

You can:
| Name | Lysophosphatidic acid receptor 3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | LPAR3 |
| Synonym | Lysophosphatidic acid receptor Edg-7 LPA3 receptor LPA-3 LPA receptor 3 endothelial differentiation gene 7, lysophosphatidic acid G-protein-coupled receptor 7 [ Show all ] |
| Disease | Fibrosis |
| Length | 353 |
| Amino acid sequence | MNECHYDKHMDFFYNRSNTDTVDDWTGTKLVIVLCVGTFFCLFIFFSNSLVIAAVIKNRKFHFPFYYLLANLAAADFFAGIAYVFLMFNTGPVSKTLTVNRWFLRQGLLDSSLTASLTNLLVIAVERHMSIMRMRVHSNLTKKRVTLLILLVWAIAIFMGAVPTLGWNCLCNISACSSLAPIYSRSYLVFWTVSNLMAFLIMVVVYLRIYVYVKRKTNVLSPHTSGSISRRRTPMKLMKTVMTVLGAFVVCWTPGLVVLLLDGLNCRQCGVQHVKRWFLLLALLNSVVNPIIYSYKDEDMYGTMKKMICCFSQENPERRPSRIPSTVLSRSDTGSQYIEDSISQGAVCNKSTS |
| UniProt | Q9UBY5 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9UBY5 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9UBY5. |
| BioLiP | N/A |
| Therapeutic Target Database | T95923 |
| ChEMBL | CHEMBL3250 |
| IUPHAR | 274 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 3425 | 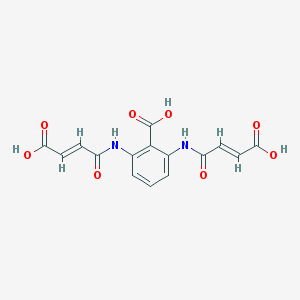 CHEMBL485119 CHEMBL485119 | C15H12N2O8 | 348.267 | 8 / 5 | -0.1 | Yes |
| 553284 | 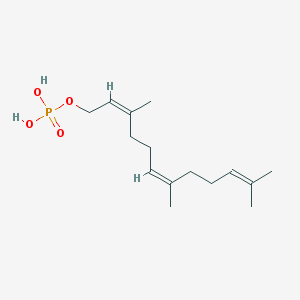 Farnesyl monophosphate Farnesyl monophosphate | C15H27O4P | 302.351 | 4 / 2 | 3.7 | Yes |
| 17213 | 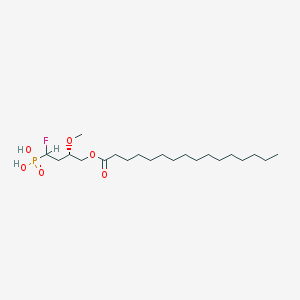 CHEMBL193196 CHEMBL193196 | C21H42FO6P | 440.533 | 7 / 2 | 6.4 | No |
| 17532 | 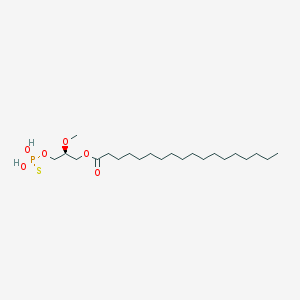 CHEMBL357053 CHEMBL357053 | C22H45O6PS | 468.63 | 7 / 2 | 8.4 | No |
| 17541 | 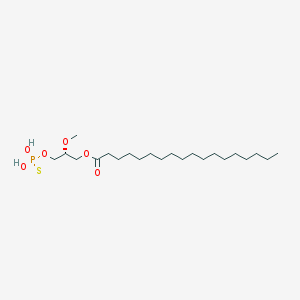 CHEMBL153043 CHEMBL153043 | C22H45O6PS | 468.63 | 7 / 2 | 8.4 | No |
