

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 | |
| | | | | | | | | | | | | | | | | | | | | | | | | |
| MQMADAATIATMNKAAGGDKLAELFSLVPDLLEAANTSGNASLQLPDLWWELGLELPDGAPPGHPPGSGGAESADTEARVRILISVVYWVVCALGLAGNLLVLYLMKSMQGWRKSSINLFVTNLALTDFQFVLTLPFWAVENALDFKWPFGKAMCKIVSMVTSMNMYASVFFLTAMSVTRYHSVASALKSHRTRGHGRGDCCGRSLGDSCCFSAKALCVWIWALAALASLPSAIFSTTVKVMGEELCLVRFPDKLLGRDRQFWLGLYHSQKVLLGFVLPLGIIILCYLLLVRFIADRRAAGTKGGAAVAGGRPTGASARRLSKVTKSVTIVVLSFFLCWLPNQALTTWSILIKFNAVPFSQEYFLCQVYAFPVSVCLAHSNSCLNPVLYCLVRREFRKALKSLLWRIASPSITSMRPFTATTKPEHEDQGLQAPAPPHAAAEPDLLYYPPGVVVYSGGRYDLLPSSSAY | |
| CCCCCCCCHHHHCCCCCCCCHHHHHHCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCC | |
| 9866555335421234772301044148514323788888777886533345777888888889888776544442657898489999999999998899978856515888888999999999999999999879999999768975387589999999999999999999999999727672300742003677644443221001013678999999999999999699987548986995689403786322310689999999999999999999999999999999995356876554310000001566532011222467899999999977999999999999845789628999999999999999999999899999997599999999999545267873457887777788878887778999876789863348994433798767778765679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 | |
| | | | | | | | | | | | | | | | | | | | | | | | | |
| MQMADAATIATMNKAAGGDKLAELFSLVPDLLEAANTSGNASLQLPDLWWELGLELPDGAPPGHPPGSGGAESADTEARVRILISVVYWVVCALGLAGNLLVLYLMKSMQGWRKSSINLFVTNLALTDFQFVLTLPFWAVENALDFKWPFGKAMCKIVSMVTSMNMYASVFFLTAMSVTRYHSVASALKSHRTRGHGRGDCCGRSLGDSCCFSAKALCVWIWALAALASLPSAIFSTTVKVMGEELCLVRFPDKLLGRDRQFWLGLYHSQKVLLGFVLPLGIIILCYLLLVRFIADRRAAGTKGGAAVAGGRPTGASARRLSKVTKSVTIVVLSFFLCWLPNQALTTWSILIKFNAVPFSQEYFLCQVYAFPVSVCLAHSNSCLNPVLYCLVRREFRKALKSLLWRIASPSITSMRPFTATTKPEHEDQGLQAPAPPHAAAEPDLLYYPPGVVVYSGGRYDLLPSSSAY | |
| 6434444343324443344314301420231033122223230323322232323232313333343333133431330000000210020002013201100000000342310000000000010100000000000000025332100100000000012100100000000000000000000020232233333333333333212100100000000000000000000020253742110101013433443310020011001010012212100020000000101313334344434334333444443331000000000000010031100000000002303203332322200100000000000200011000000004400410140023114442343343344444444444344534454435441322111002143132320246357 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 | | | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCHHHHCCCCCCCCHHHHHHCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCC