

You can:
| Name | Somatostatin receptor type 3 |
|---|---|
| Species | Mus musculus (Mouse) |
| Gene | Sstr3 |
| Synonym | SSR-28 SS3R SS3-R SS-3-R SRIF1C [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 428 |
| Amino acid sequence | MATVTYPSSEPTTLDPGNASSTWPLDTTLGNTSAGASLTGLAVSGILISLVYLVVCVVGLLGNSLVIYVVLRHTSSPSVTSVYILNLALADELFMLGLPFLAAQNALSYWPFGSLMCRLVMAVDGINQFTSIFCLTVMSVDRYLAVVHPTRSARWRTAPVARTVSAAVWVASAVVVLPVVVFSGVPRGMSTCHMQWPEPAAAWRTAFIIYTAALGFFGPLLVICLCYLLIVVKVRSTTRRVRAPSCQWVQAPACQRRRRSERRVTRMVVAVVALFVLCWMPFYLLNIVNVVCPLPEEPAFFGLYFLVVALPYANSCANPILYGFLSYRFKQGFRRILLRPSRRIRSQEPGSGPPEKTEEEEDEEEEERREEEERRMQRGQEMNGRLSQIAQAGTSGQQPRPCTGTAKEQQLLPQEATAGDKASTLSHL |
| UniProt | P30935 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2238 |
| IUPHAR | 357 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 16299 | 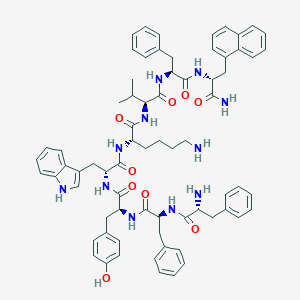 BDBM84629 BDBM84629 | C71H81N11O9 | 1232.5 | 11 / 12 | 8.1 | No |
| 19011 | 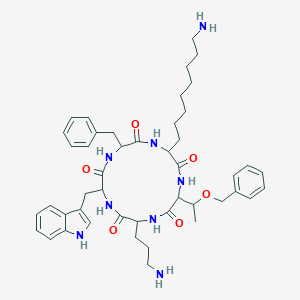 SA V SA V | C46H62N8O6 | 823.052 | 8 / 8 | 4.6 | No |
| 28610 | 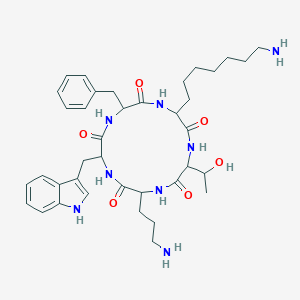 SA II SA II | C38H54N8O6 | 718.9 | 8 / 9 | 2.6 | No |
| 39292 | 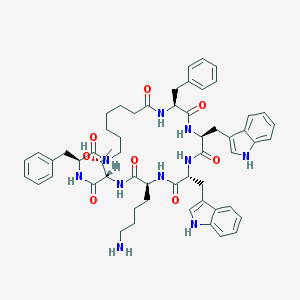 L-362855 L-362855 | C57H70N10O8 | 1023.25 | 9 / 11 | 5.7 | No |
| 46044 | 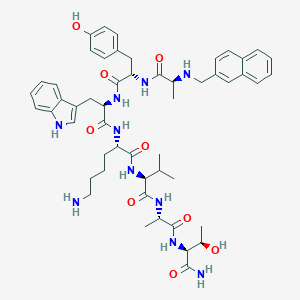 BDBM82458 BDBM82458 | C52H68N10O9 | 977.177 | 11 / 12 | 3.4 | No |
| 50022 | 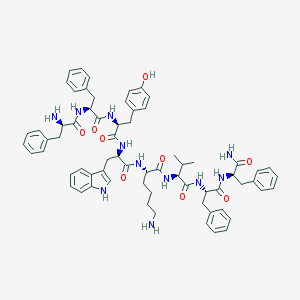 CHEMBL268112 CHEMBL268112 | C67H79N11O9 | 1182.44 | 11 / 12 | 5.6 | No |
