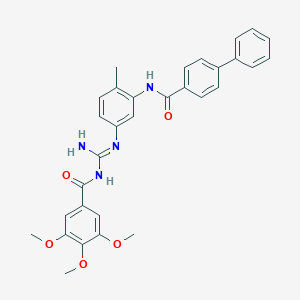 MRT-83 MRT-83 | C31H30N4O5 | 538.604 | 6 / 3 | 5.5 | No |
| 536680 |  CHEMBL560932 CHEMBL560932 | C21H25FN4O | 368.456 | 3 / 1 | 4.2 | Yes |
| 27405 |  CHEMBL3126694 CHEMBL3126694 | C23H21Cl2N3O5S | 522.397 | 5 / 3 | 4.2 | No |
| 27457 |  CHEMBL1822299 CHEMBL1822299 | C28H36N6O4 | 520.634 | 7 / 2 | 3.8 | No |
| 27608 |  CHEMBL1823585 CHEMBL1823585 | C24H22ClN5O2 | 447.923 | 5 / 1 | 4.6 | Yes |
| 536702 | 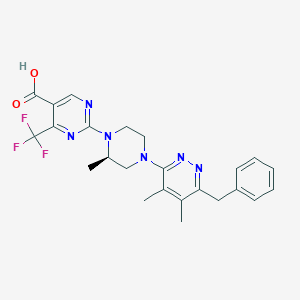 SCHEMBL1830420 SCHEMBL1830420 | C24H25F3N6O2 | 486.499 | 11 / 1 | 4.1 | No |
| 28119 | 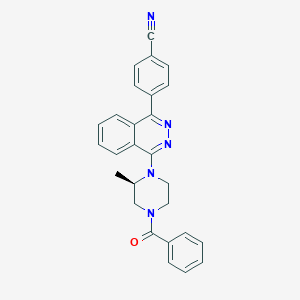 CHEMBL1084736 CHEMBL1084736 | C27H23N5O | 433.515 | 5 / 0 | 4.3 | Yes |
| 459466 | 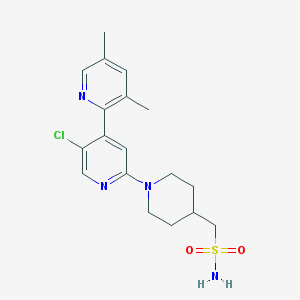 CHEMBL3696776 CHEMBL3696776 | C18H23ClN4O2S | 394.918 | 6 / 1 | 2.7 | Yes |
| 536737 | 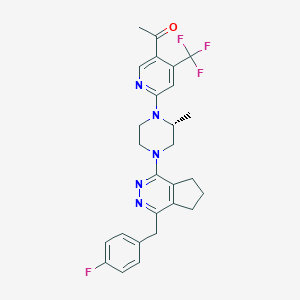 CHEMBL3919524 CHEMBL3919524 | C27H27F4N5O | 513.541 | 10 / 0 | 5.1 | No |
| 29576 |  CHEMBL3262645 CHEMBL3262645 | C24H22ClN7O | 459.938 | 6 / 1 | 2.7 | Yes |
| 29701 |  BDBM50350339 BDBM50350339 | C25H23FN6O3 | 474.496 | 5 / 2 | 2.2 | Yes |
| 29800 |  Vismodegib Vismodegib | C19H14Cl2N2O3S | 421.292 | 4 / 1 | 3.8 | Yes |
| 30703 |  CHEMBL2031291 CHEMBL2031291 | C31H30N4O5 | 538.604 | 6 / 3 | 5.5 | No |
| 30783 | 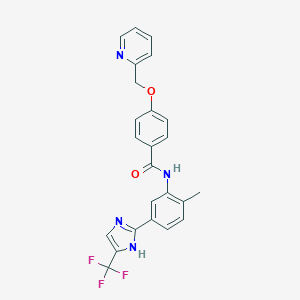 CHEMBL2059863 CHEMBL2059863 | C24H19F3N4O2 | 452.437 | 7 / 2 | 4.3 | Yes |
| 536771 | 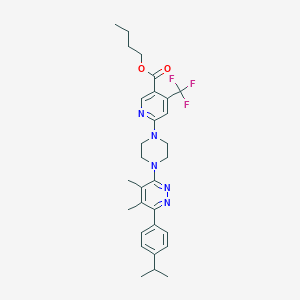 CHEMBL3959511 CHEMBL3959511 | C30H36F3N5O2 | 555.646 | 10 / 0 | 6.7 | No |
