

You can:
| Name | P2Y purinoceptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | P2RY2 |
| Synonym | Purinergic receptor purinergic receptor P2Y P2Y2 receptor P2Y2 P2Y purinoceptor 2 [ Show all ] |
| Disease | Dry eye disease Constipation Cystic fibrosis Lung cancer |
| Length | 377 |
| Amino acid sequence | MAADLGPWNDTINGTWDGDELGYRCRFNEDFKYVLLPVSYGVVCVPGLCLNAVALYIFLCRLKTWNASTTYMFHLAVSDALYAASLPLLVYYYARGDHWPFSTVLCKLVRFLFYTNLYCSILFLTCISVHRCLGVLRPLRSLRWGRARYARRVAGAVWVLVLACQAPVLYFVTTSARGGRVTCHDTSAPELFSRFVAYSSVMLGLLFAVPFAVILVCYVLMARRLLKPAYGTSGGLPRAKRKSVRTIAVVLAVFALCFLPFHVTRTLYYSFRSLDLSCHTLNAINMAYKVTRPLASANSCLDPVLYFLAGQRLVRFARDAKPPTGPSPATPARRRLGLRRSDRTDMQRIEDVLGSSEDSRRTESTPAGSENTKDIRL |
| UniProt | P41231 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P41231 |
| 3D structure model | This predicted structure model is from GPCR-EXP P41231. |
| BioLiP | N/A |
| Therapeutic Target Database | T93515 |
| ChEMBL | CHEMBL4398 |
| IUPHAR | 324 |
| DrugBank | BE0002401 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 1037 | 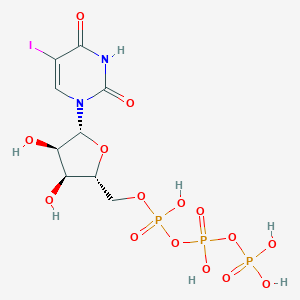 CHEMBL220398 CHEMBL220398 | C9H14IN2O15P3 | 610.036 | 15 / 7 | -5.5 | No |
| 553257 | 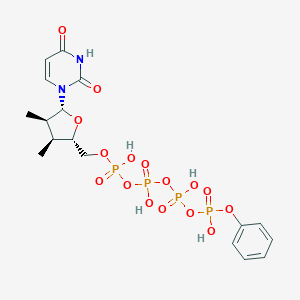 MRS2768 MRS2768 | C17H24N2O16P4 | 636.272 | 16 / 5 | -2.1 | No |
| 3492 | 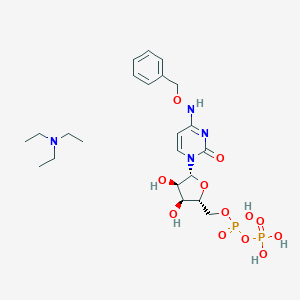 CHEMBL1084612 CHEMBL1084612 | C22H36N4O12P2 | 610.494 | 13 / 6 | N/A | No |
| 3880 | 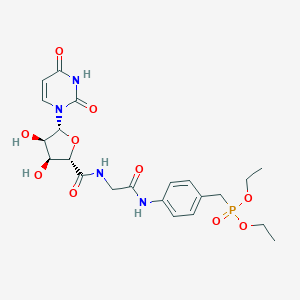 PSB-6426 PSB-6426 | C22H29N4O10P | 540.466 | 10 / 5 | -1.6 | No |
| 4530 | 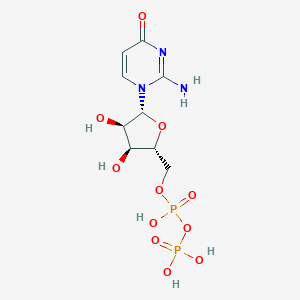 CHEMBL1767415 CHEMBL1767415 | C9H15N3O11P2 | 403.177 | 11 / 6 | -4.5 | No |
