

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAADLGPWNDTINGTWDGDELGYRCRFNEDFKYVLLPVSYGVVCVPGLCLNAVALYIFLCRLKTWNASTTYMFHLAVSDALYAASLPLLVYYYARGDHWPFSTVLCKLVRFLFYTNLYCSILFLTCISVHRCLGVLRPLRSLRWGRARYARRVAGAVWVLVLACQAPVLYFVTTSARGGRVTCHDTSAPELFSRFVAYSSVMLGLLFAVPFAVILVCYVLMARRLLKPAYGTSGGLPRAKRKSVRTIAVVLAVFALCFLPFHVTRTLYYSFRSLDLSCHTLNAINMAYKVTRPLASANSCLDPVLYFLAGQRLVRFARDAKPPTGPSPATPARRRLGLRRSDRTDMQRIEDVLGSSEDSRRTESTPAGSENTKDIRL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 97888887777767888887677788750078999999999999999998999889766416788789999999999999999989999999966997667867899999999999999999999999970476625030223655025888899999999999989998412162299148938898167799999999999999999999999999999999832246665533300005789999999999998578999999999984158990699999999999999999999898997881488899999998411047865565678888878988776777777777788998888898777655679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAADLGPWNDTINGTWDGDELGYRCRFNEDFKYVLLPVSYGVVCVPGLCLNAVALYIFLCRLKTWNASTTYMFHLAVSDALYAASLPLLVYYYARGDHWPFSTVLCKLVRFLFYTNLYCSILFLTCISVHRCLGVLRPLRSLRWGRARYARRVAGAVWVLVLACQAPVLYFVTTSARGGRVTCHDTSAPELFSRFVAYSSVMLGLLFAVPFAVILVCYVLMARRLLKPAYGTSGGLPRAKRKSVRTIAVVLAVFALCFLPFHVTRTLYYSFRSLDLSCHTLNAINMAYKVTRPLASANSCLDPVLYFLAGQRLVRFARDAKPPTGPSPATPARRRLGLRRSDRTDMQRIEDVLGSSEDSRRTESTPAGSENTKDIRL | |
| 76452443322223334444343404345402100002201200230131101000000122343310000000001000000000101000003443320040011100111131110000000000310000001002023223320000000001000000010100013035572211010213473131110111011020132112000200000001013134445444543110000000000000000123101000010013123341413300200110010001300021000000004501410240034334444344444444444444444434434544554544643445574464363 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MAADLGPWNDTINGTWDGDELGYRCRFNEDFKYVLLPVSYGVVCVPGLCLNAVALYIFLCRLKTWNASTTYMFHLAVSDALYAASLPLLVYYYARGDHWPFSTVLCKLVRFLFYTNLYCSILFLTCISVHRCLGVLRPLRSLRWGRARYARRVAGAVWVLVLACQAPVLYFVTTSARGGRVTCHDTSAPELFSRFVAYSSVMLGLLFAVPFAVILVCYVLMARRLLKPAYGTSGGLPRAKRKSVRTIAVVLAVFALCFLPFHVTRTLYYSFRSLDLSCHTLNAINMAYKVTRPLASANSCLDPVLYFLAGQRLVRFARDAKPPTGPSPATPARRRLGLRRSDRTDMQRIEDVLGSSEDSRRTESTPAGSENTKDIRL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.26 | 0.25 | 0.85 | 2.83 | Download | EAQAAAEQLKTTRNAYIQKYLGSPGARSSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.37 | 0.33 | 