| 52236 | 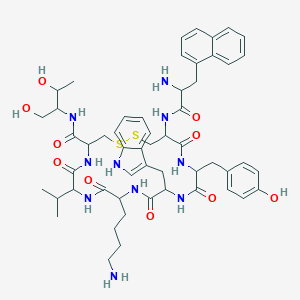 BDBM84620 BDBM84620 | C54H70N10O10S2 | 1083.33 | 14 / 13 | 2.9 | No |
| 443976 | 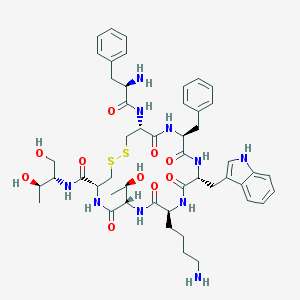 CHEMBL3350037 CHEMBL3350037 | C49H66N10O10S2 | 1019.25 | 14 / 13 | 1.0 | No |
| 58025 | 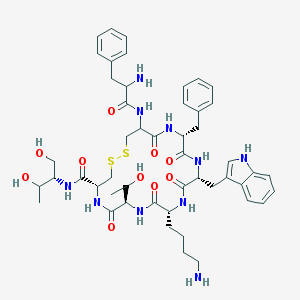 Octreotide Octreotide | C49H66N10O10S2 | 1019.25 | 14 / 13 | 1.0 | No |
| 58032 | 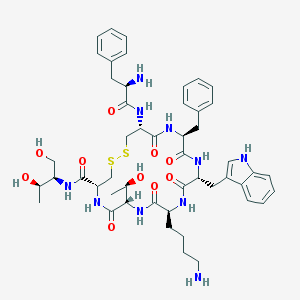 Octreotide Octreotide | C49H66N10O10S2 | 1019.25 | 14 / 13 | 1.0 | No |
| 553529 | 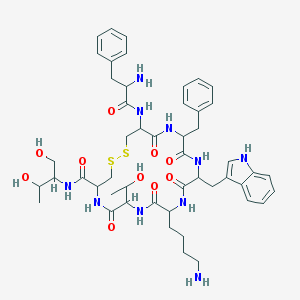 Octreotide Octreotide | C49H66N10O10S2 | 1019.25 | 14 / 13 | 1.0 | No |
| 59018 | 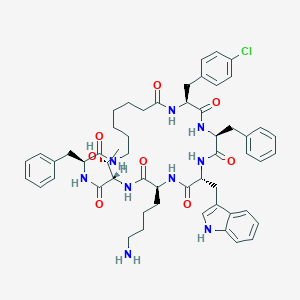 BDBM82466 BDBM82466 | C55H68ClN9O8 | 1018.65 | 9 / 10 | 6.2 | No |
| 61295 | 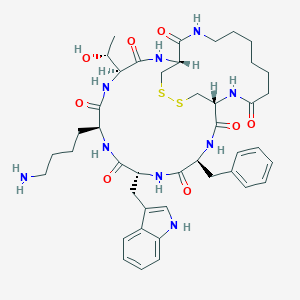 CHEMBL2369699 CHEMBL2369699 | C43H59N9O8S2 | 894.12 | 11 / 10 | 2.1 | No |
| 65482 | 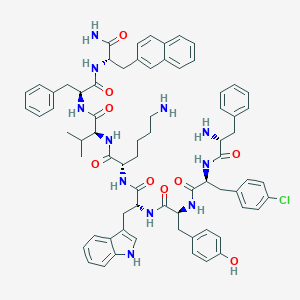 BDBM84640 BDBM84640 | C71H80ClN11O9 | 1266.94 | 11 / 12 | 8.8 | No |
| 68362 | 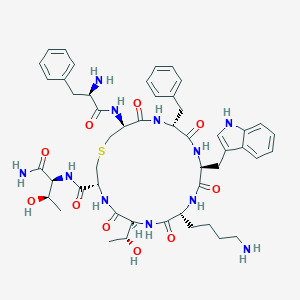 CHEMBL436783 CHEMBL436783 | C49H65N11O10S | 1000.19 | 13 / 13 | 0.5 | No |
| 78636 | 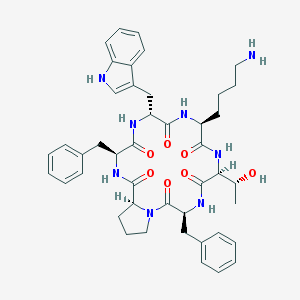 CHEMBL421362 CHEMBL421362 | C44H54N8O7 | 806.965 | 8 / 8 | 3.4 | No |
| 85792 | 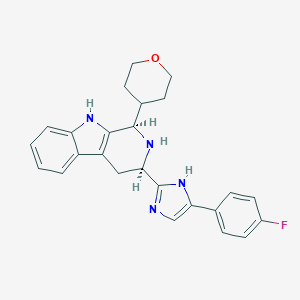 CHEMBL2204941 CHEMBL2204941 | C25H25FN4O | 416.5 | 4 / 3 | 3.6 | Yes |
| 93068 | 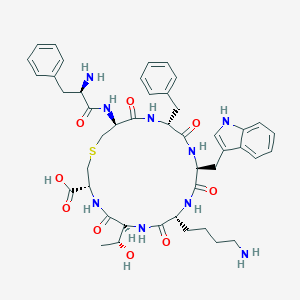 CHEMBL69379 CHEMBL69379 | C45H57N9O9S | 900.065 | 12 / 11 | -0.2 | No |