| 17546 | 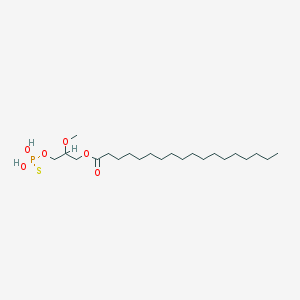 CHEMBL2017139 CHEMBL2017139 | C22H45O6PS | 468.63 | 7 / 2 | 8.4 | No |
| 20485 | 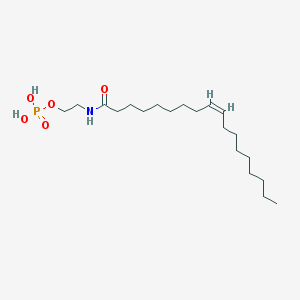 NAEPA NAEPA | C20H40NO5P | 405.516 | 5 / 3 | 5.2 | No |
| 25663 | 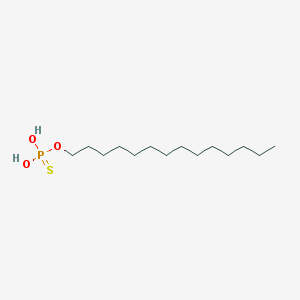 CHEMBL189296 CHEMBL189296 | C14H31O3PS | 310.433 | 4 / 2 | 6.8 | No |
| 25976 | 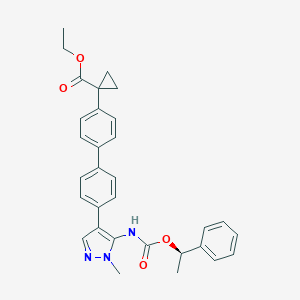 CHEMBL2182062 CHEMBL2182062 | C31H31N3O4 | 509.606 | 5 / 1 | 5.8 | No |
| 29256 | 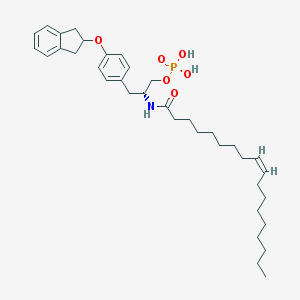 CHEMBL313413 CHEMBL313413 | C36H54NO6P | 627.803 | 6 / 3 | 9.3 | No |
| 29833 | 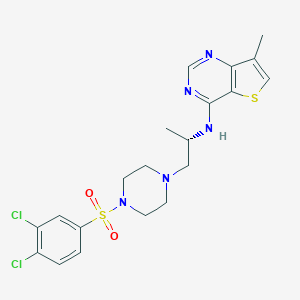 LPA2 antagonist 1 LPA2 antagonist 1 | C20H23Cl2N5O2S2 | 500.457 | 8 / 1 | 4.3 | No |
| 442851 | 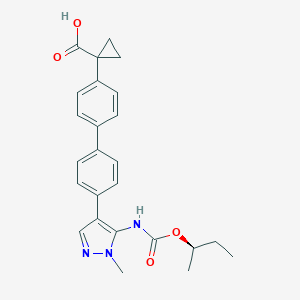 CHEMBL2182024 CHEMBL2182024 | C25H27N3O4 | 433.508 | 5 / 2 | 4.5 | Yes |
| 33971 | 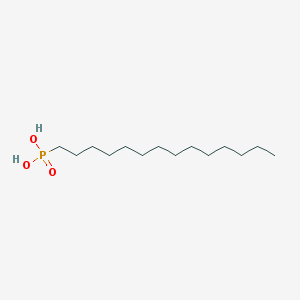 Tetradecylphosphonic acid Tetradecylphosphonic acid | C14H31O3P | 278.373 | 3 / 2 | 5.1 | No |
| 34950 | 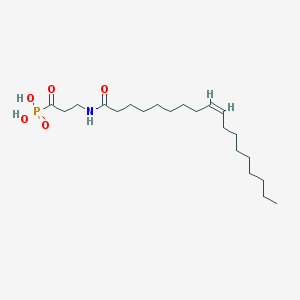 CHEMBL117529 CHEMBL117529 | C21H40NO5P | 417.527 | 5 / 3 | 4.9 | Yes |
| 43194 | 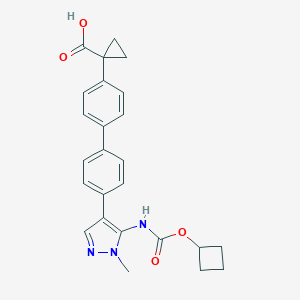 CHEMBL2182026 CHEMBL2182026 | C25H25N3O4 | 431.492 | 5 / 2 | 4.1 | Yes |
| 47586 | 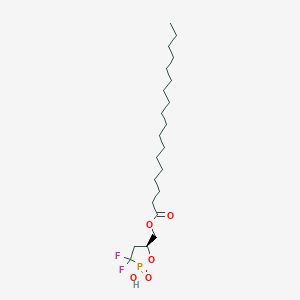 CHEMBL215722 CHEMBL215722 | C22H41F2O5P | 454.536 | 7 / 1 | 8.2 | No |
| 49425 | 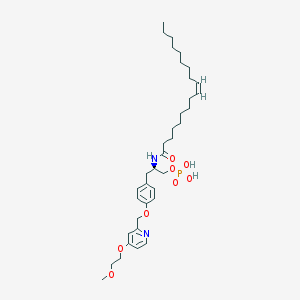 CHEMBL182446 CHEMBL182446 | C36H57N2O8P | 676.832 | 9 / 3 | 7.5 | No |
| 49613 | 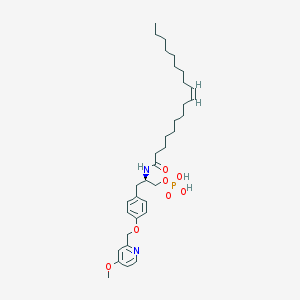 CHEMBL265967 CHEMBL265967 | C34H53N2O7P | 632.779 | 8 / 3 | 7.7 | No |
| 50272 | 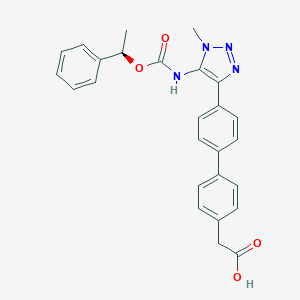 CHEMBL2182042 CHEMBL2182042 | C26H24N4O4 | 456.502 | 6 / 2 | 4.2 | Yes |