| 4768 | 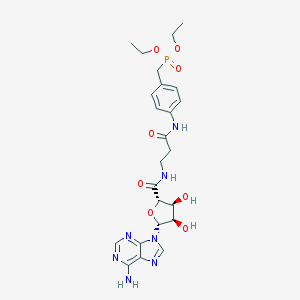 CHEMBL517017 CHEMBL517017 | C24H32N7O8P | 577.535 | 12 / 5 | -1.3 | No |
| 5441 | 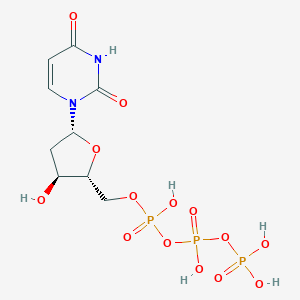 dUTP dUTP | C9H15N2O14P3 | 468.14 | 14 / 6 | -5.5 | No |
| 5500 | 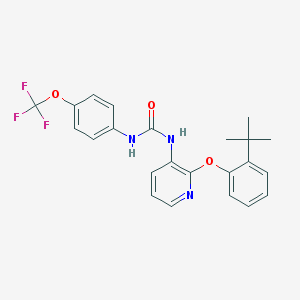 BPTU BPTU | C23H22F3N3O3 | 445.442 | 7 / 2 | 6.1 | No |
| 6220 | 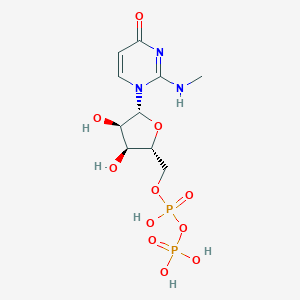 CHEMBL1767416 CHEMBL1767416 | C10H17N3O11P2 | 417.204 | 11 / 6 | -4.1 | No |
| 441970 | 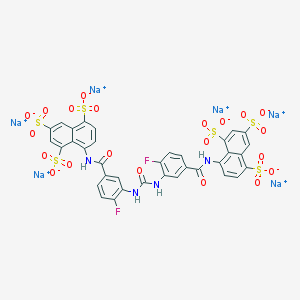 CHEMBL268784 CHEMBL268784 | C35H18F2N4Na6O21S6 | 1198.83 | 23 / 4 | N/A | No |
| 7014 | 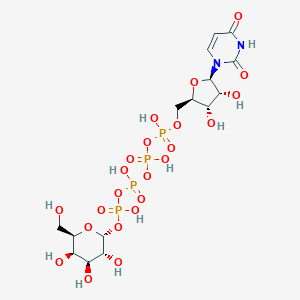 CHEMBL444212 CHEMBL444212 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 7017 | 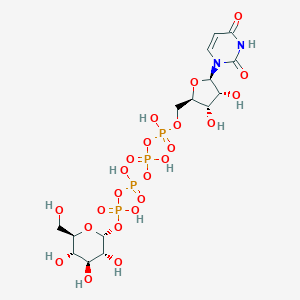 CHEMBL499138 CHEMBL499138 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 7021 | 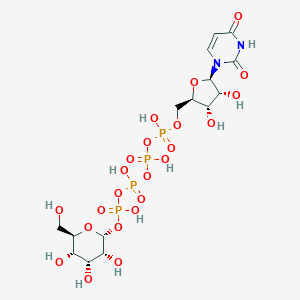 CHEMBL1784885 CHEMBL1784885 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 7025 | 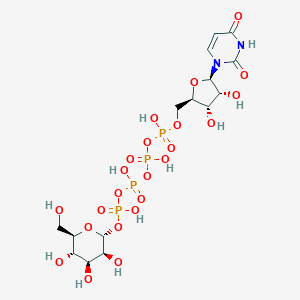 CHEMBL1784886 CHEMBL1784886 | C15H26N2O23P4 | 726.259 | 23 / 11 | -8.5 | No |
| 7533 | 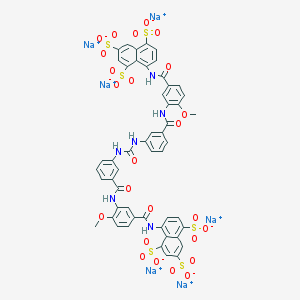 CHEMBL413910 CHEMBL413910 | C51H34N6Na6O25S6 | 1461.15 | 25 / 6 | N/A | No |
| 12385 | 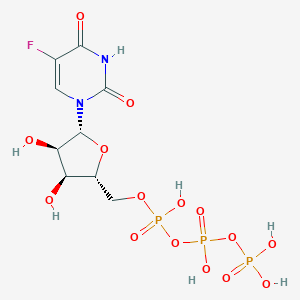 FUTP FUTP | C9H14FN2O15P3 | 502.13 | 16 / 7 | -5.8 | No |
| 12536 | 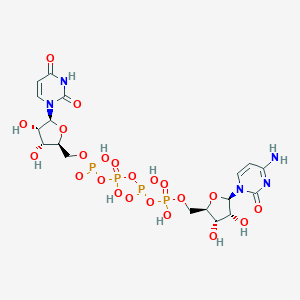 CHEMBL1162198 CHEMBL1162198 | C18H27N5O22P4 | 789.322 | 22 / 10 | -9.4 | No |
| 12613 | 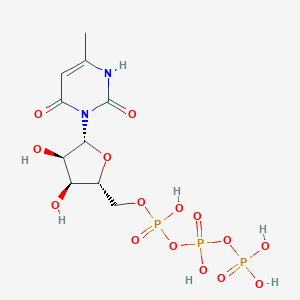 CHEMBL413841 CHEMBL413841 | C10H17N2O15P3 | 498.166 | 15 / 7 | -5.8 | No |
| 14782 | 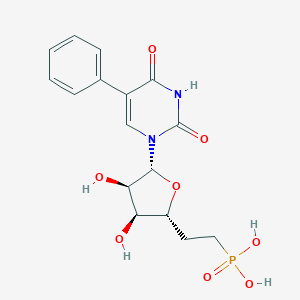 CHEMBL1957438 CHEMBL1957438 | C16H19N2O8P | 398.308 | 8 / 5 | -2.0 | Yes |