| 96176 | 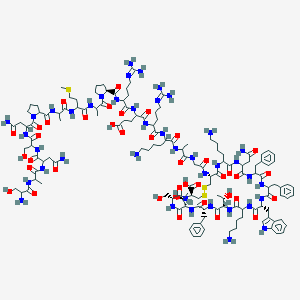 BDBM82460 BDBM82460 | C137H207N41O39S3 | 3148.59 | 48 / 44 | -16.0 | No |
| 96186 | 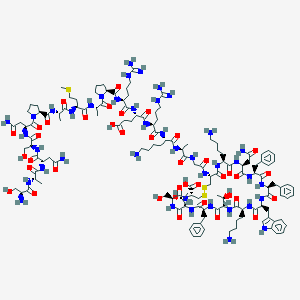 Somatostatin 28 Somatostatin 28 | C137H207N41O39S3 | 3148.59 | 48 / 46 | -15.2 | No |
| 106549 | 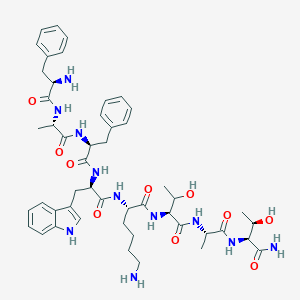 BDBM84625 BDBM84625 | C49H67N11O10 | 970.142 | 12 / 13 | -0.5 | No |
| 106954 | 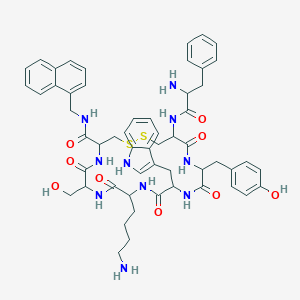 BDBM84623 BDBM84623 | C55H64N10O9S2 | 1073.3 | 13 / 12 | 3.8 | No |
| 112369 | 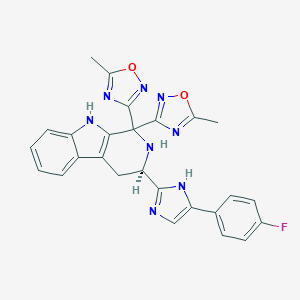 CHEMBL2204940 CHEMBL2204940 | C26H21FN8O2 | 496.506 | 9 / 3 | 3.4 | Yes |
| 116986 | 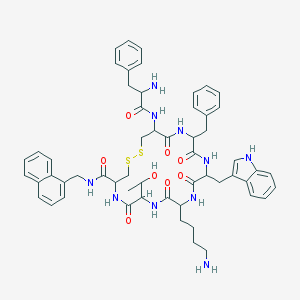 BDBM84628 BDBM84628 | C56H66N10O8S2 | 1071.33 | 12 / 11 | 4.6 | No |
| 118924 | 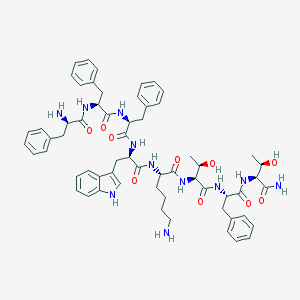 BIM 23052 BIM 23052 | C61H75N11O10 | 1122.34 | 12 / 13 | 2.7 | No |
| 120863 | 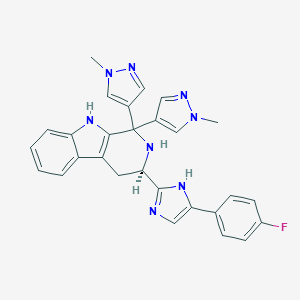 CHEMBL2204939 CHEMBL2204939 | C28H25FN8 | 492.562 | 5 / 3 | 2.8 | Yes |
| 123394 | 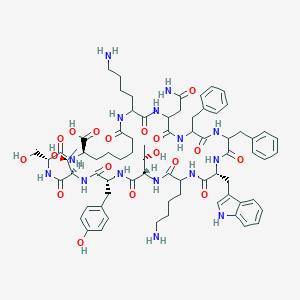 BDBM82467 BDBM82467 | C73H99N15O18 | 1474.68 | 20 / 20 | -0.8 | No |
| 128026 | 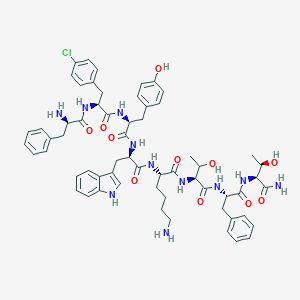 BDBM84624 BDBM84624 | C61H74ClN11O11 | 1172.78 | 13 / 14 | 3.7 | No |
| 148745 | 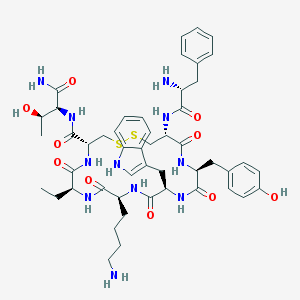 BIM-23023 BIM-23023 | C49H65N11O10S2 | 1032.25 | 14 / 13 | 0.8 | No |