| 30995 | 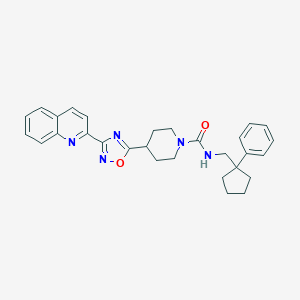 CHEMBL1822295 CHEMBL1822295 | C29H31N5O2 | 481.6 | 5 / 1 | 5.3 | No |
| 536799 | 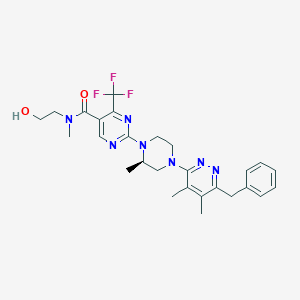 CHEMBL3903125 CHEMBL3903125 | C27H32F3N7O2 | 543.595 | 11 / 1 | 3.3 | No |
| 31907 |  CHEMBL2160067 CHEMBL2160067 | C28H25ClN6O | 496.999 | 6 / 2 | 5.6 | No |
| 32191 |  CHEMBL3262638 CHEMBL3262638 | C25H30ClN7O | 480.013 | 6 / 1 | 2.6 | Yes |
| 32381 |  CID 56673458 CID 56673458 | C23H19Cl2F3N4O3 | 527.325 | 6 / 1 | 4.1 | No |
| 32893 |  CHEMBL3262642 CHEMBL3262642 | C25H22ClFN6O | 476.94 | 6 / 1 | 3.9 | Yes |
| 34321 |  CHEMBL1823618 CHEMBL1823618 | C23H28N4O | 376.504 | 5 / 0 | 5.1 | No |
| 459528 |  SCHEMBL2714478 SCHEMBL2714478 | C19H23ClN4O | 358.87 | 4 / 0 | 3.2 | Yes |
| 35109 |  CHEMBL495875 CHEMBL495875 | C25H24N6O | 424.508 | 6 / 1 | 3.2 | Yes |
| 36231 |  CHEMBL1823842 CHEMBL1823842 | C23H26N4O3 | 406.486 | 6 / 0 | 4.4 | Yes |
| 459542 | 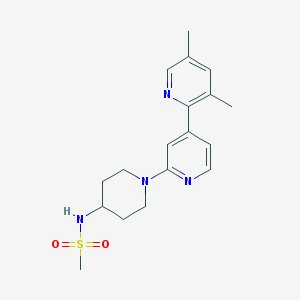 CHEMBL3696800 CHEMBL3696800 | C18H24N4O2S | 360.476 | 6 / 1 | 2.3 | Yes |
| 36578 | 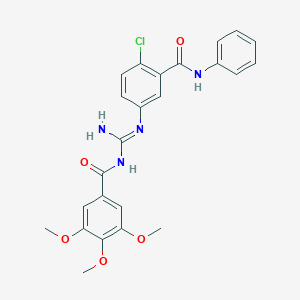 CHEMBL2031295 CHEMBL2031295 | C24H23ClN4O5 | 482.921 | 6 / 3 | 4.2 | Yes |
| 548319 | 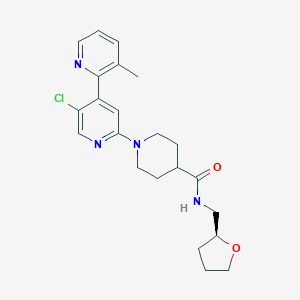 CHEMBL3891652 CHEMBL3891652 | C22H27ClN4O2 | 414.934 | 5 / 1 | 3.1 | Yes |
| 558398 | 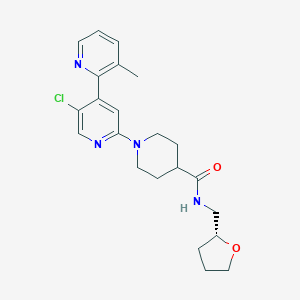 SCHEMBL2712532 SCHEMBL2712532 | C22H27ClN4O2 | 414.934 | 5 / 1 | 3.1 | Yes |
| 467429 |  SCHEMBL2713676 SCHEMBL2713676 | C21H25ClN4O2 | 400.907 | 5 / 1 | 2.7 | Yes |
| 459548 |  CHEMBL3696783 CHEMBL3696783 | C18H20ClN5O2S | 405.901 | 7 / 1 | 2.3 | Yes |