| 16608 | 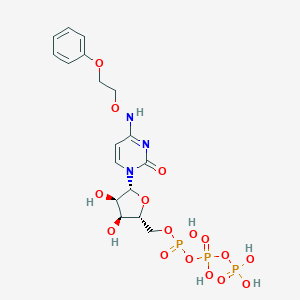 CHEMBL3261361 CHEMBL3261361 | C17H24N3O16P3 | 619.305 | 16 / 7 | -3.3 | No |
| 17590 | 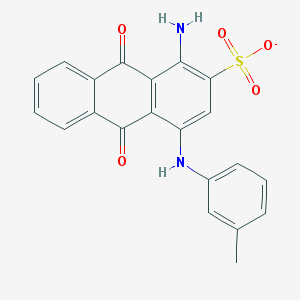 CHEMBL445413 CHEMBL445413 | C21H15N2O5S- | 407.42 | 7 / 2 | 3.9 | Yes |
| 18301 | 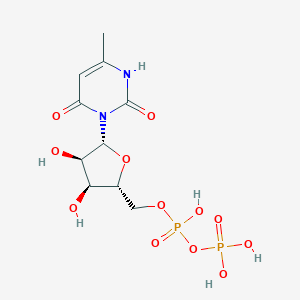 CHEMBL386096 CHEMBL386096 | C10H16N2O12P2 | 418.188 | 12 / 6 | -4.7 | No |
| 18895 | 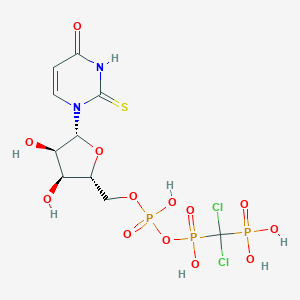 CHEMBL519835 CHEMBL519835 | C10H15Cl2N2O13P3S | 567.112 | 14 / 7 | -4.4 | No |
| 19507 | 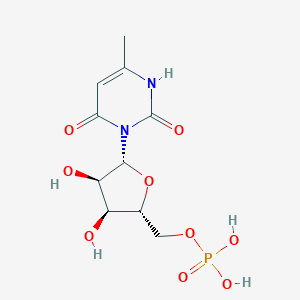 CHEMBL215263 CHEMBL215263 | C10H15N2O9P | 338.209 | 9 / 5 | -3.6 | Yes |
| 21439 | 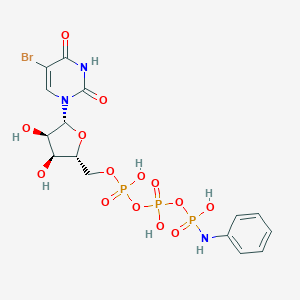 CHEMBL1765120 CHEMBL1765120 | C15H19BrN3O14P3 | 638.149 | 15 / 7 | -2.3 | No |
| 22715 | 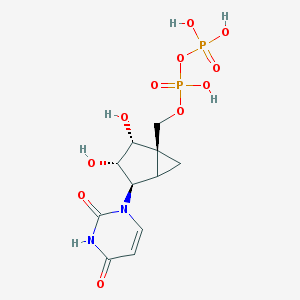 CHEMBL215811 CHEMBL215811 | C11H16N2O11P2 | 414.2 | 11 / 6 | -4.6 | No |
| 22746 | 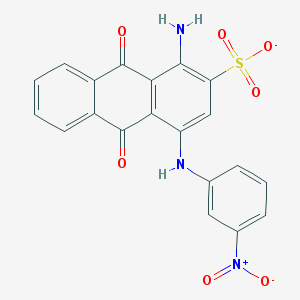 CHEMBL404659 CHEMBL404659 | C20H12N3O7S- | 438.39 | 9 / 2 | 3.3 | Yes |
| 23818 | 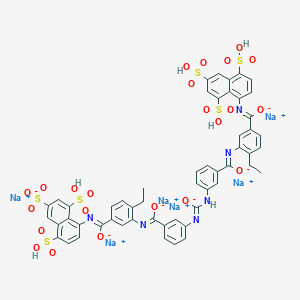 CID 136825090 CID 136825090 | C53H38N6Na6O23S6 | 1457.2 | 28 / 6 | N/A | No |
| 24479 |  CHEMBL1162183 CHEMBL1162183 | C26H33N5O22P4 | 891.458 | 22 / 8 | -6.5 | No |
| 25571 |  CHEMBL1957440 CHEMBL1957440 | C16H18FN2O8P | 416.298 | 9 / 5 | -1.9 | Yes |
| 26412 |  CHEMBL1672105 CHEMBL1672105 | C23H13Cl2N6O5S- | 556.354 | 11 / 3 | 5.3 | No |
| 26468 |  CHEMBL393101 CHEMBL393101 | C12H16NO15P3 | 507.173 | 15 / 6 | -4.7 | No |
| 26846 |  PSB 06126 PSB 06126 | C24H15N2NaO5S | 466.443 | 7 / 2 | N/A | N/A |
| 27553 |  2-thio-UDP 2-thio-UDP | C9H14N2O11P2S | 420.222 | 12 / 6 | -4.1 | No |