| 160208 | 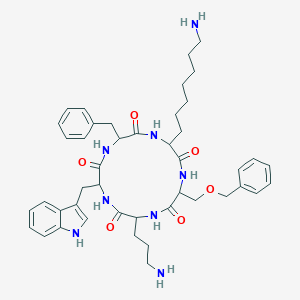 BDBM84644 BDBM84644 | C44H58N8O6 | 794.998 | 8 / 8 | 3.6 | No |
| 166170 | 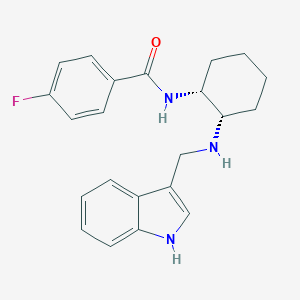 CHEMBL3287613 CHEMBL3287613 | C22H24FN3O | 365.452 | 3 / 3 | 3.8 | Yes |
| 166627 | 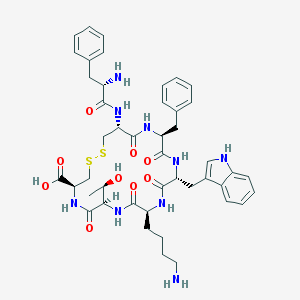 CHEMBL421493 CHEMBL421493 | C45H57N9O9S2 | 932.125 | 13 / 11 | -0.2 | No |
| 174018 | 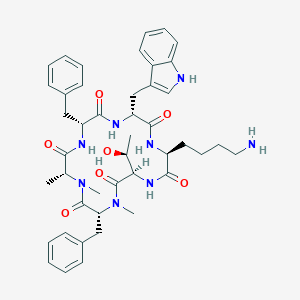 BDBM82454 BDBM82454 | C44H56N8O7 | 808.981 | 8 / 7 | 3.4 | No |
| 174353 | 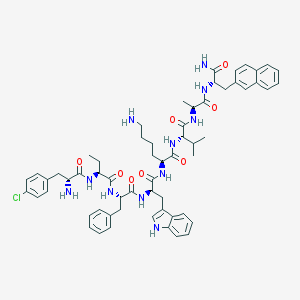 BDBM84617 BDBM84617 | C60H74ClN11O8 | 1112.77 | 10 / 11 | 6.5 | No |
| 183481 | 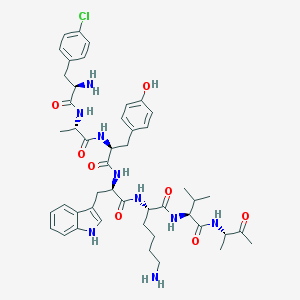 BDBM84616 BDBM84616 | C47H62ClN9O8 | 916.518 | 10 / 10 | 2.5 | No |
| 448942 | 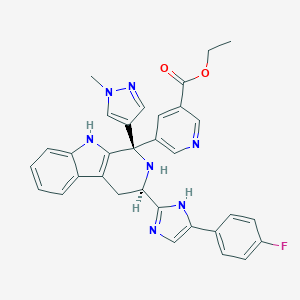 CHEMBL3323076 CHEMBL3323076 | C32H28FN7O2 | 561.621 | 7 / 3 | 3.6 | No |
| 448944 | 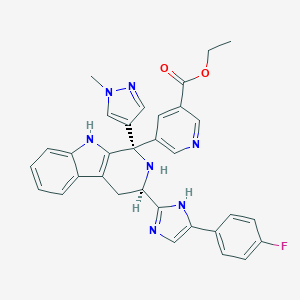 CHEMBL3323077 CHEMBL3323077 | C32H28FN7O2 | 561.621 | 7 / 3 | 3.6 | No |
| 186165 | 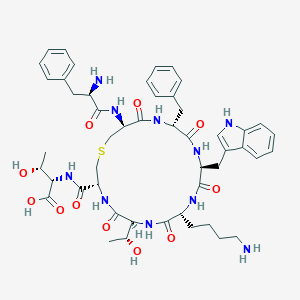 CHEMBL266469 CHEMBL266469 | C49H64N10O11S | 1001.17 | 14 / 13 | -1.1 | No |
| 200931 | 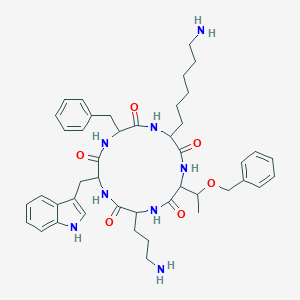 SA IV SA IV | C44H58N8O6 | 794.998 | 8 / 8 | 3.5 | No |
| 211331 | 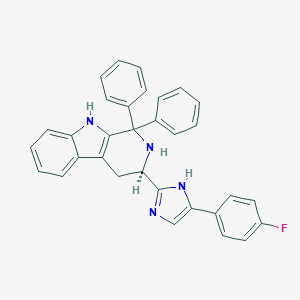 CHEMBL2204934 CHEMBL2204934 | C32H25FN4 | 484.578 | 3 / 3 | 6.1 | No |
| 223708 | 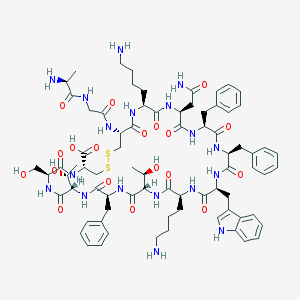 SOMATOSTATIN SOMATOSTATIN | C76H104N18O19S2 | 1637.9 | 24 / 22 | -3.1 | No |
