

You can:
| Name | Relaxin-3 receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | RXFP3 |
| Synonym | GPCR135 RXFPR3 RXFP3 RLN3R1 RLN3 receptor 1 [ Show all ] |
| Disease | N/A |
| Length | 469 |
| Amino acid sequence | MQMADAATIATMNKAAGGDKLAELFSLVPDLLEAANTSGNASLQLPDLWWELGLELPDGAPPGHPPGSGGAESADTEARVRILISVVYWVVCALGLAGNLLVLYLMKSMQGWRKSSINLFVTNLALTDFQFVLTLPFWAVENALDFKWPFGKAMCKIVSMVTSMNMYASVFFLTAMSVTRYHSVASALKSHRTRGHGRGDCCGRSLGDSCCFSAKALCVWIWALAALASLPSAIFSTTVKVMGEELCLVRFPDKLLGRDRQFWLGLYHSQKVLLGFVLPLGIIILCYLLLVRFIADRRAAGTKGGAAVAGGRPTGASARRLSKVTKSVTIVVLSFFLCWLPNQALTTWSILIKFNAVPFSQEYFLCQVYAFPVSVCLAHSNSCLNPVLYCLVRREFRKALKSLLWRIASPSITSMRPFTATTKPEHEDQGLQAPAPPHAAAEPDLLYYPPGVVVYSGGRYDLLPSSSAY |
| UniProt | Q9NSD7 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9NSD7 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9NSD7. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL1628472 |
| IUPHAR | 353 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 3469 |  BDBM50383001 BDBM50383001 | C129H207N43O33S2 | 2952.46 | 41 / 44 | -9.9 | No |
| 18070 |  BDBM50383000 BDBM50383000 | C138H218N46O34 | 3065.55 | 41 / 45 | -9.7 | No |
| 51684 |  CID 70683690 CID 70683690 | C173H274N52O48S4 | 3978.65 | 60 / 62 | -14.5 | No |
| 62486 |  BDBM50383002 BDBM50383002 | C142H225N47O36 | 3166.66 | 43 / 47 | -10.6 | No |
| 553640 | 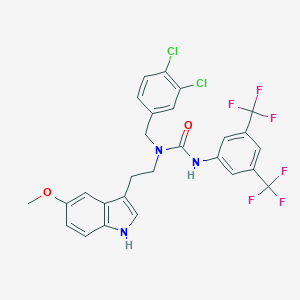 135PAM1 135PAM1 | C27H21Cl2F6N3O2 | 604.374 | 8 / 2 | 7.7 | No |
| 107416 | 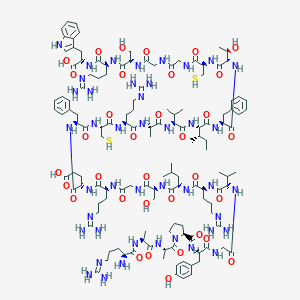 BDBM50382996 BDBM50382996 | C133H207N43O34S2 | 3016.51 | 42 / 45 | -9.6 | No |
| 113478 | 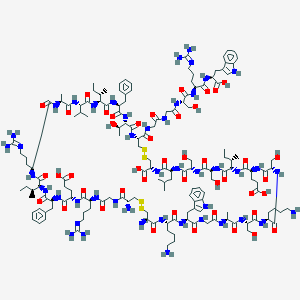 BDBM50382993 BDBM50382993 | C154H239N45O44S4 | 3553.12 | 55 / 52 | -15.7 | No |
| 179484 | 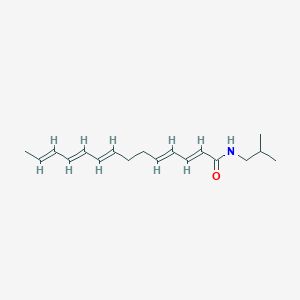 N-Isobutyl-2,4,8,10,12-tetradecapentaenamide N-Isobutyl-2,4,8,10,12-tetradecapentaenamide | C18H27NO | 273.42 | 1 / 1 | 4.9 | Yes |
| 212148 |  BDBM50382992 BDBM50382992 | C166H262N50O46S4 | 3822.47 | 58 / 56 | -16.0 | No |
| 340575 |  BDBM50382995 BDBM50382995 | C128H205N43O33S4 | 3002.56 | 43 / 46 | -9.9 | No |
| 344778 |  BDBM50382998 BDBM50382998 | C139H220N46O34 | 3079.58 | 41 / 45 | -9.3 | No |
| 347613 |  CID 70692144 CID 70692144 | C139H220N44O35 | 3067.56 | 42 / 50 | -9.8 | No |
| 397430 |  CID 70694195 CID 70694195 | C136H213N43O35 | 3010.47 | 41 / 49 | -7.3 | No |
| 405866 |  CID 70687964 CID 70687964 | C140H222N46O35 | 3109.6 | 42 / 52 | -7.7 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417