| 27723 |  CHEMBL1957445 CHEMBL1957445 | C18H19N2O8PS | 454.39 | 9 / 5 | -1.0 | Yes |
| 29163 |  CHEMBL1162203 CHEMBL1162203 | C25H31FN6O23P4 | 926.435 | 24 / 11 | -7.8 | No |
| 30282 | 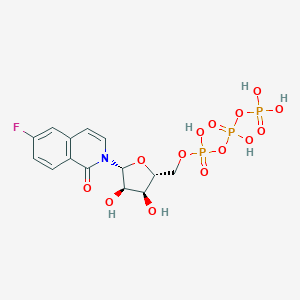 CHEMBL241320 CHEMBL241320 | C14H17FNO14P3 | 535.203 | 15 / 6 | -3.7 | No |
| 30315 | 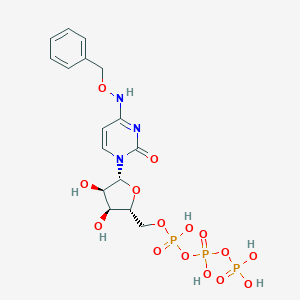 CHEMBL1784892 CHEMBL1784892 | C16H22N3O15P3 | 589.279 | 15 / 7 | -3.7 | No |
| 30389 | 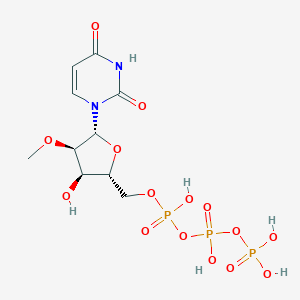 CHEMBL220337 CHEMBL220337 | C10H17N2O15P3 | 498.166 | 15 / 6 | -5.3 | No |
| 30505 | 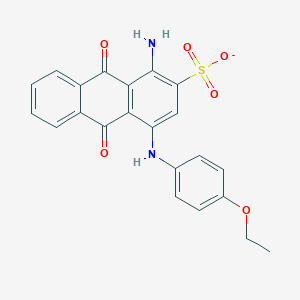 CHEMBL271881 CHEMBL271881 | C22H17N2O6S- | 437.446 | 8 / 2 | 3.8 | Yes |
| 31288 | 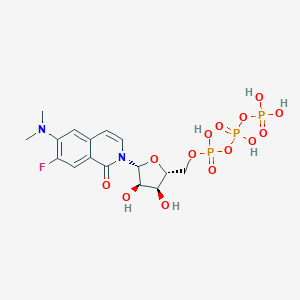 CHEMBL239573 CHEMBL239573 | C16H22FN2O14P3 | 578.272 | 16 / 6 | -3.6 | No |
| 32087 | 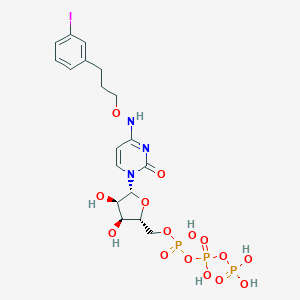 CHEMBL3261365 CHEMBL3261365 | C18H25IN3O15P3 | 743.23 | 15 / 7 | -3.1 | No |
| 32090 | 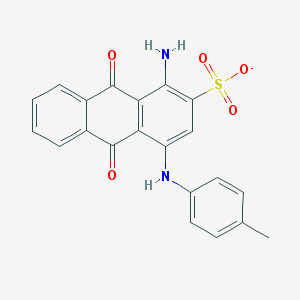 CHEMBL404450 CHEMBL404450 | C21H15N2O5S- | 407.42 | 7 / 2 | 3.9 | Yes |
| 32485 | 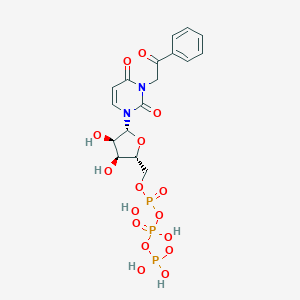 CHEMBL385488 CHEMBL385488 | C17H21N2O16P3 | 602.274 | 16 / 6 | -4.5 | No |
| 33911 | 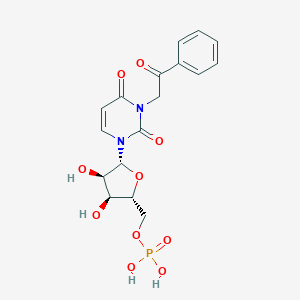 CHEMBL215381 CHEMBL215381 | C17H19N2O10P | 442.317 | 10 / 4 | -2.4 | Yes |
| 34089 | 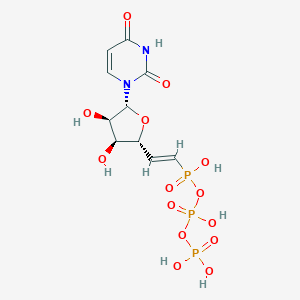 CHEMBL523850 CHEMBL523850 | C10H15N2O14P3 | 480.151 | 14 / 7 | -6.1 | No |
| 35578 | 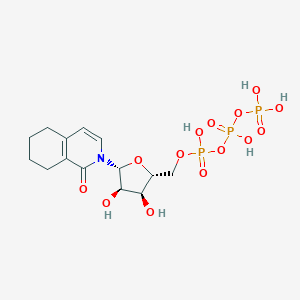 CHEMBL239814 CHEMBL239814 | C14H22NO14P3 | 521.244 | 14 / 6 | -4.0 | No |
| 36210 | 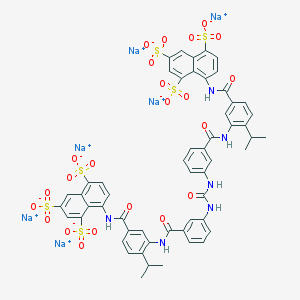 CHEMBL405500 CHEMBL405500 | C55H42N6Na6O23S6 | 1485.26 | 23 / 6 | N/A | No |