| 223726 | 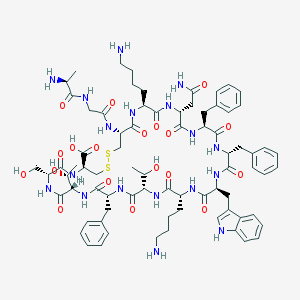 SOMATOSTATIN SOMATOSTATIN | C76H104N18O19S2 | 1637.9 | 24 / 22 | -3.1 | No |
| 554414 | 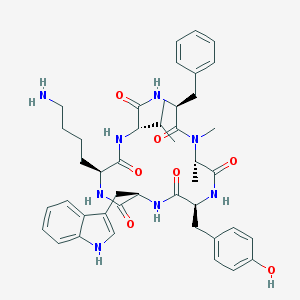 seglitide seglitide | C44H56N8O7 | 808.981 | 8 / 8 | 3.9 | No |
| 228996 | 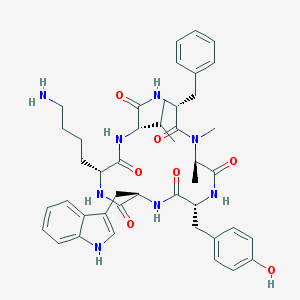 CHEMBL413535 CHEMBL413535 | C44H56N8O7 | 808.981 | 8 / 8 | 3.9 | No |
| 229006 | 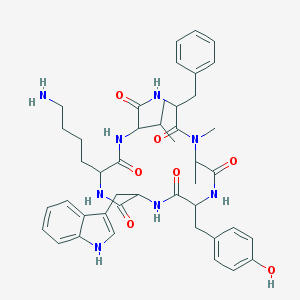 BDBM81766 BDBM81766 | C44H56N8O7 | 808.981 | 8 / 8 | 3.9 | No |
| 231524 | 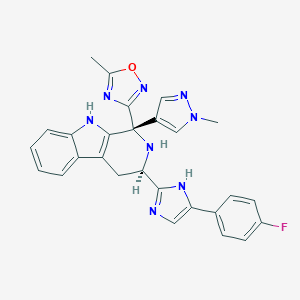 MK-4256 MK-4256 | C27H23FN8O | 494.534 | 7 / 3 | 3.1 | Yes |
| 231528 | 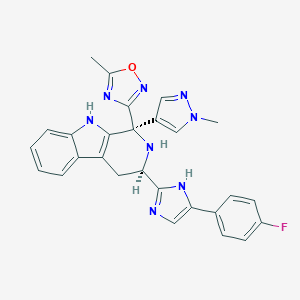 CHEMBL2204936 CHEMBL2204936 | C27H23FN8O | 494.534 | 7 / 3 | 3.1 | Yes |
| 231530 | 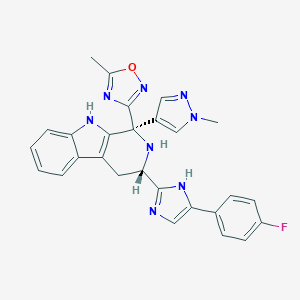 CHEMBL2204937 CHEMBL2204937 | C27H23FN8O | 494.534 | 7 / 3 | 3.1 | Yes |
| 231533 | 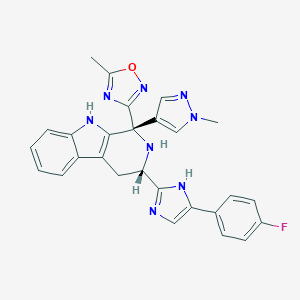 CHEMBL2204938 CHEMBL2204938 | C27H23FN8O | 494.534 | 7 / 3 | 3.1 | Yes |
| 259590 | 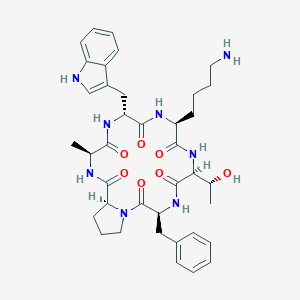 CHEMBL3349593 CHEMBL3349593 | C38H50N8O7 | 730.867 | 8 / 8 | 1.8 | No |
| 261031 | 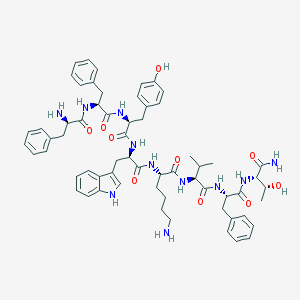 BIM-23058 BIM-23058 | C62H77N11O10 | 1136.36 | 12 / 13 | 4.7 | No |
| 268941 | 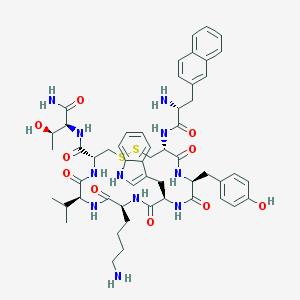 Lanreotide Lanreotide | C54H69N11O10S2 | 1096.33 | 14 / 13 | 2.5 | No |
| 554609 | 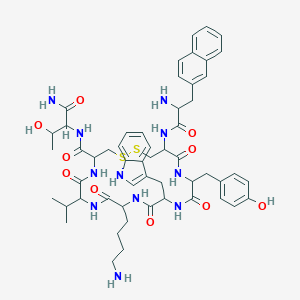 Lanreotide acetate Lanreotide acetate | C54H69N11O10S2 | 1096.33 | 14 / 13 | 2.5 | No |