| 51245 | 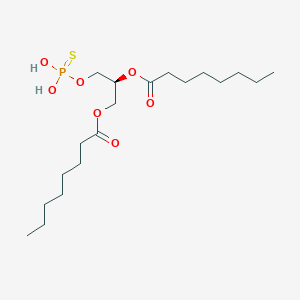 CHEMBL202242 CHEMBL202242 | C19H37O7PS | 440.532 | 8 / 2 | 6.0 | No |
| 51248 | 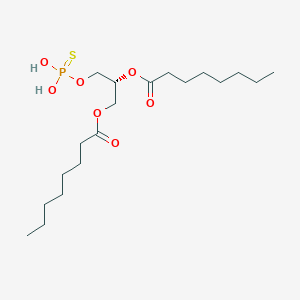 CHEMBL202361 CHEMBL202361 | C19H37O7PS | 440.532 | 8 / 2 | 6.0 | No |
| 537349 | 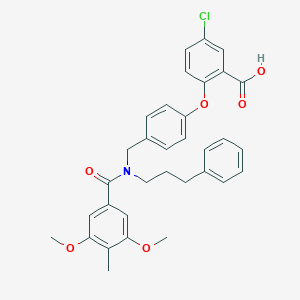 CHEMBL3975893 CHEMBL3975893 | C33H32ClNO6 | 574.07 | 6 / 1 | 7.2 | No |
| 57810 | 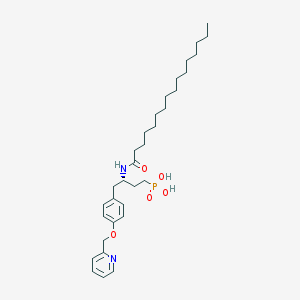 CHEMBL360064 CHEMBL360064 | C32H51N2O5P | 574.743 | 6 / 3 | 7.9 | No |
| 60442 | 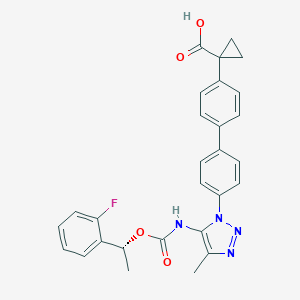 CHEMBL2182030 CHEMBL2182030 | C28H25FN4O4 | 500.53 | 7 / 2 | 5.1 | No |
| 65690 | 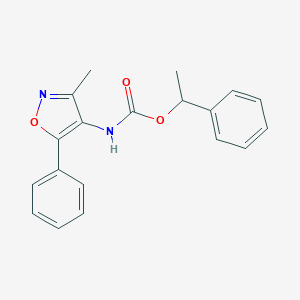 CHEMBL404575 CHEMBL404575 | C19H18N2O3 | 322.364 | 4 / 1 | 3.9 | Yes |
| 67556 | 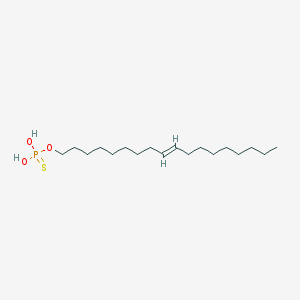 CHEMBL187459 CHEMBL187459 | C18H37O3PS | 364.525 | 4 / 2 | 8.1 | No |
| 71930 | 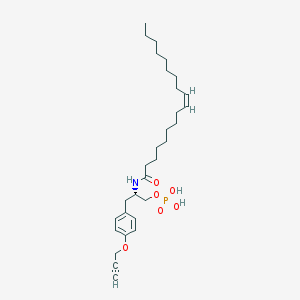 CHEMBL328474 CHEMBL328474 | C30H48NO6P | 549.689 | 6 / 3 | 7.3 | No |
| 72937 | 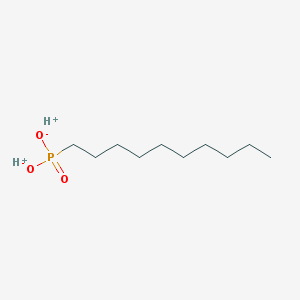 SCHEMBL8438208 SCHEMBL8438208 | C10H23O3P | 222.265 | 3 / 2 | N/A | N/A |
| 75268 | 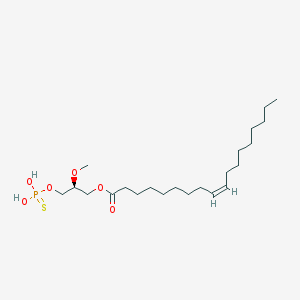 (2S)-OMPT (2S)-OMPT | C22H43O6PS | 466.614 | 7 / 2 | 7.5 | No |
| 472301 | 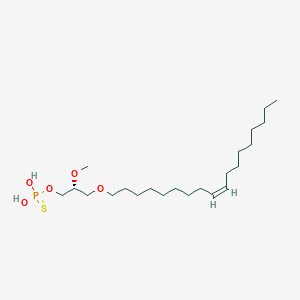 CHEMBL3621961 CHEMBL3621961 | C22H45O5PS | 452.631 | 6 / 2 | 7.8 | No |
| 76230 | 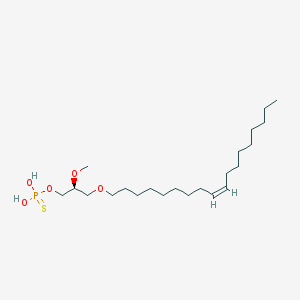 alkyl OMPT alkyl OMPT | C22H45O5PS | 452.631 | 6 / 2 | 7.8 | No |
| 77231 | 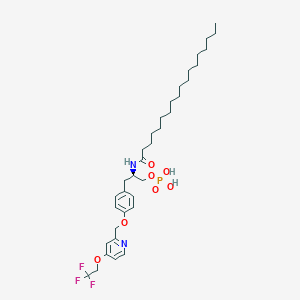 CHEMBL3218459 CHEMBL3218459 | C35H54F3N2O7P | 702.793 | 11 / 3 | 9.7 | No |
| 77519 | 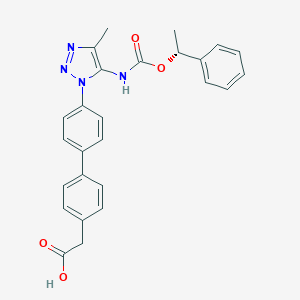 CHEMBL2182028 CHEMBL2182028 | C26H24N4O4 | 456.502 | 6 / 2 | 4.6 | Yes |