| 37984 | 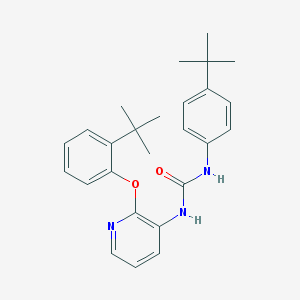 CHEMBL2333771 CHEMBL2333771 | C26H31N3O2 | 417.553 | 3 / 2 | 6.6 | No |
| 38248 | 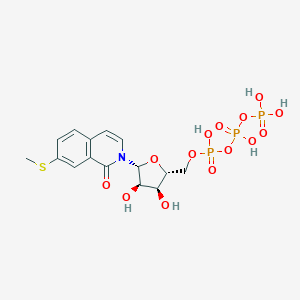 CHEMBL240203 CHEMBL240203 | C15H20NO14P3S | 563.299 | 15 / 6 | -3.3 | No |
| 38813 | 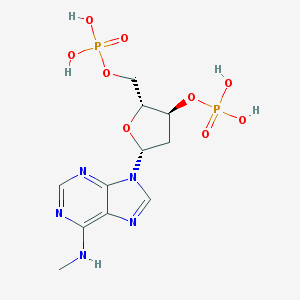 CHEMBL129841 CHEMBL129841 | C11H17N5O9P2 | 425.231 | 13 / 5 | -2.9 | No |
| 42142 | 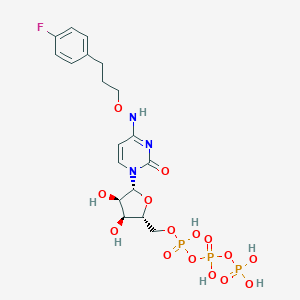 CHEMBL3261363 CHEMBL3261363 | C18H25FN3O15P3 | 635.324 | 16 / 7 | -3.6 | No |
| 459594 | 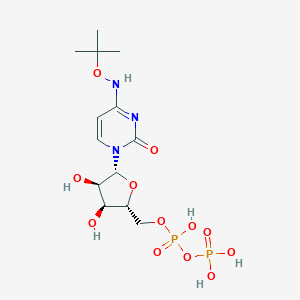 CHEMBL1084296 CHEMBL1084296 | C13H23N3O12P2 | 475.284 | 12 / 6 | -3.1 | No |
| 42488 | 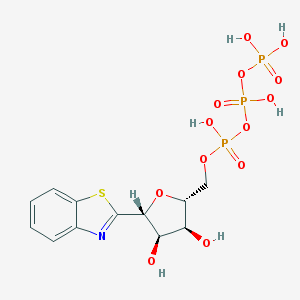 CHEMBL241481 CHEMBL241481 | C12H16NO13P3S | 507.235 | 15 / 6 | -3.3 | No |
| 43949 | 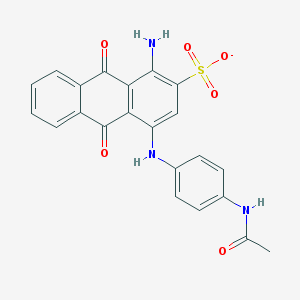 CHEMBL271688 CHEMBL271688 | C22H16N3O6S- | 450.445 | 8 / 3 | 2.7 | Yes |
| 44383 | 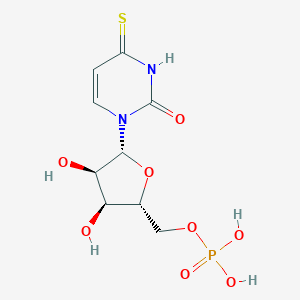 CHEMBL1230331 CHEMBL1230331 | C9H13N2O8PS | 340.243 | 9 / 5 | -3.0 | Yes |
| 47984 | 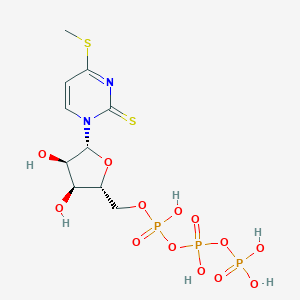 CHEMBL482472 CHEMBL482472 | C10H17N2O13P3S2 | 530.288 | 15 / 6 | -4.3 | No |
| 49429 | 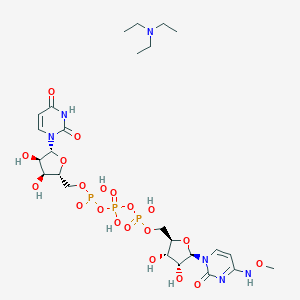 CHEMBL1083256 CHEMBL1083256 | C25H43N6O20P3 | 840.562 | 21 / 9 | N/A | No |
| 49772 | 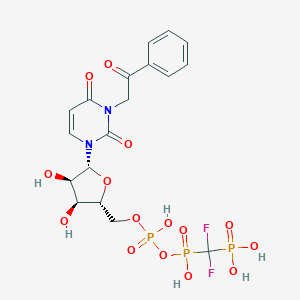 CHEMBL1765118 CHEMBL1765118 | C18H21F2N2O15P3 | 636.283 | 17 / 6 | -3.9 | No |
| 51490 | 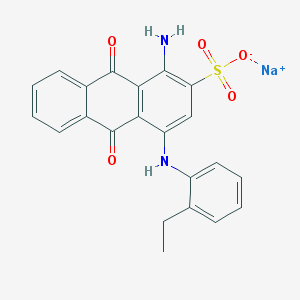 CHEMBL402238 CHEMBL402238 | C22H17N2NaO5S | 444.437 | 7 / 2 | N/A | N/A |
| 53513 | 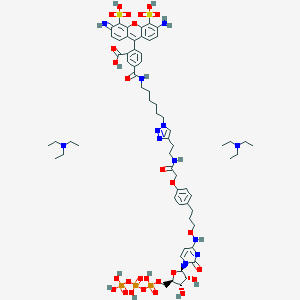 CHEMBL3261377 CHEMBL3261377 | C63H89N12O27P3S2 | 1603.5 | 33 / 14 | N/A | No |