| 37287 |  CHEMBL2063139 CHEMBL2063139 | C27H42 | 366.633 | 0 / 0 | 9.0 | No |
| 459552 |  SCHEMBL2712050 SCHEMBL2712050 | C18H21ClN4O2 | 360.842 | 5 / 0 | 2.9 | Yes |
| 37462 |  CID 56659615 CID 56659615 | C28H26N4O3 | 466.541 | 3 / 1 | 3.7 | Yes |
| 37942 |  CHEMBL1084730 CHEMBL1084730 | C29H28N4O | 448.57 | 4 / 0 | 5.5 | No |
| 38180 |  CHEMBL473896 CHEMBL473896 | C31H27F3N2O | 500.565 | 5 / 2 | 7.1 | No |
| 39123 |  CHEMBL1822446 CHEMBL1822446 | C25H32N6O2 | 448.571 | 6 / 2 | 3.0 | Yes |
| 39375 | 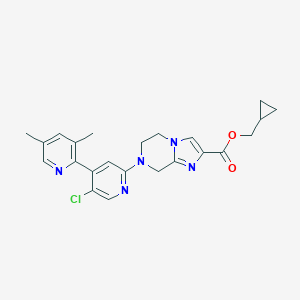 CHEMBL3262647 CHEMBL3262647 | C23H24ClN5O2 | 437.928 | 6 / 0 | 3.5 | Yes |
| 40169 | 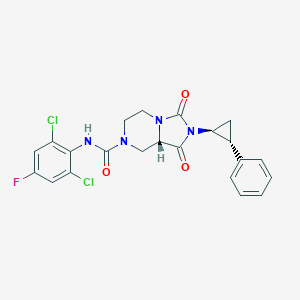 CHEMBL1813101 CHEMBL1813101 | C22H19Cl2FN4O3 | 477.317 | 4 / 1 | 3.3 | Yes |
| 40170 | 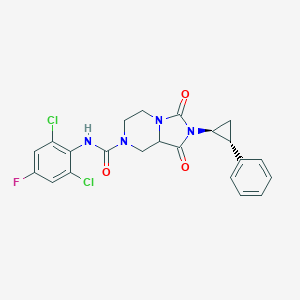 CID 56673457 CID 56673457 | C22H19Cl2FN4O3 | 477.317 | 4 / 1 | 3.3 | Yes |
| 537031 | 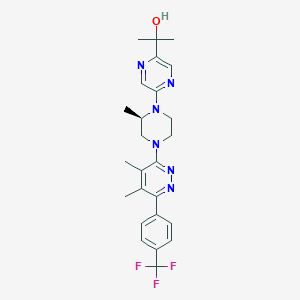 CHEMBL3959587 CHEMBL3959587 | C25H29F3N6O | 486.543 | 10 / 1 | 3.5 | Yes |
| 41264 | 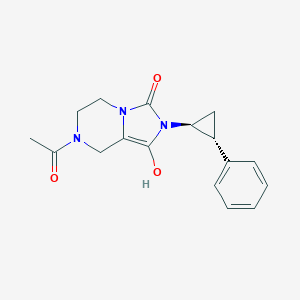 BDBM50349302 BDBM50349302 | C17H19N3O3 | 313.357 | 3 / 1 | 0.4 | Yes |
| 41914 | 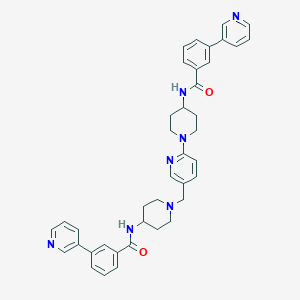 CHEMBL2160208 CHEMBL2160208 | C40H41N7O2 | 651.815 | 7 / 2 | 5.3 | No |
| 41952 | 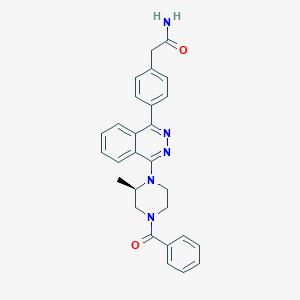 CHEMBL1209190 CHEMBL1209190 | C28H27N5O2 | 465.557 | 5 / 1 | 3.4 | Yes |
| 42144 | 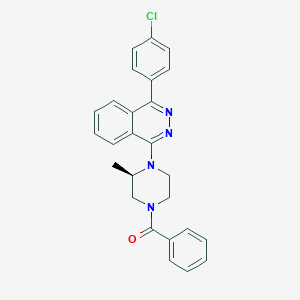 CHEMBL1086467 CHEMBL1086467 | C26H23ClN4O | 442.947 | 4 / 0 | 5.2 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417