| 79843 | 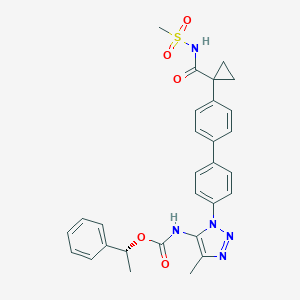 CHEMBL2182037 CHEMBL2182037 | C29H29N5O5S | 559.641 | 7 / 2 | 4.3 | No |
| 79956 | 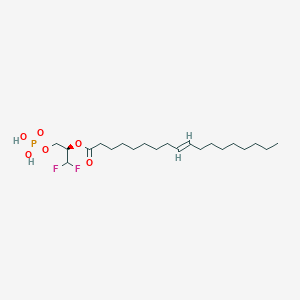 CHEMBL192861 CHEMBL192861 | C21H39F2O6P | 456.508 | 8 / 2 | 6.7 | No |
| 82533 | 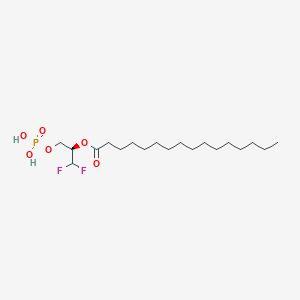 CHEMBL195383 CHEMBL195383 | C19H37F2O6P | 430.47 | 8 / 2 | 6.6 | No |
| 86135 | 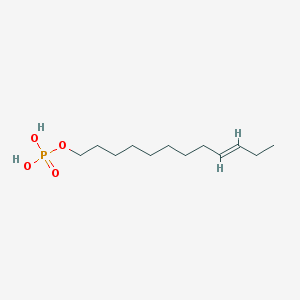 CHEMBL190349 CHEMBL190349 | C12H25O4P | 264.302 | 4 / 2 | 3.1 | Yes |
| 86327 | 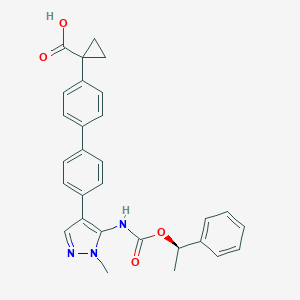 CHEMBL2182063 CHEMBL2182063 | C29H27N3O4 | 481.552 | 5 / 2 | 5.1 | No |
| 86863 | 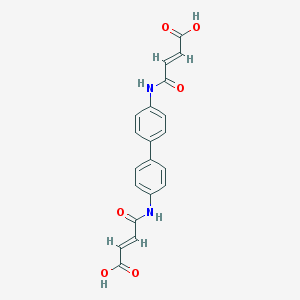 36840-10-5 36840-10-5 | C20H16N2O6 | 380.356 | 6 / 4 | 1.4 | Yes |
| 89072 | 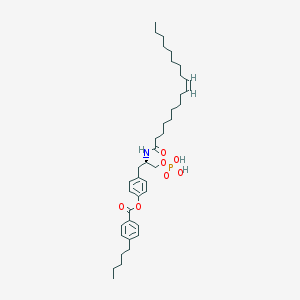 CHEMBL314553 CHEMBL314553 | C39H60NO7P | 685.883 | 7 / 3 | 11.1 | No |
| 89771 | 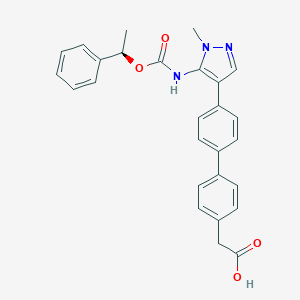 CHEMBL2182061 CHEMBL2182061 | C27H25N3O4 | 455.514 | 5 / 2 | 4.8 | Yes |
| 91233 | 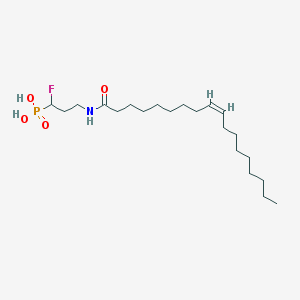 CHEMBL117663 CHEMBL117663 | C21H41FNO4P | 421.534 | 5 / 3 | 6.1 | No |
| 93763 | 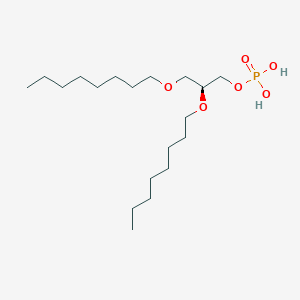 CHEMBL203986 CHEMBL203986 | C19H41O6P | 396.505 | 6 / 2 | 5.1 | No |
| 93765 | 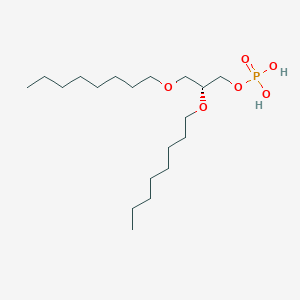 CHEMBL202243 CHEMBL202243 | C19H41O6P | 396.505 | 6 / 2 | 5.1 | No |
| 96493 | 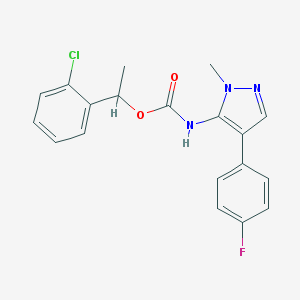 CHEMBL2182057 CHEMBL2182057 | C19H17ClFN3O2 | 373.812 | 4 / 1 | 4.4 | Yes |
| 96551 | 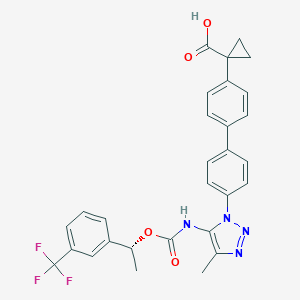 CHEMBL2182032 CHEMBL2182032 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.9 | No |
| 100155 | 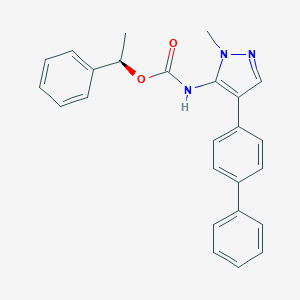 CHEMBL2182055 CHEMBL2182055 | C25H23N3O2 | 397.478 | 3 / 1 | 5.3 | No |