| 53629 | 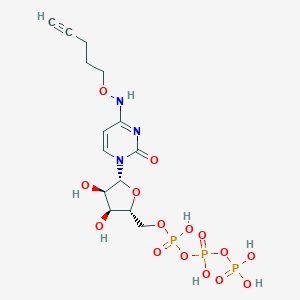 CHEMBL3261373 CHEMBL3261373 | C14H22N3O15P3 | 565.257 | 15 / 7 | -4.2 | No |
| 58255 | 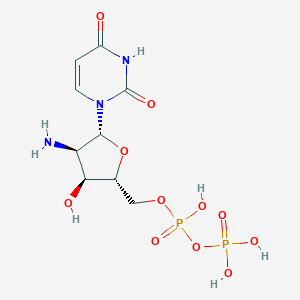 CHEMBL215911 CHEMBL215911 | C9H15N3O11P2 | 403.177 | 12 / 6 | -7.3 | No |
| 58526 | 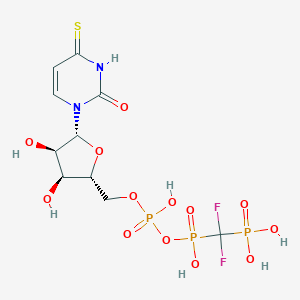 CHEMBL1765119 CHEMBL1765119 | C10H15F2N2O13P3S | 534.209 | 16 / 7 | -4.6 | No |
| 60005 | 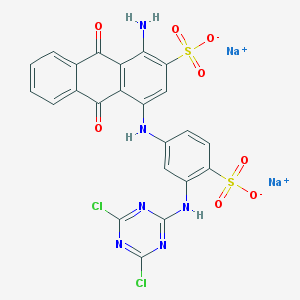 Azure KX Azure KX | C23H12Cl2N6Na2O8S2 | 681.383 | 14 / 3 | N/A | No |
| 62391 | 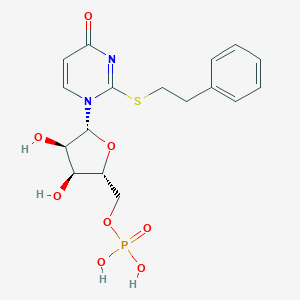 CHEMBL1767421 CHEMBL1767421 | C17H21N2O8PS | 444.395 | 9 / 4 | 0.1 | Yes |
| 65131 | 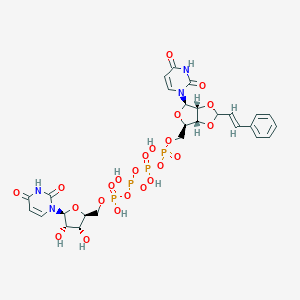 CHEMBL1162157 CHEMBL1162157 | C27H32N4O23P4 | 904.453 | 23 / 8 | -6.0 | No |
| 68554 | 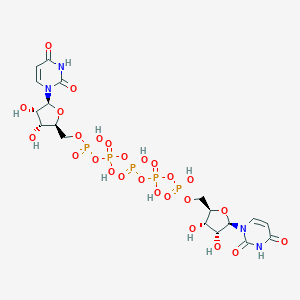 CHEMBL2113401 CHEMBL2113401 | C18H27N4O26P5 | 870.285 | 26 / 11 | -10.2 | No |
| 68908 | 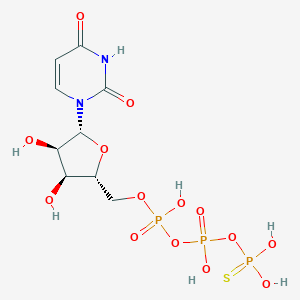 UTP-Gamma-S UTP-Gamma-S | C9H15N2O14P3S | 500.2 | 15 / 7 | -4.1 | No |
| 459854 | 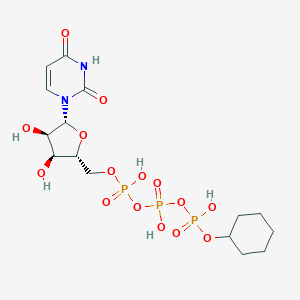 CHEMBL1083258 CHEMBL1083258 | C15H25N2O15P3 | 566.285 | 15 / 6 | -3.5 | No |
| 72312 | 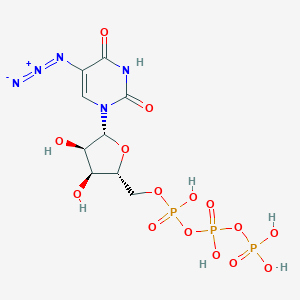 CHEMBL219779 CHEMBL219779 | C9H14N5O15P3 | 525.152 | 17 / 7 | -5.0 | No |
| 72460 | 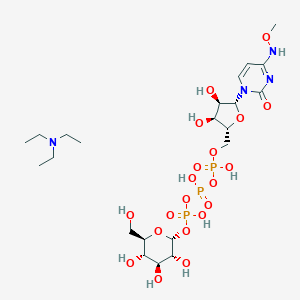 CHEMBL1083264 CHEMBL1083264 | C22H43N4O20P3 | 776.515 | 21 / 10 | N/A | No |
| 73967 | 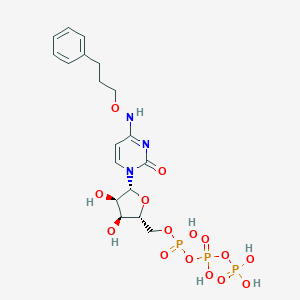 MRS-4062 MRS-4062 | C18H26N3O15P3 | 617.333 | 15 / 7 | -3.7 | No |
| 75411 | 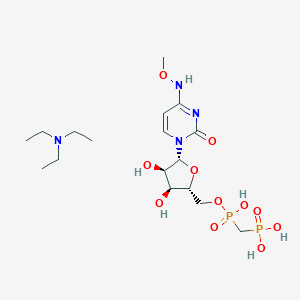 CHEMBL1084613 CHEMBL1084613 | C17H34N4O11P2 | 532.424 | 12 / 6 | N/A | No |