| 276243 | 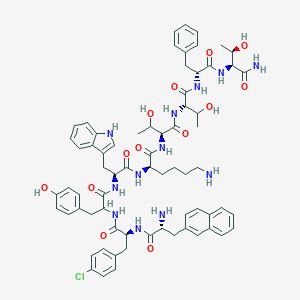 BDBM84619 BDBM84619 | C69H83ClN12O13 | 1323.94 | 15 / 16 | 4.1 | No |
| 293362 | 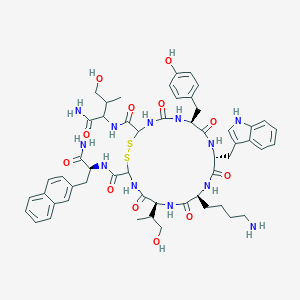 BDBM82462 BDBM82462 | C54H68N12O12S2 | 1141.33 | 15 / 15 | 1.5 | No |
| 499334 | 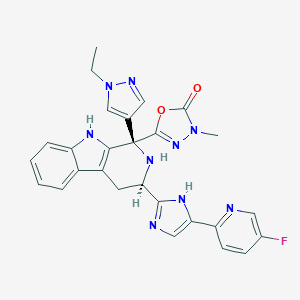 CHEMBL3582336 CHEMBL3582336 | C27H24FN9O2 | 525.548 | 8 / 3 | 2.0 | No |
| 300105 | 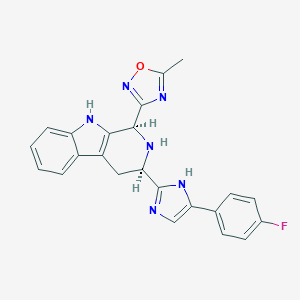 CHEMBL2204932 CHEMBL2204932 | C23H19FN6O | 414.444 | 6 / 3 | 3.3 | Yes |
| 300106 | 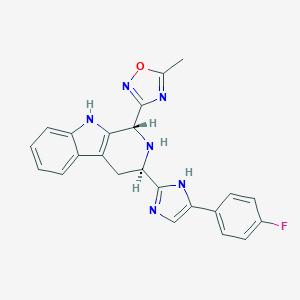 CHEMBL2204933 CHEMBL2204933 | C23H19FN6O | 414.444 | 6 / 3 | 3.3 | Yes |
| 310854 |  CHEMBL1076624 CHEMBL1076624 | C34H44F2N4O6 | 642.745 | 11 / 2 | N/A | No |
| 454098 | 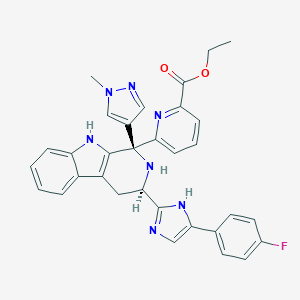 CHEMBL3323072 CHEMBL3323072 | C32H28FN7O2 | 561.621 | 7 / 3 | 4.0 | No |
| 454100 |  CHEMBL3323073 CHEMBL3323073 | C32H28FN7O2 | 561.621 | 7 / 3 | 4.0 | No |
| 454255 |  CHEMBL3323081 CHEMBL3323081 | C30H24FN7O2 | 533.567 | 7 / 4 | 1.0 | No |
| 454258 |  CHEMBL3323080 CHEMBL3323080 | C30H24FN7O2 | 533.567 | 7 / 4 | 1.0 | No |
| 316659 |  BIM-23027 BIM-23027 | C43H54N8O7 | 794.954 | 8 / 8 | 3.5 | No |
| 454594 |  CHEMBL3323084 CHEMBL3323084 | C30H24FN7O2 | 533.567 | 7 / 4 | 0.6 | No |
| 454596 |  CHEMBL3323085 CHEMBL3323085 | C30H24FN7O2 | 533.567 | 7 / 4 | 0.6 | No |
| 325274 |  Vapreotide Vapreotide | C57H70N12O9S2 | 1131.38 | 13 / 13 | 3.6 | No |
| 332963 |  BDBM82461 BDBM82461 | C59H73N11O9 | 1080.3 | 11 / 12 | 5.0 | No |