| 445645 | 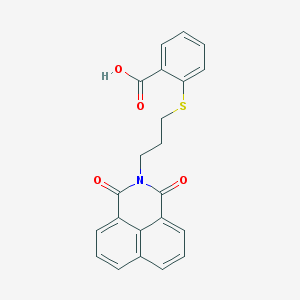 325850-81-5 325850-81-5 | C22H17NO4S | 391.441 | 5 / 1 | 4.2 | Yes |
| 102485 | 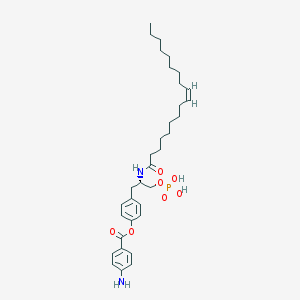 CHEMBL94245 CHEMBL94245 | C34H51N2O7P | 630.763 | 8 / 4 | 8.0 | No |
| 104953 | 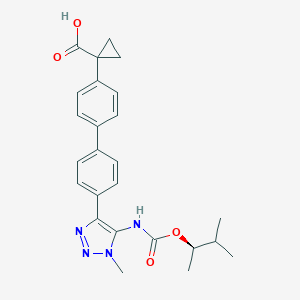 CHEMBL2182045 CHEMBL2182045 | C25H28N4O4 | 448.523 | 6 / 2 | 4.5 | Yes |
| 111766 | 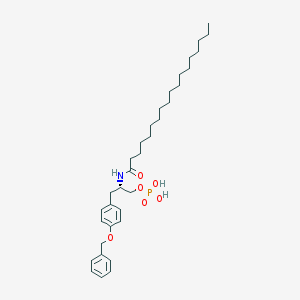 CHEMBL191411 CHEMBL191411 | C34H54NO6P | 603.781 | 6 / 3 | 9.7 | No |
| 113883 | 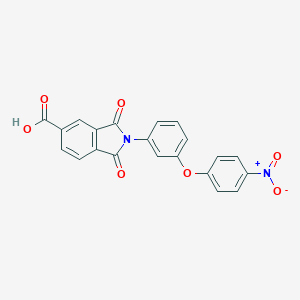 CHEMBL604677 CHEMBL604677 | C21H12N2O7 | 404.334 | 7 / 1 | 3.3 | Yes |
| 114528 | 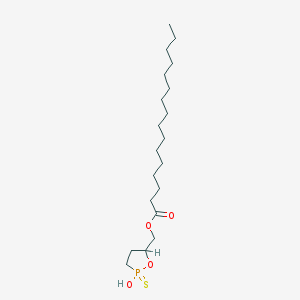 CHEMBL211465 CHEMBL211465 | C20H39O4PS | 406.562 | 5 / 1 | 8.1 | No |
| 117460 | 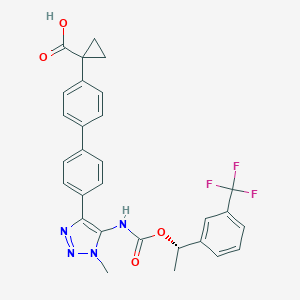 CHEMBL2182047 CHEMBL2182047 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.5 | No |
| 117463 | 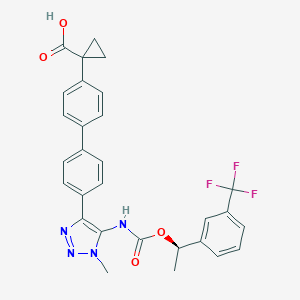 CHEMBL2182046 CHEMBL2182046 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.5 | No |
| 538875 | 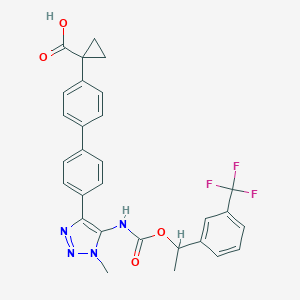 CHEMBL3969018 CHEMBL3969018 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.5 | No |
| 122948 | 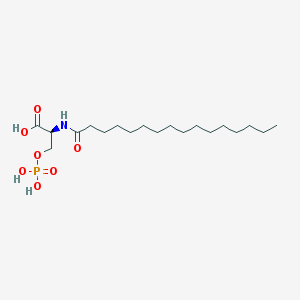 L-NASPA L-NASPA | C19H38NO7P | 423.487 | 7 / 4 | 4.7 | Yes |
| 124786 | 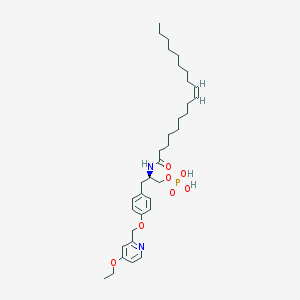 CHEMBL183143 CHEMBL183143 | C35H55N2O7P | 646.806 | 8 / 3 | 8.0 | No |
| 127642 | 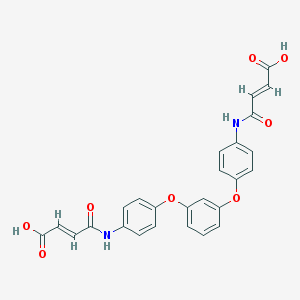 CHEMBL482498 CHEMBL482498 | C26H20N2O8 | 488.452 | 8 / 4 | 2.9 | Yes |
| 127778 | 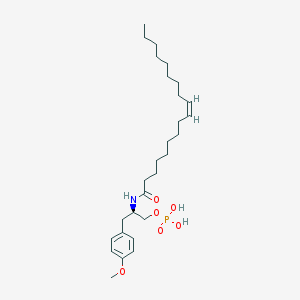 CHEMBL94011 CHEMBL94011 | C28H48NO6P | 525.667 | 6 / 3 | 7.2 | No |