| 75474 | 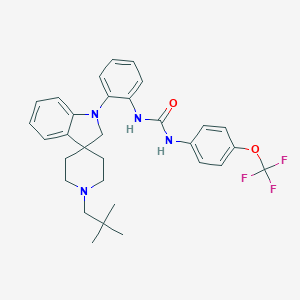 CHEMBL3104636 CHEMBL3104636 | C31H35F3N4O2 | 552.642 | 7 / 2 | 7.4 | No |
| 75726 | 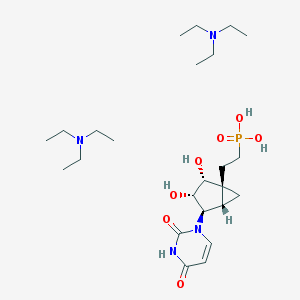 CHEMBL1957448 CHEMBL1957448 | C24H47N4O7P | 534.635 | 9 / 5 | N/A | No |
| 75981 | 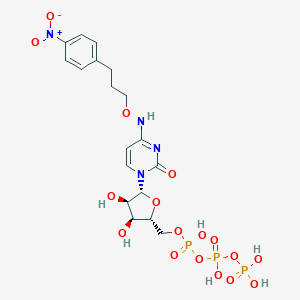 CHEMBL3261368 CHEMBL3261368 | C18H25N4O17P3 | 662.33 | 17 / 7 | -3.9 | No |
| 76440 | 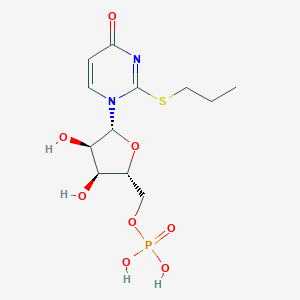 CHEMBL1767419 CHEMBL1767419 | C12H19N2O8PS | 382.324 | 9 / 4 | -1.0 | Yes |
| 77912 | 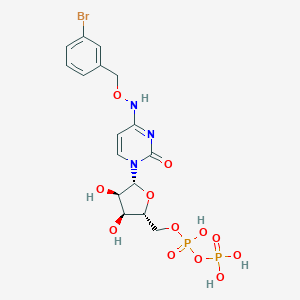 CHEMBL3220046 CHEMBL3220046 | C16H20BrN3O12P2 | 588.197 | 12 / 6 | -2.7 | No |
| 78869 | 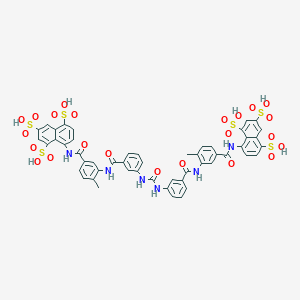 suramin suramin | C51H40N6O23S6 | 1297.26 | 23 / 12 | 1.5 | No |
| 79698 | 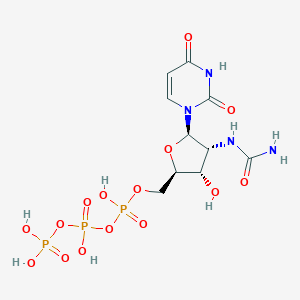 CHEMBL484295 CHEMBL484295 | C10H17N4O15P3 | 526.18 | 15 / 8 | -6.4 | No |
| 82203 | 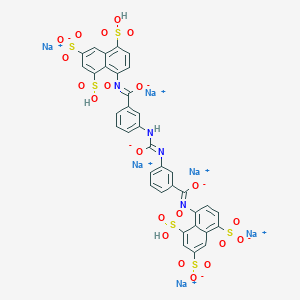 CID 136775315 CID 136775315 | C35H20N4Na6O21S6 | 1162.85 | 24 / 4 | N/A | No |
| 82496 | 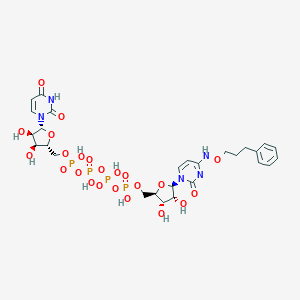 CHEMBL3261379 CHEMBL3261379 | C27H37N5O23P4 | 923.5 | 23 / 10 | -6.7 | No |
| 83773 | 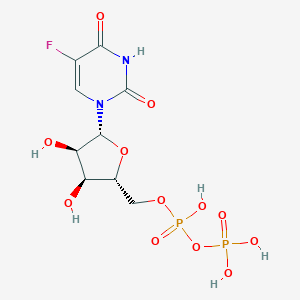 5-fluorouridine diphosphate 5-fluorouridine diphosphate | C9H13FN2O12P2 | 422.151 | 13 / 6 | -4.7 | No |
| 459972 | 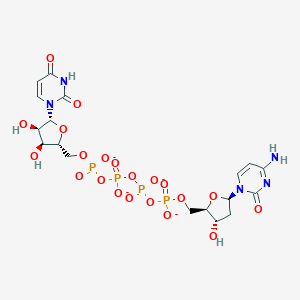 CHEMBL1767407 CHEMBL1767407 | C18H23N5O21P4-4 | 769.291 | 21 / 5 | -8.8 | No |
| 84174 | 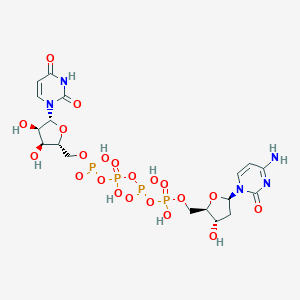 denufosol denufosol | C18H27N5O21P4 | 773.323 | 21 / 9 | -8.4 | No |
| 85063 | 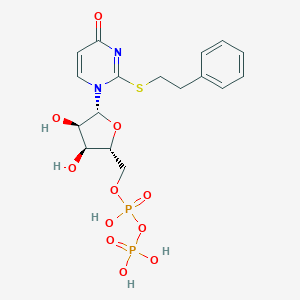 CHEMBL1767413 CHEMBL1767413 | C17H22N2O11P2S | 524.374 | 12 / 5 | -1.9 | No |