| 333293 | 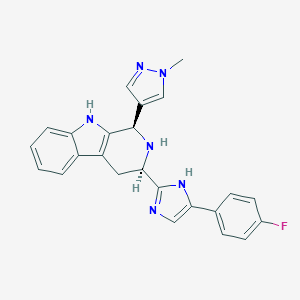 CHEMBL2204942 CHEMBL2204942 | C24H21FN6 | 412.472 | 4 / 3 | 3.0 | Yes |
| 333295 | 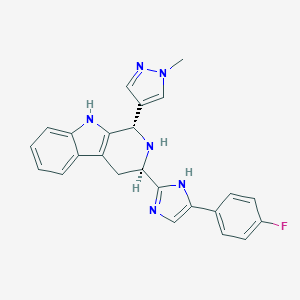 CHEMBL2204931 CHEMBL2204931 | C24H21FN6 | 412.472 | 4 / 3 | 3.0 | Yes |
| 455090 | 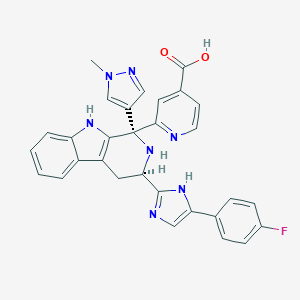 CHEMBL3323083 CHEMBL3323083 | C30H24FN7O2 | 533.567 | 7 / 4 | 0.7 | No |
| 455091 | 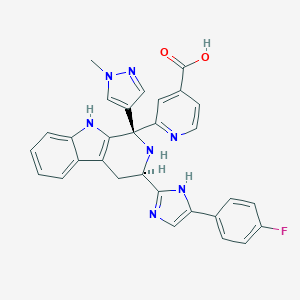 CHEMBL3323082 CHEMBL3323082 | C30H24FN7O2 | 533.567 | 7 / 4 | 0.7 | No |
| 455395 | 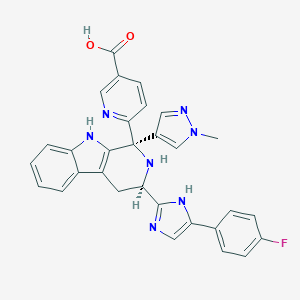 CHEMBL3323087 CHEMBL3323087 | C30H24FN7O2 | 533.567 | 7 / 4 | 0.7 | No |
| 455397 | 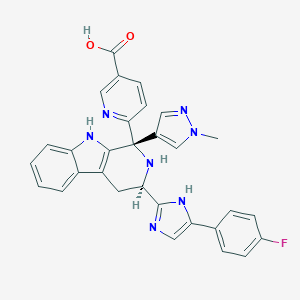 CHEMBL3323086 CHEMBL3323086 | C30H24FN7O2 | 533.567 | 7 / 4 | 0.7 | No |
| 345534 | 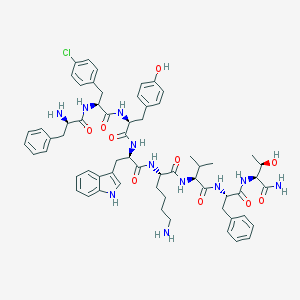 SCHEMBL1579410 SCHEMBL1579410 | C62H76ClN11O10 | 1170.81 | 12 / 13 | 5.3 | No |
| 345925 | 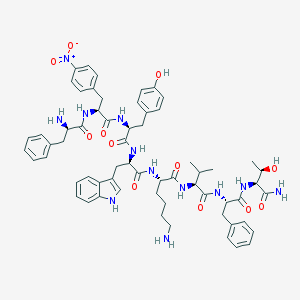 CHEMBL3350185 CHEMBL3350185 | C62H76N12O12 | 1181.36 | 14 / 13 | 4.5 | No |
| 346566 | 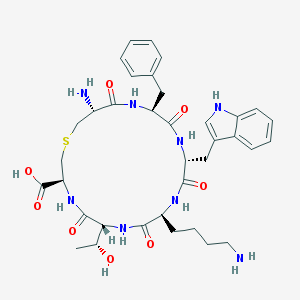 CHEMBL306702 CHEMBL306702 | C36H48N8O8S | 752.888 | 11 / 10 | -1.6 | No |
| 348761 | 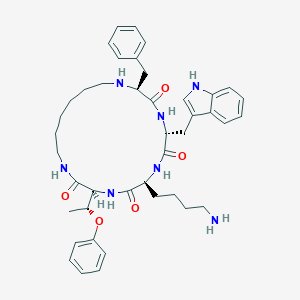 cycloantagonist SA cycloantagonist SA | C42H55N7O5 | 737.946 | 7 / 7 | 4.8 | No |
| 357453 | 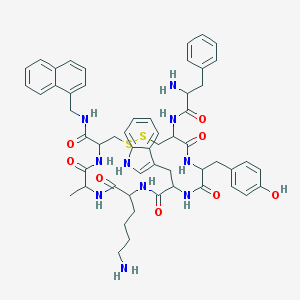 BDBM84626 BDBM84626 | C55H64N10O8S2 | 1057.3 | 12 / 11 | 4.3 | No |
| 361204 | 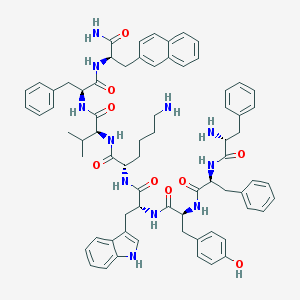 BIM 23056 BIM 23056 | C71H81N11O9 | 1232.5 | 11 / 12 | 8.1 | No |
