

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNECHYDKHMDFFYNRSNTDTVDDWTGTKLVIVLCVGTFFCLFIFFSNSLVIAAVIKNRKFHFPFYYLLANLAAADFFAGIAYVFLMFNTGPVSKTLTVNRWFLRQGLLDSSLTASLTNLLVIAVERHMSIMRMRVHSNLTKKRVTLLILLVWAIAIFMGAVPTLGWNCLCNISACSSLAPIYSRSYLVFWTVSNLMAFLIMVVVYLRIYVYVKRKTNVLSPHTSGSISRRRTPMKLMKTVMTVLGAFVVCWTPGLVVLLLDGLNCRQCGVQHVKRWFLLLALLNSVVNPIIYSYKDEDMYGTMKKMICCFSQENPERRPSRIPSTVLSRSDTGSQYIEDSISQGAVCNKSTS | |
| CCCCCCCCCSSCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 98755777443135898887689985399999999999999999998887121367258889699999999999999999999999999865040353685464879999999999999999999805035257888875689999999999999999999999763751689976503213566125866799999999999999999999999755652165400466788999999999999999999999999999999778977618999999999999998859999827999999999994878999876688888887767777998877776677888888899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNECHYDKHMDFFYNRSNTDTVDDWTGTKLVIVLCVGTFFCLFIFFSNSLVIAAVIKNRKFHFPFYYLLANLAAADFFAGIAYVFLMFNTGPVSKTLTVNRWFLRQGLLDSSLTASLTNLLVIAVERHMSIMRMRVHSNLTKKRVTLLILLVWAIAIFMGAVPTLGWNCLCNISACSSLAPIYSRSYLVFWTVSNLMAFLIMVVVYLRIYVYVKRKTNVLSPHTSGSISRRRTPMKLMKTVMTVLGAFVVCWTPGLVVLLLDGLNCRQCGVQHVKRWFLLLALLNSVVNPIIYSYKDEDMYGTMKKMICCFSQENPERRPSRIPSTVLSRSDTGSQYIEDSISQGAVCNKSTS | |
| 74434234222112132243335434201000000011200220332330000000103300100000000000000010111111000001222210111000100000000000030000001000000010214431233000000010122023202100000012342620000001232110000021133112100100120011003204403535544443333411000000000000200311110000000002421002000000002002300230001001064003001100001123446544443444334434444543554344454346647 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCSSCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNECHYDKHMDFFYNRSNTDTVDDWTGTKLVIVLCVGTFFCLFIFFSNSLVIAAVIKNRKFHFPFYYLLANLAAADFFAGIAYVFLMFNTGPVSKTLTVNRWFLRQGLLDSSLTASLTNLLVIAVERHMSIMRMRVHSNLTKKRVTLLILLVWAIAIFMGAVPTLGWNCLCNISACSSLAPIYSRSYLVFWTVSNLMAFLIMVVVYLRIYVYVKRKTNVLSPHTSGSISRRRTPMKLMKTVMTVLGAFVVCWTPGLVVLLLDGLNCRQCGVQHVKRWFLLLALLNSVVNPIIYSYKDEDMYGTMKKMICCFSQENPERRPSRIPSTVLSRSDTGSQYIEDSISQGAVCNKSTS | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.25 | 0.23 | 0.82 | 3.02 | Download | -------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAG-------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------------------------------- | |||||||||||||||||||
| 2 | 4z34A | 0.51 | 0.47 | 0.86 | 4.06 | Download | -PQCFYNESIAFFYNRSGKHLATEWN-TVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWNAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCC-PQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQIL--------------------------------------------- | |||||||||||||||||||
