

You can:
| Name | Putative P2Y purinoceptor 10 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | P2RY10 |
| Synonym | P2Y-like receptor P2RY10 purinergic receptor P2Y purinergic receptor P2Y, G-protein coupled 10 purinergic receptor P2Y10 [ Show all ] |
| Disease | N/A |
| Length | 339 |
| Amino acid sequence | MANLDKYTETFKMGSNSTSTAEIYCNVTNVKFQYSLYATTYILIFIPGLLANSAALWVLCRFISKKNKAIIFMINLSVADLAHVLSLPLRIYYYISHHWPFQRALCLLCFYLKYLNMYASICFLTCISLQRCFFLLKPFRARDWKRRYDVGISAAIWIVVGTACLPFPILRSTDLNNNKSCFADLGYKQMNAVALVGMITVAELAGFVIPVIIIAWCTWKTTISLRQPPMAFQGISERQKALRMVFMCAAVFFICFTPYHINFIFYTMVKETIISSCPVVRIALYFHPFCLCLASLCCLLDPILYYFMASEFRDQLSRHGSSVTRSRLMSKESGSSMIG |
| UniProt | O00398 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | O00398 |
| 3D structure model | This predicted structure model is from GPCR-EXP O00398. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3562166 |
| IUPHAR | 165 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 557914 | 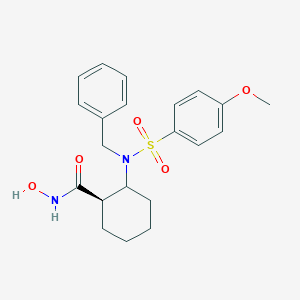 CHEMBL81113 CHEMBL81113 | C21H26N2O5S | 418.508 | 6 / 2 | 2.8 | Yes |
| 467468 | 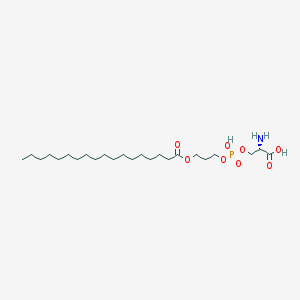 CHEMBL567724 CHEMBL567724 | C24H48NO8P | 509.621 | 9 / 3 | 4.0 | No |