| 129050 | 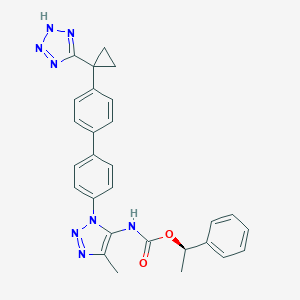 CHEMBL2182038 CHEMBL2182038 | C28H26N8O2 | 506.57 | 7 / 2 | 5.0 | No |
| 130629 | 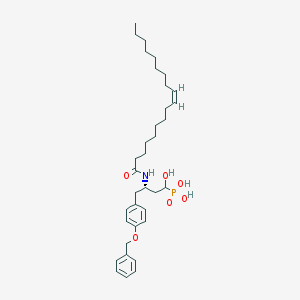 CID 44392848 CID 44392848 | C35H54NO6P | 615.792 | 6 / 4 | 8.5 | No |
| 135411 | 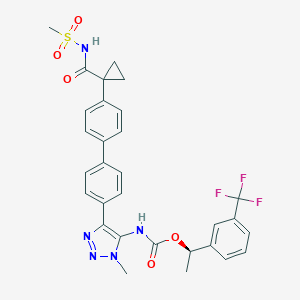 CHEMBL2182050 CHEMBL2182050 | C30H28F3N5O5S | 627.639 | 10 / 2 | 4.8 | No |
| 138417 | 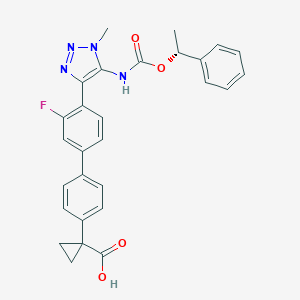 CHEMBL2182044 CHEMBL2182044 | C28H25FN4O4 | 500.53 | 7 / 2 | 4.7 | No |
| 140723 | 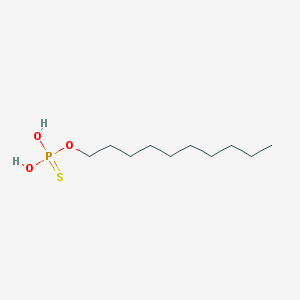 Thiophosphoric acid decyl ester Thiophosphoric acid decyl ester | C10H23O3PS | 254.325 | 4 / 2 | 4.7 | Yes |
| 141050 | 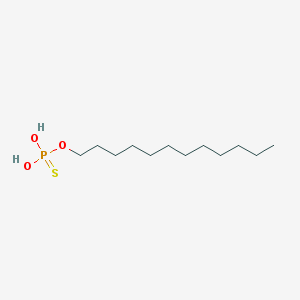 dodecyl-thiophosphate dodecyl-thiophosphate | C12H27O3PS | 282.379 | 4 / 2 | 5.8 | No |
| 141527 | 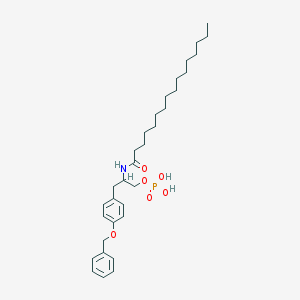 CHEMBL441066 CHEMBL441066 | C32H50NO6P | 575.727 | 6 / 3 | 8.6 | No |
| 141740 | 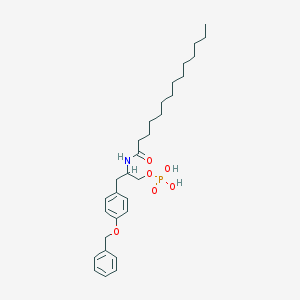 CHEMBL432821 CHEMBL432821 | C30H46NO6P | 547.673 | 6 / 3 | 7.5 | No |
| 144143 | 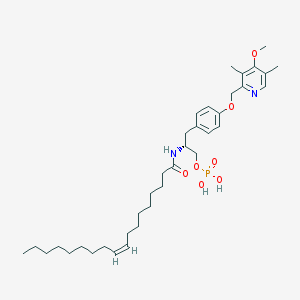 CHEMBL362053 CHEMBL362053 | C36H57N2O7P | 660.833 | 8 / 3 | 8.4 | No |
| 144146 | 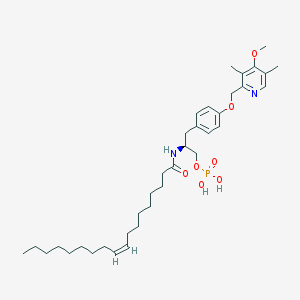 CHEMBL181917 CHEMBL181917 | C36H57N2O7P | 660.833 | 8 / 3 | 8.4 | No |
| 145190 | 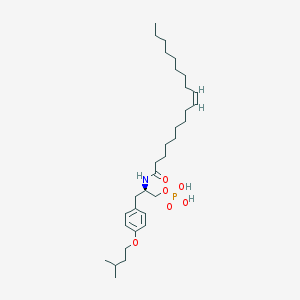 CHEMBL329422 CHEMBL329422 | C32H56NO6P | 581.775 | 6 / 3 | 8.9 | No |
| 145192 |  CHEMBL92593 CHEMBL92593 | C32H56NO6P | 581.775 | 6 / 3 | 8.9 | No |
| 146422 | 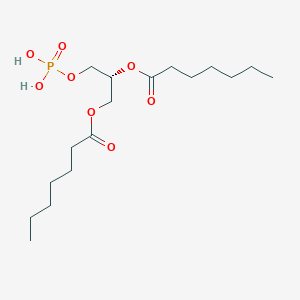 CHEMBL482505 CHEMBL482505 | C17H33O8P | 396.417 | 8 / 2 | 3.2 | Yes |
| 146755 | 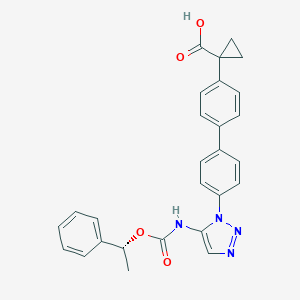 CHEMBL2182040 CHEMBL2182040 | C27H24N4O4 | 468.513 | 6 / 2 | 4.6 | Yes |