| 3 | 4z34A | 0.51 | 0.45 | 0.82 | 3.35 | Download | EPQCFYNESIAFFYNRSGKHLATEWNT-VSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAH------------------YLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVLAY-EKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG-------------------------------------------- | |||||||||||||||||||
| 4 | 4z34 | 0.49 | 0.47 | 0.87 | 1.59 | Download | EPQCFYNESIAFFYNRSGKHLATEWN-TVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFR-MQLHTMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWRKAMKKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV-LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG-------------------------------------------- | |||||||||||||||||||
| 5 | 4z34 | 0.49 | 0.47 | 0.86 | 1.18 | Download | -NEPQCNESIAFFYNRSGKHLATEWN-TVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTG-PNTRRLTSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETRKQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV-LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG-------------------------------------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.25 | 0.23 | 0.81 | 3.22 | Download | --------------NFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAG-------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------------------------------- | |||||||||||||||||||
| 7 | 4z34 | 0.50 | 0.47 | 0.86 | 1.82 | Download | --QCFYNESIAFFYNRSGKHLATEWN-TVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLERKAMKDFQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV-LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG-------------------------------------------- | |||||||||||||||||||
| 8 | 1u19A | 0.17 | 0.22 | 0.94 | 4.58 | Download | MNGTEGPNFYVPFSNKTGVVRSPFEAPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEA----AAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA------------------ | |||||||||||||||||||
| 9 | 5tjvA | 0.26 | 0.23 | 0.82 | 3.32 | Download | -------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKA--GKRAMS-----FSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------------------------------- | |||||||||||||||||||
| 10 | 5g53A | 0.23 | 0.22 | 0.79 | 3.58 | Download | -----------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS-TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCCGEGQVACFEDVVPMNYMVYFNFFVLVPLLLMLGVYLRIFLAARRQLKQM---------TLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLENLY------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
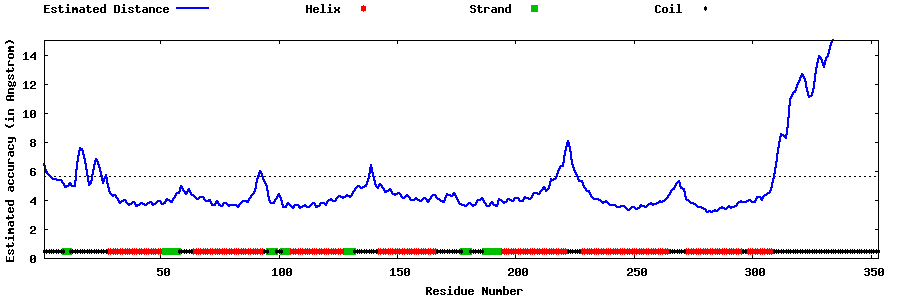
| Generated 3D models | Estimated local accuracy of models | ||
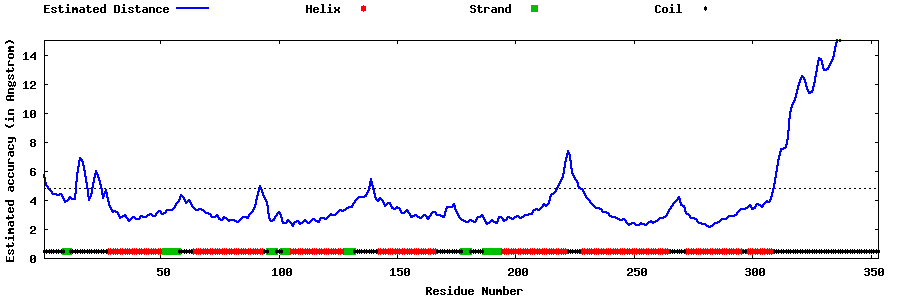 | |||
|
|
|||
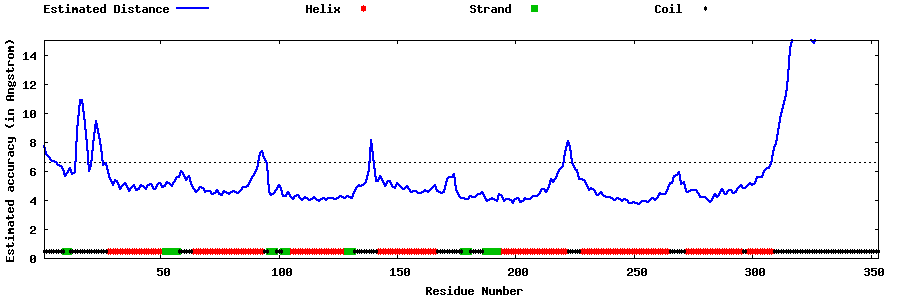 | |||
|
| |||
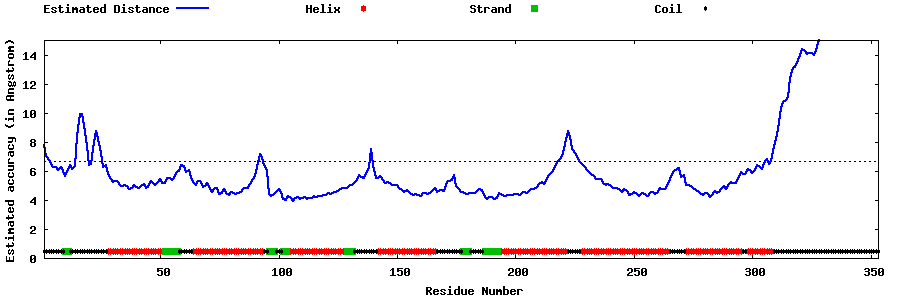 | |||
|
| |||
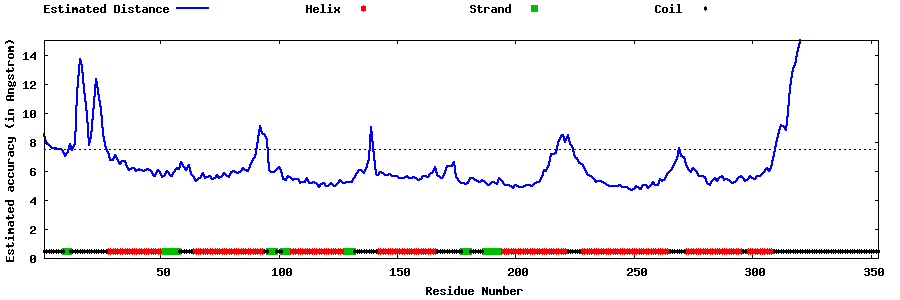 | |||
|
| |||
 | |||
|
| |||