| 468170 | 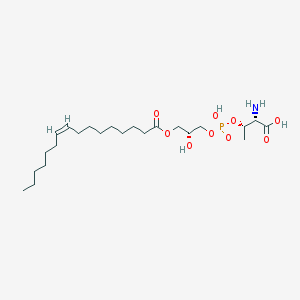 CHEMBL3577163 CHEMBL3577163 | C23H44NO9P | 509.577 | 10 / 4 | 1.5 | No |
| 469559 | 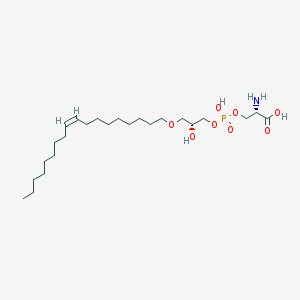 CHEMBL3577155 CHEMBL3577155 | C24H48NO8P | 509.621 | 9 / 4 | 2.5 | No |
| 469612 |  CHEMBL3577149 CHEMBL3577149 | C22H42NO8P | 479.551 | 9 / 3 | 2.0 | Yes |
| 553610 |  Sphingosine 1-phosphate Sphingosine 1-phosphate | C18H38NO5P | 379.478 | 6 / 4 | 1.9 | Yes |
| 471763 |  CHEMBL3577142 CHEMBL3577142 | C24H45FNO8P | 525.595 | 10 / 3 | 3.2 | No |
| 471766 |  CHEMBL3577141 CHEMBL3577141 | C24H45FNO8P | 525.595 | 10 / 3 | 3.2 | No |
| 472223 | 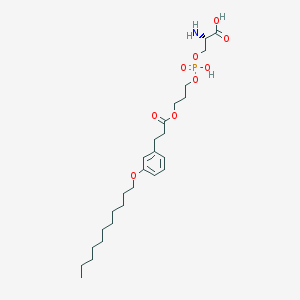 CHEMBL3577180 CHEMBL3577180 | C26H44NO9P | 545.61 | 10 / 3 | 2.5 | No |
| 473005 |  CHEMBL3577177 CHEMBL3577177 | C22H36NO9P | 489.502 | 10 / 3 | 0.4 | Yes |
| 473837 | 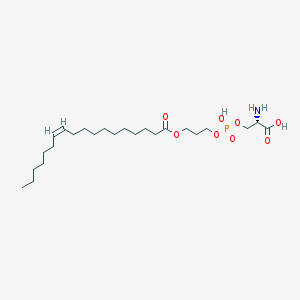 CHEMBL3577154 CHEMBL3577154 | C24H46NO8P | 507.605 | 9 / 3 | 3.1 | No |
| 474594 | 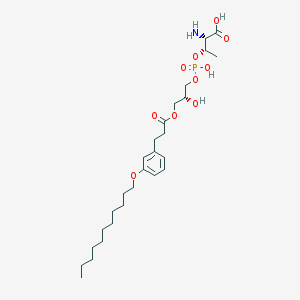 CHEMBL3577263 CHEMBL3577263 | C27H46NO10P | 575.636 | 11 / 4 | 2.0 | No |
| 475156 | 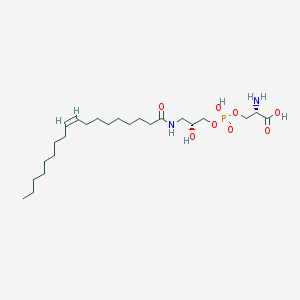 CHEMBL3577156 CHEMBL3577156 | C24H47N2O8P | 522.62 | 9 / 5 | 1.6 | No |
| 476068 | 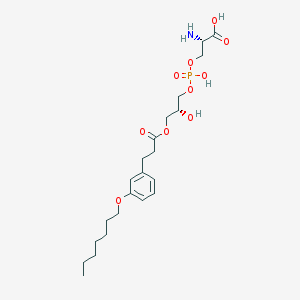 CHEMBL3577168 CHEMBL3577168 | C22H36NO10P | 505.501 | 11 / 4 | -0.6 | No |