MQMADAATIATMNKAAGGDKLAELFSLVPDLLEAANTSGNASLQLPDLWWELGLELPDGAPPGHPPGSGGAESADTEARVRILISVVYWVVCALGLAGNLLVLYLMKSMQGWRKSSINLFVTNLALTDFQFVLTLPFWAVENALDFKWPFGKAMCKIVSMVTSMNMYASVFFLTAMSVTRYHSVASALKSHRTRGHGRGDCCGRSLGDSCCFSAKALCVWIWALAALASLPSAIFSTTVKVMGEELCLVRFPDKLLGRDRQFWLGLYHSQKVLLGFVLPLGIIILCYLLLVRFIADRRAAGTKGGAAVAGGRPTGASARRLSKVTKSVTIVVLSFFLCWLPNQALTTWSILIKFNAVPFSQEYFLCQVYAFPVSVCLAHSNSCLNPVLYCLVRREFRKALKSLLWRIASPSITSMRPFTATTKPEHEDQGLQAPAPPHAAAEPDLLYYPPGVVVYSGGRYDLLPSSSAY | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.25 | 0.25 | 0.79 | 2.52 | Download | LDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK-TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRT---------------PAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSP-----SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------ | |||||||||||||||||||
| 2 | 5ungA | 0.26 | 0.23 | 0.77 | 4.06 | Download | LEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSGSCSQKPSDKHL--------DAIPILYYIIFVIGFLVNIVVVTLFCCQKGPK-KVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFLSQRRNPWQ----------------ASYIVPLVWCMACLSSLPTFYFRDVRTIEGVNACIMAFPP----EKYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSYG--------------KNRITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVIN-SCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR---------------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.30 | 0.22 | 0.64 | 3.27 | Download | --------------------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK-TATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTP---------------AKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFP-----SPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVR-----------LLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV------DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------ | |||||||||||||||||||
| 4 | 2ziy | 0.18 | 0.20 | 0.78 | 1.51 | Download | -------------------------DLRDN--ETWWY-NPSIIVH----------------------PHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKS-LQTPANMFIINLAFSDFTFSLVFPLMTISCF-LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMS--H-------------RRAFIMIIFVWLWSVLWAIGPIFGWGAYLEGVLCNCSFDYISRD------STTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV---------TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKET-EDDKDAETEIPAGE----SSDAA---PSADAAQMKE-------------- | |||||||||||||||||||
| 5 | 4djh | 0.28 | 0.24 | 0.63 | 1.17 | Download | ----------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK-TATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPL---------------KAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDD----YSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFGVIQTPDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---A---------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.26 | 0.25 | 0.78 | 3.25 | Download | -PKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEG-----KVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK-TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRT---------------PAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSP-----SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG-----------SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------ | |||||||||||||||||||
| 7 | 4djh | 0.29 | 0.24 | 0.63 | 1.73 | Download | ------------------------------------------------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK-TATNIYIFNLALADALVTTTMPFQST-VYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLK---------------AKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDD----DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFRTVTTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF---------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.31 | 0.22 | 0.65 | 4.77 | Download | ------------------------------------------------------------------GSPG-ARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK-TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRT---------------PAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSP-----SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------ | |||||||||||||||||||
| 9 | 5unfA | 0.26 | 0.23 | 0.77 | 3.01 | Download | WETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMDALKLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGP-KKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFP----------------------WQASYIVPLVWCMACLSSLPTFYFRDVRTIEGVNACIMAFPPE----KYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSYG--------------KNRITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVIN-SCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR---------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.31 | 0.21 | 0.63 | 2.37 | Download | -----------------------------------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKM-KTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRT---------------PRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHP-----TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS-----------KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
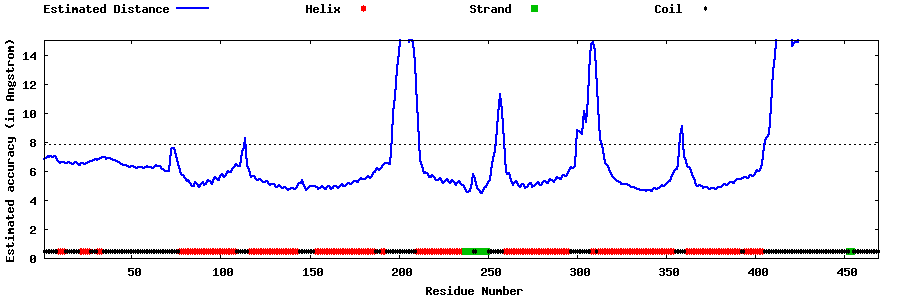
| Generated 3D models | Estimated local accuracy of models | ||
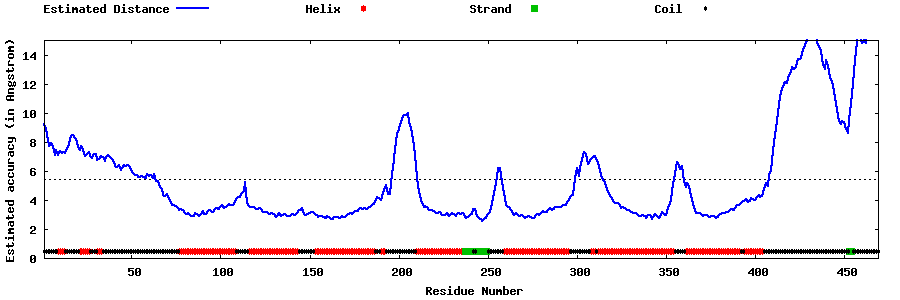 | |||
|
|
|||
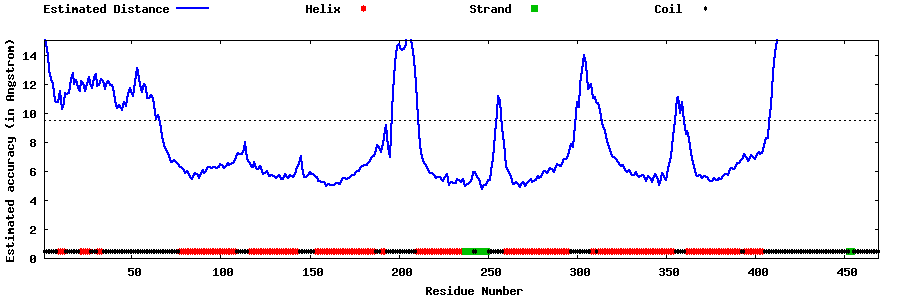 | |||
|
| |||
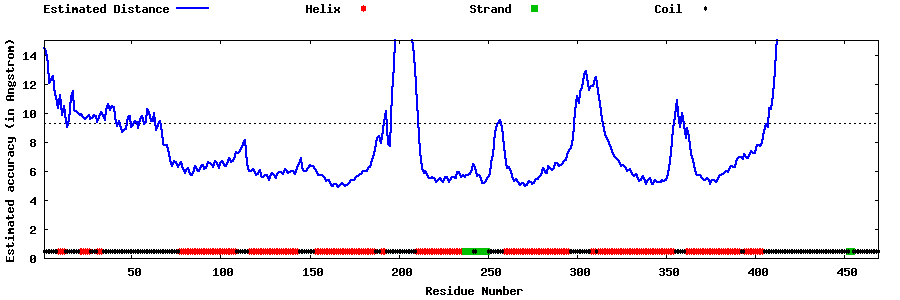 | |||
|
| |||
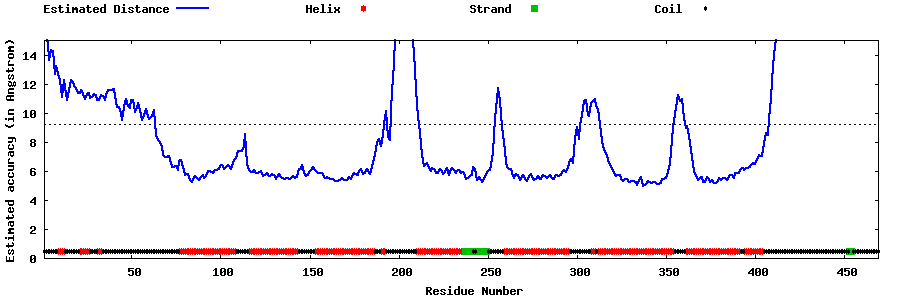 | |||
|
| |||
 | |||
|
| |||