0.80 | 4.21 | Download | -------------------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVKNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYEEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR---------------------------------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.20 | 0.19 | 0.78 | 3.76 | Download | ---------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRM-----KEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.17 | 0.21 | 0.93 | 1.52 | Download | ---DLRDNETWWYNPSIIVHPWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVFPLMTISCF-LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGYTLEGVLCNCSFDYISRD-STTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEW-------VTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKET---------EDDK--D-AET-EIPAGESSDAAPSADAAQMKE | |||||||||||||||||||
| 5 | 4xnv | 0.38 | 0.32 | 0.78 | 1.17 | Download | ---------------------SFKCATKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYTEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.27 | 0.24 | 0.79 | 3.29 | Download | ----------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.18 | 0.21 | 0.76 | 1.66 | Download | -----------------------------DAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYLEGVLCNCSFDYISRDS-TTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVT-------PYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVL---------------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.20 | 0.19 | 0.77 | 4.81 | Download | ---------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE---------KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.20 | 0.19 | 0.78 | 3.36 | Download | ---------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLR-----MKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.29 | 0.25 | 0.78 | 4.11 | Download | -----------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKAL-----ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
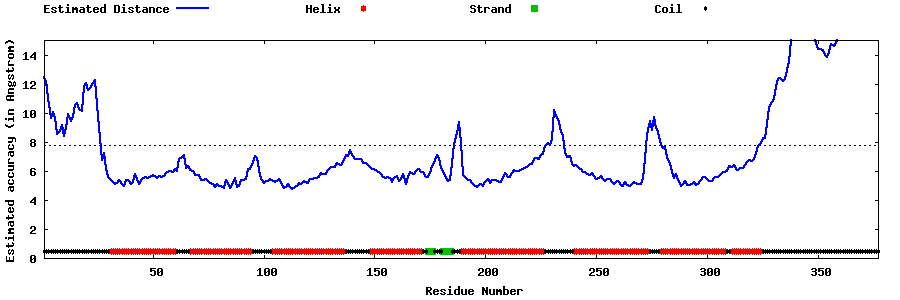
| Generated 3D models | Estimated local accuracy of models | ||
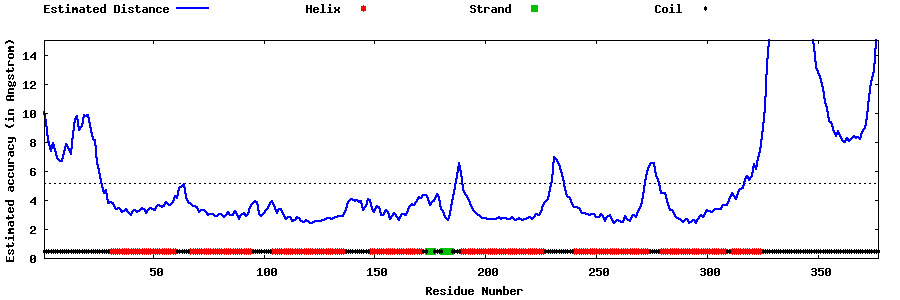 | |||
|
|
|||
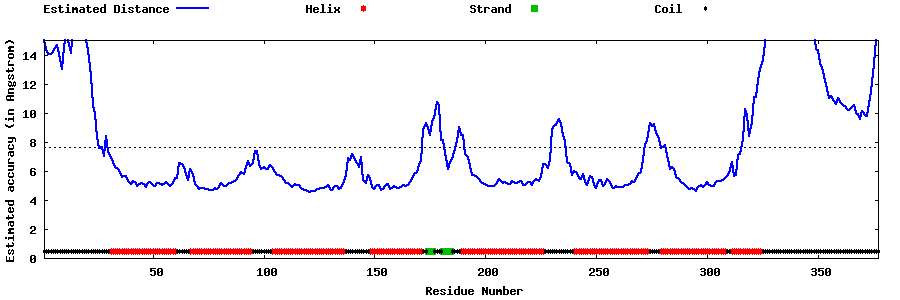 | |||
|
| |||
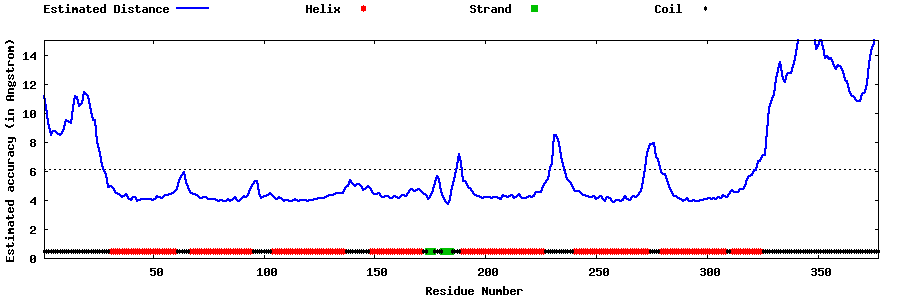 | |||
|
| |||
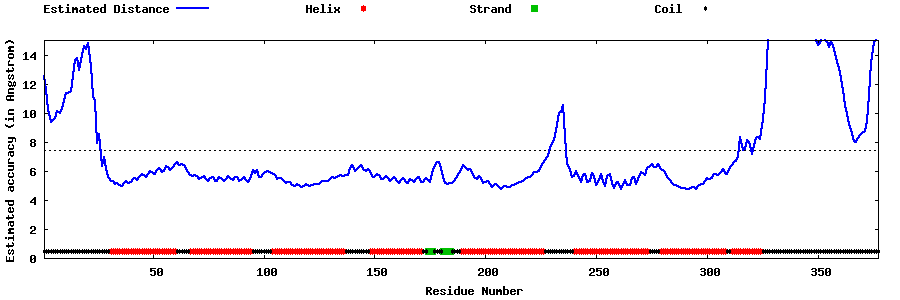 | |||
|
| |||
 | |||
|
| |||