| 476221 | 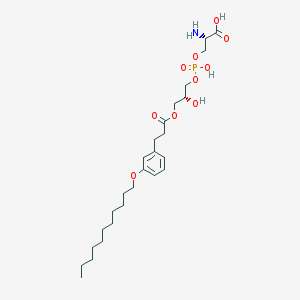 CHEMBL3577172 CHEMBL3577172 | C26H44NO10P | 561.609 | 11 / 4 | 1.6 | No |
| 476770 | 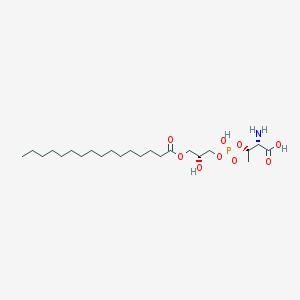 CHEMBL3577164 CHEMBL3577164 | C23H46NO9P | 511.593 | 10 / 4 | 2.4 | No |
| 478842 | 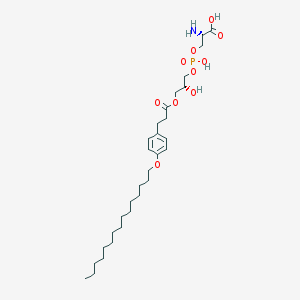 CHEMBL3577176 CHEMBL3577176 | C30H52NO10P | 617.717 | 11 / 4 | 3.7 | No |
| 478850 | 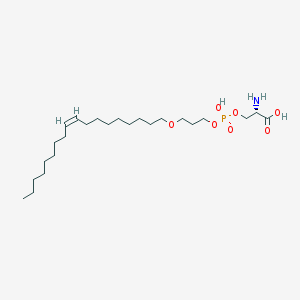 CHEMBL3577157 CHEMBL3577157 | C24H48NO7P | 493.622 | 8 / 3 | 3.5 | Yes |
| 479994 | 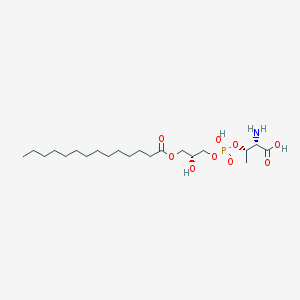 CHEMBL3577165 CHEMBL3577165 | C21H42NO9P | 483.539 | 10 / 4 | 1.3 | Yes |
| 480369 | 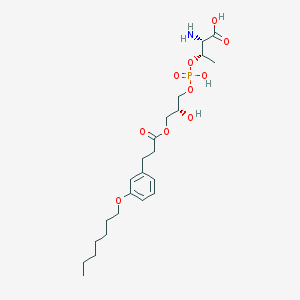 CHEMBL3577264 CHEMBL3577264 | C23H38NO10P | 519.528 | 11 / 4 | -0.2 | No |
| 482812 | 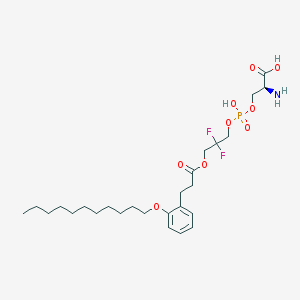 CHEMBL3577262 CHEMBL3577262 | C26H42F2NO9P | 581.591 | 12 / 3 | 2.9 | No |
| 483139 | 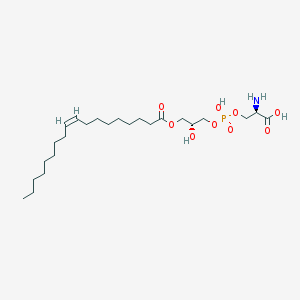 CHEMBL3577160 CHEMBL3577160 | C24H46NO9P | 523.604 | 10 / 4 | 2.1 | No |
| 483142 | 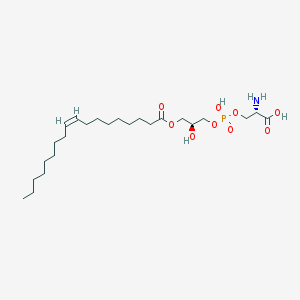 CHEMBL3577138 CHEMBL3577138 | C24H46NO9P | 523.604 | 10 / 4 | 2.1 | No |
| 483145 | 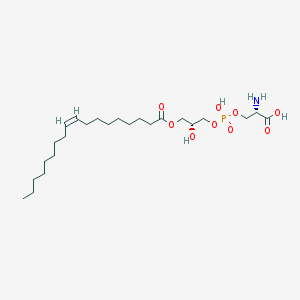 CHEBI:52649 CHEBI:52649 | C24H46NO9P | 523.604 | 10 / 4 | 2.1 | No |