| 151959 | 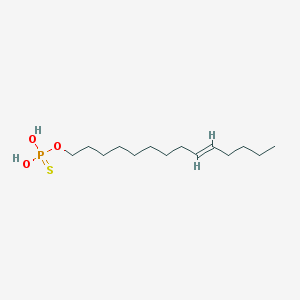 CHEMBL187402 CHEMBL187402 | C14H29O3PS | 308.417 | 4 / 2 | 5.9 | No |
| 153097 | 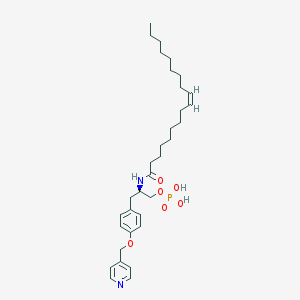 CHEMBL360399 CHEMBL360399 | C33H51N2O6P | 602.753 | 7 / 3 | 7.7 | No |
| 153098 | 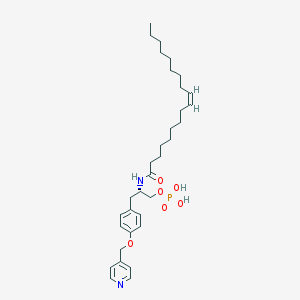 CHEMBL360397 CHEMBL360397 | C33H51N2O6P | 602.753 | 7 / 3 | 7.7 | No |
| 159720 | 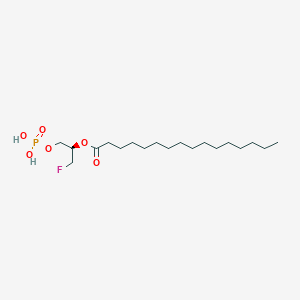 CHEMBL190771 CHEMBL190771 | C19H38FO6P | 412.479 | 7 / 2 | 6.1 | No |
| 162858 | 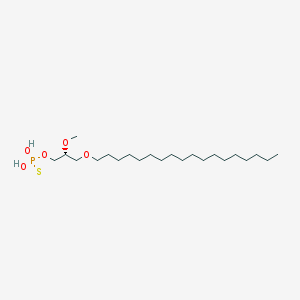 CHEMBL2335048 CHEMBL2335048 | C22H47O5PS | 454.647 | 6 / 2 | 8.8 | No |
| 162863 | 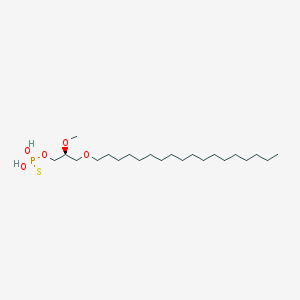 CHEMBL2335047 CHEMBL2335047 | C22H47O5PS | 454.647 | 6 / 2 | 8.8 | No |
| 162866 | 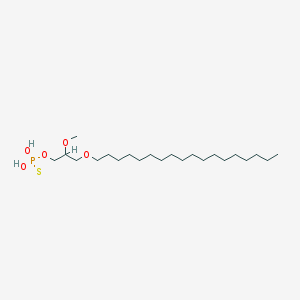 CHEMBL2335052 CHEMBL2335052 | C22H47O5PS | 454.647 | 6 / 2 | 8.8 | No |
| 163620 | 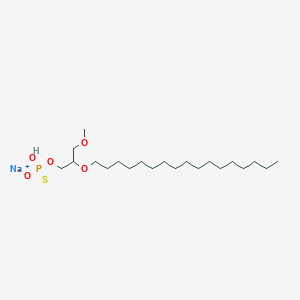 CHEMBL2335050 CHEMBL2335050 | C21H44NaO5PS | 462.602 | 6 / 1 | N/A | N/A |
| 164229 | 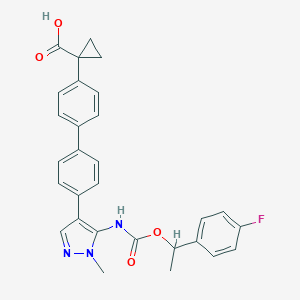 CHEMBL2182065 CHEMBL2182065 | C29H26FN3O4 | 499.542 | 6 / 2 | 5.2 | No |
| 164313 | 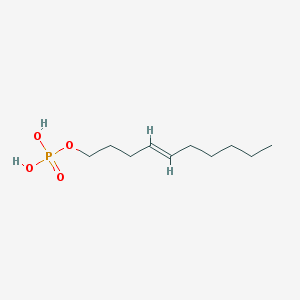 CHEMBL188081 CHEMBL188081 | C10H21O4P | 236.248 | 4 / 2 | 2.2 | Yes |
| 562568 | 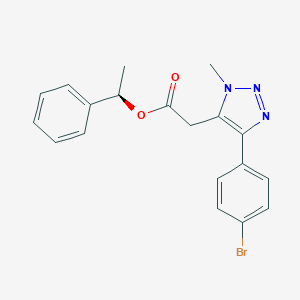 SCHEMBL16706497 SCHEMBL16706497 | C19H18BrN3O2 | 400.276 | 4 / 0 | 3.8 | Yes |
| 550008 | 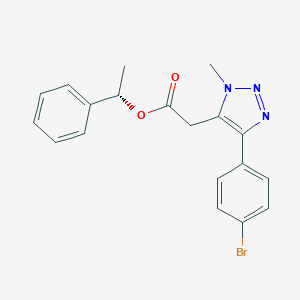 CHEMBL3976990 CHEMBL3976990 | C19H18BrN3O2 | 400.276 | 4 / 0 | 3.8 | Yes |
| 168845 | 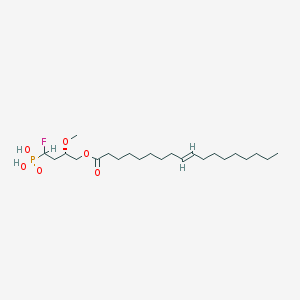 alpha-fluoromethylenephosphonate alpha-fluoromethylenephosphonate | C23H44FO6P | 466.571 | 7 / 2 | 6.6 | No |