| 459996 | 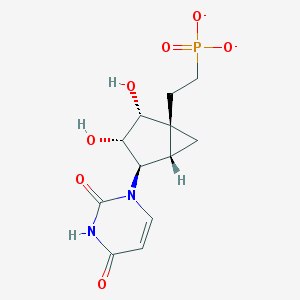 CHEMBL1957448 CHEMBL1957448 | C12H15N2O7P-2 | 330.233 | 7 / 3 | -3.3 | Yes |
| 87798 | 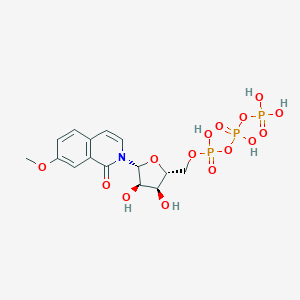 CHEMBL399018 CHEMBL399018 | C15H20NO15P3 | 547.238 | 15 / 6 | -3.9 | No |
| 88247 | 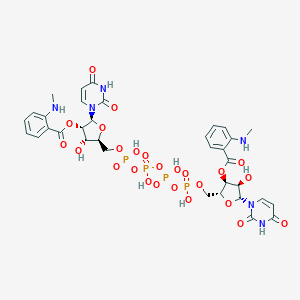 CHEMBL1162181 CHEMBL1162181 | C34H40N6O25P4 | 1056.61 | 27 / 10 | -3.6 | No |
| 90105 | 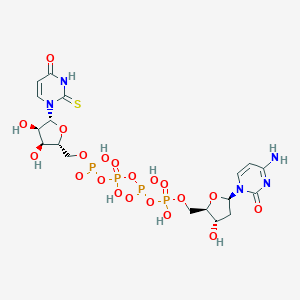 CHEMBL509434 CHEMBL509434 | C18H27N5O20P4S | 789.384 | 21 / 9 | -7.8 | No |
| 90540 | 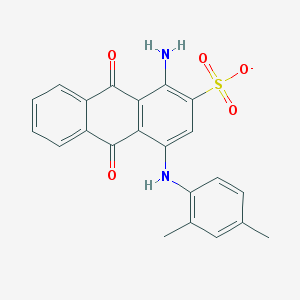 CHEMBL256688 CHEMBL256688 | C22H17N2O5S- | 421.447 | 7 / 2 | 4.2 | Yes |
| 90563 | 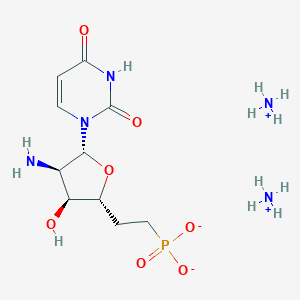 CHEMBL1957446 CHEMBL1957446 | C10H22N5O7P | 355.288 | 8 / 5 | N/A | N/A |
| 91002 | 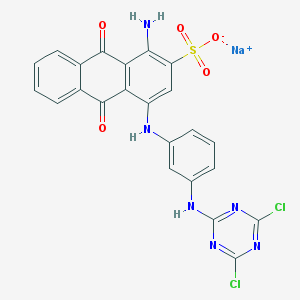 PSB-10211 PSB-10211 | C23H13Cl2N6NaO5S | 579.344 | 11 / 3 | N/A | No |
| 91123 | 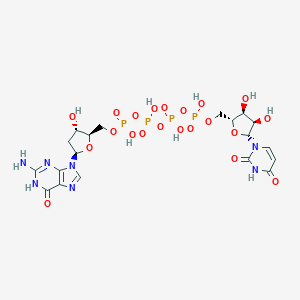 CHEMBL505403 CHEMBL505403 | C19H27N7O21P4 | 813.348 | 23 / 10 | -8.6 | No |
| 94099 | 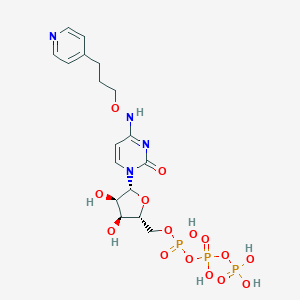 CHEMBL3261371 CHEMBL3261371 | C17H25N4O15P3 | 618.321 | 16 / 7 | -4.8 | No |
| 94789 | 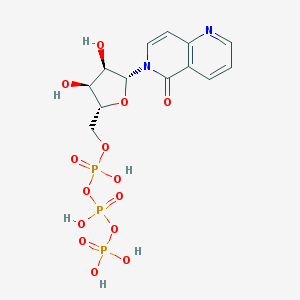 CHEMBL428387 CHEMBL428387 | C13H17N2O14P3 | 518.2 | 15 / 6 | -4.9 | No |
| 460091 | 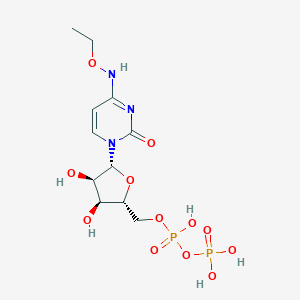 CHEMBL1084295 CHEMBL1084295 | C11H19N3O12P2 | 447.23 | 12 / 6 | -3.7 | No |
| 98052 | 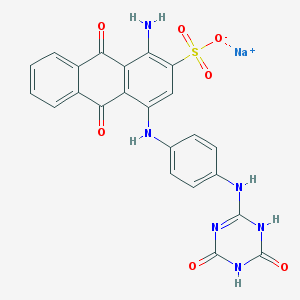 CHEMBL1672106 CHEMBL1672106 | C23H15N6NaO7S | 542.458 | 9 / 5 | N/A | No |
| 99039 | 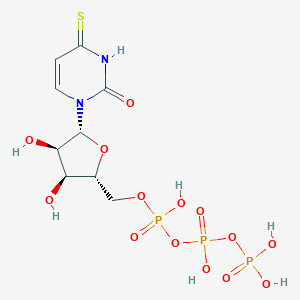 4-Thio-utp 4-Thio-utp | C9H15N2O14P3S | 500.2 | 15 / 7 | -5.2 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417