| 485205 | 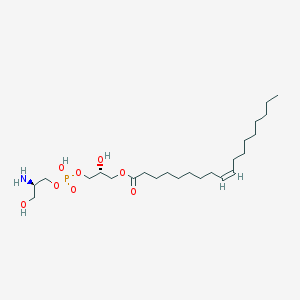 CHEMBL3577159 CHEMBL3577159 | C24H48NO8P | 509.621 | 9 / 4 | 1.9 | No |
| 486132 | 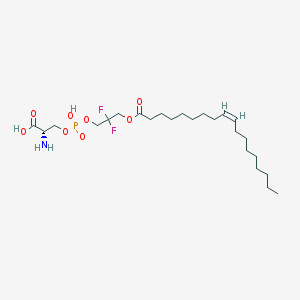 CHEMBL3577143 CHEMBL3577143 | C24H44F2NO8P | 543.586 | 11 / 3 | 3.4 | No |
| 487038 | 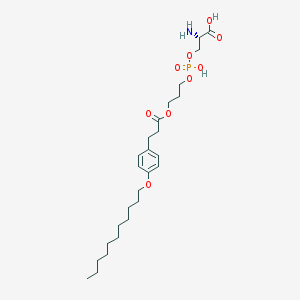 CHEMBL3577181 CHEMBL3577181 | C26H44NO9P | 545.61 | 10 / 3 | 2.5 | No |
| 488333 | 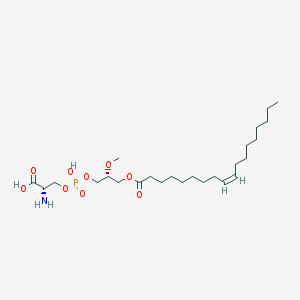 CHEMBL3577139 CHEMBL3577139 | C25H48NO9P | 537.631 | 10 / 3 | 2.7 | No |
| 488958 | 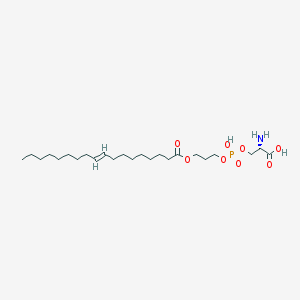 CHEMBL3577153 CHEMBL3577153 | C24H46NO8P | 507.605 | 9 / 3 | 3.1 | No |
| 488959 | 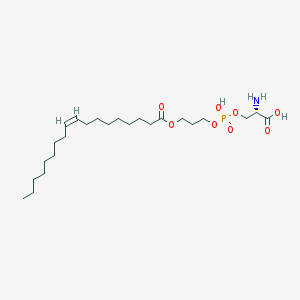 CHEMBL3577137 CHEMBL3577137 | C24H46NO8P | 507.605 | 9 / 3 | 3.1 | No |
| 490918 | 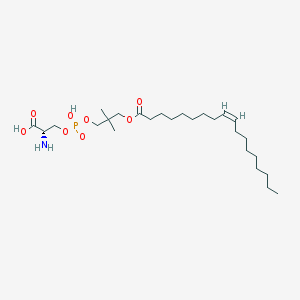 CHEMBL3577140 CHEMBL3577140 | C26H50NO8P | 535.659 | 9 / 3 | 3.9 | No |
| 492361 | 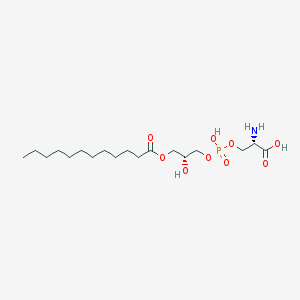 CHEMBL3577147 CHEMBL3577147 | C18H36NO9P | 441.458 | 10 / 4 | -0.2 | Yes |
| 494196 | 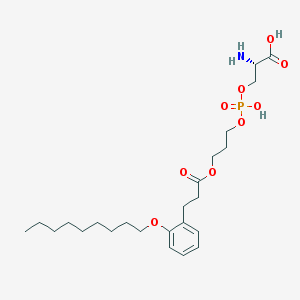 CHEMBL3577178 CHEMBL3577178 | C24H40NO9P | 517.556 | 10 / 3 | 1.5 | No |
| 494805 | 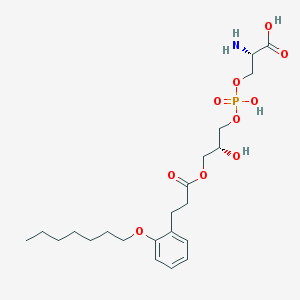 CHEMBL3577167 CHEMBL3577167 | C22H36NO10P | 505.501 | 11 / 4 | -0.6 | No |