| 361647 |  BDBM84621 BDBM84621 | C61H69N11O9S2 | 1164.41 | 13 / 12 | 6.1 | No |
| 555097 |  SS14 SS14 | C63H88N16O16S2 | 1389.61 | 21 / 19 | -4.1 | No |
| 381824 |  CHEMBL2069502 CHEMBL2069502 | C25H26N4O | 398.51 | 3 / 3 | 3.5 | Yes |
| 383355 |  BDBM84630 BDBM84630 | C56H66N10O9S2 | 1087.32 | 13 / 12 | 4.2 | No |
| 390656 | 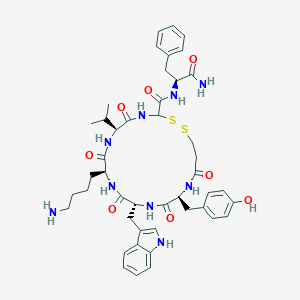 BDBM82456 BDBM82456 | C45H57N9O8S2 | 916.126 | 11 / 10 | 3.0 | No |
| 394982 | 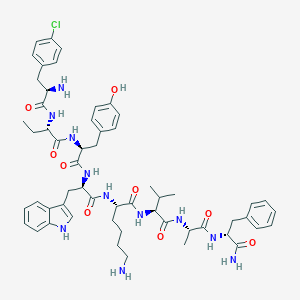 BDBM84627 BDBM84627 | C56H72ClN11O9 | 1078.71 | 11 / 12 | 4.2 | No |
| 400289 | 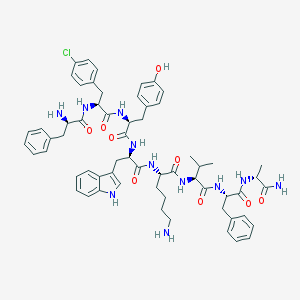 CHEMBL264562 CHEMBL264562 | C61H74ClN11O9 | 1140.78 | 11 / 12 | 5.1 | No |
| 457597 | 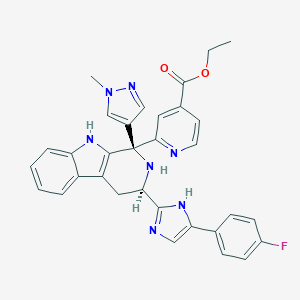 CHEMBL3323074 CHEMBL3323074 | C32H28FN7O2 | 561.621 | 7 / 3 | 3.6 | No |
| 457599 | 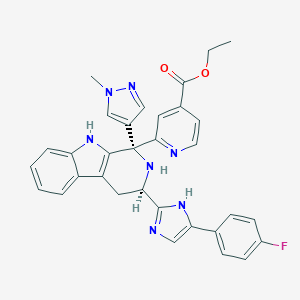 CHEMBL3323075 CHEMBL3323075 | C32H28FN7O2 | 561.621 | 7 / 3 | 3.6 | No |
| 457963 | 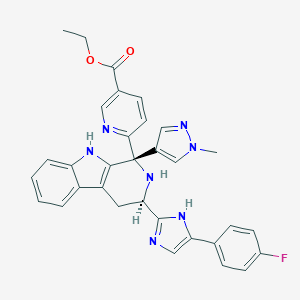 CHEMBL3323078 CHEMBL3323078 | C32H28FN7O2 | 561.621 | 7 / 3 | 3.6 | No |
| 457965 | 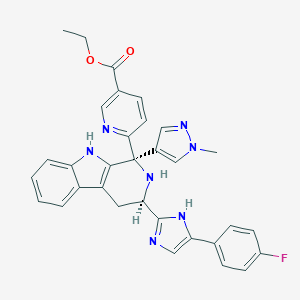 CHEMBL3323079 CHEMBL3323079 | C32H28FN7O2 | 561.621 | 7 / 3 | 3.6 | No |
| 416683 | 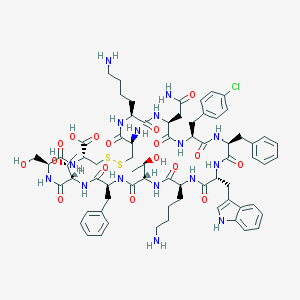 Bim-23003 Bim-23003 | C71H95ClN16O17S2 | 1544.21 | 22 / 20 | -1.6 | No |
| 435787 |  BDBM84638 BDBM84638 | C58H71N11O10 | 1082.27 | 12 / 13 | 3.4 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417