| 171084 | 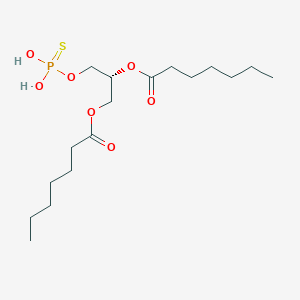 CHEMBL482506 CHEMBL482506 | C17H33O7PS | 412.478 | 8 / 2 | 4.9 | Yes |
| 172974 | 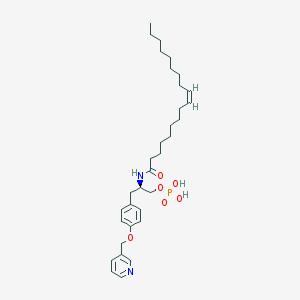 VPC32183 VPC32183 | C33H51N2O6P | 602.753 | 7 / 3 | 7.7 | No |
| 172978 | 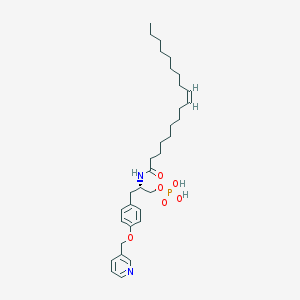 CHEMBL360727 CHEMBL360727 | C33H51N2O6P | 602.753 | 7 / 3 | 7.7 | No |
| 176340 | 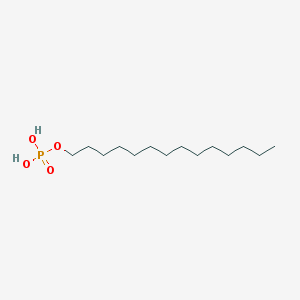 Tetradecyl dihydrogen phosphate Tetradecyl dihydrogen phosphate | C14H31O4P | 294.372 | 4 / 2 | 5.1 | No |
| 178099 | 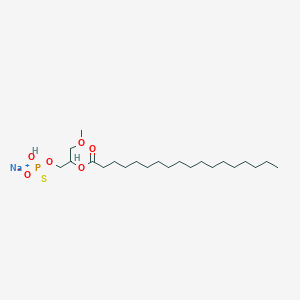 CHEMBL2335051 CHEMBL2335051 | C22H44NaO6PS | 490.612 | 7 / 1 | N/A | N/A |
| 179007 | 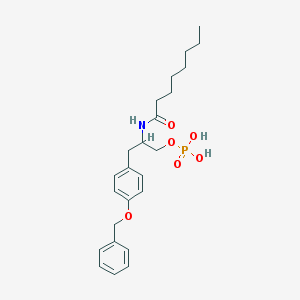 CHEMBL92452 CHEMBL92452 | C24H34NO6P | 463.511 | 6 / 3 | 4.2 | Yes |
| 180346 | 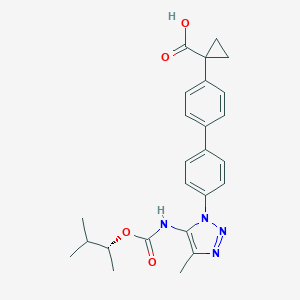 CHEMBL2182033 CHEMBL2182033 | C25H28N4O4 | 448.523 | 6 / 2 | 4.9 | Yes |
| 181599 | 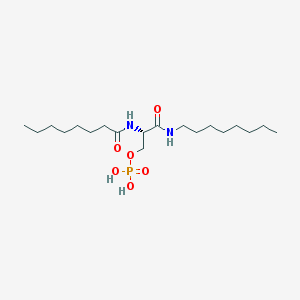 CHEMBL204037 CHEMBL204037 | C19H39N2O6P | 422.503 | 6 / 4 | 3.6 | Yes |
| 181600 | 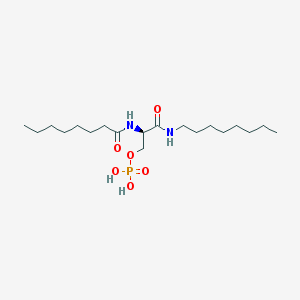 CHEMBL202251 CHEMBL202251 | C19H39N2O6P | 422.503 | 6 / 4 | 3.6 | Yes |
| 184849 | 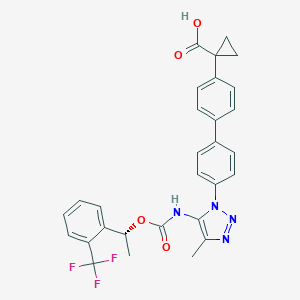 CHEMBL2182031 CHEMBL2182031 | C29H25F3N4O4 | 550.538 | 9 / 2 | 5.9 | No |
| 188631 | 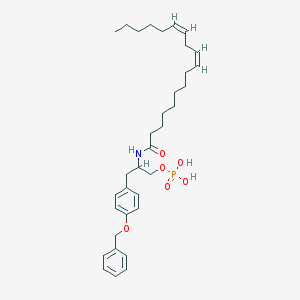 CHEMBL91168 CHEMBL91168 | C34H50NO6P | 599.749 | 6 / 3 | 8.0 | No |
| 189413 | 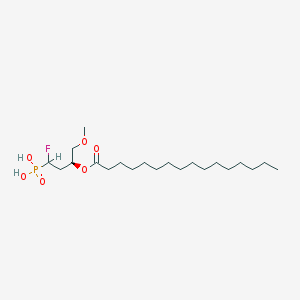 CHEMBL192107 CHEMBL192107 | C21H42FO6P | 440.533 | 7 / 2 | 6.4 | No |
| 190301 | 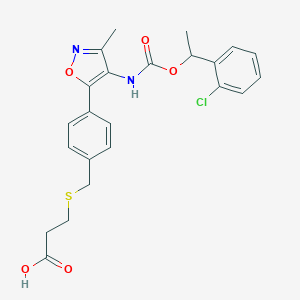 Ki16425 Ki16425 | C23H23ClN2O5S | 474.956 | 7 / 2 | 4.5 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417