| 495875 | 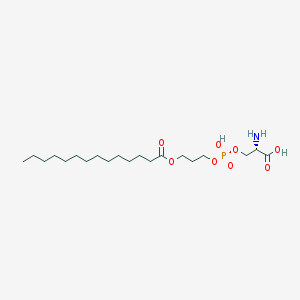 CHEMBL3577151 CHEMBL3577151 | C20H40NO8P | 453.513 | 9 / 3 | 1.9 | Yes |
| 497226 | 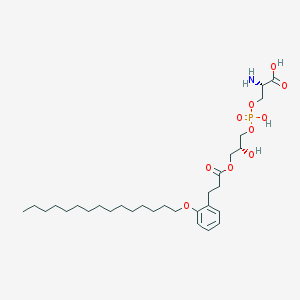 CHEMBL3577174 CHEMBL3577174 | C30H52NO10P | 617.717 | 11 / 4 | 3.7 | No |
| 497284 | 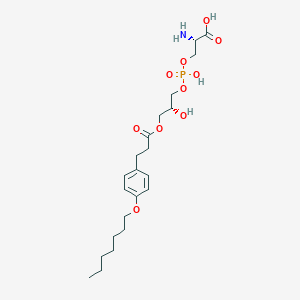 CHEMBL3577169 CHEMBL3577169 | C22H36NO10P | 505.501 | 11 / 4 | -0.6 | No |
| 498515 | 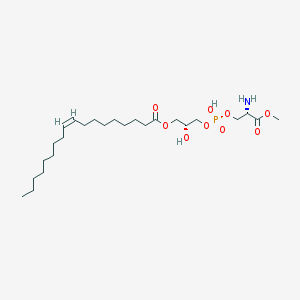 CHEMBL3577158 CHEMBL3577158 | C25H48NO9P | 537.631 | 10 / 3 | 2.5 | No |
| 499691 | 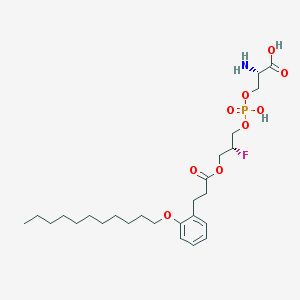 CHEMBL3577182 CHEMBL3577182 | C26H43FNO9P | 563.6 | 11 / 3 | 2.6 | No |
| 500833 | 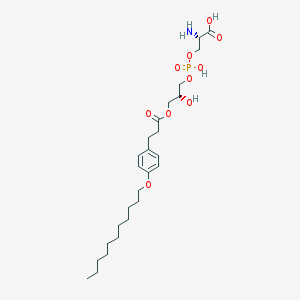 CHEMBL3577173 CHEMBL3577173 | C26H44NO10P | 561.609 | 11 / 4 | 1.6 | No |
| 566920 | 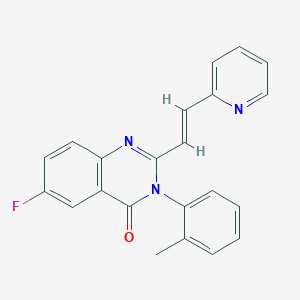 CHEMBL422227 CHEMBL422227 | C22H16FN3O | 357.388 | 4 / 0 | 3.7 | Yes |
| 501190 | 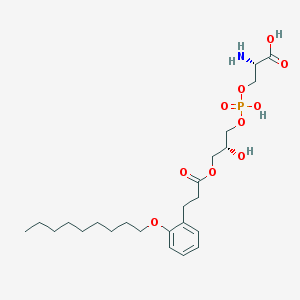 CHEMBL3577170 CHEMBL3577170 | C24H40NO10P | 533.555 | 11 / 4 | 0.5 | No |
| 567012 | 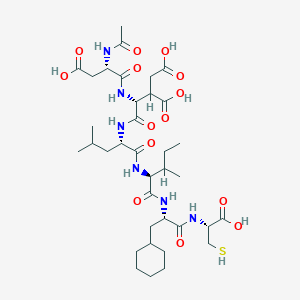 D0M2RI D0M2RI | C36H58N6O14S | 830.948 | 15 / 11 | 1.4 | No |
| 501875 | 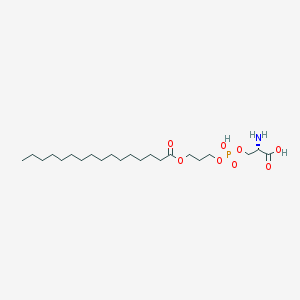 CHEMBL3577150 CHEMBL3577150 | C22H44NO8P | 481.567 | 9 / 3 | 3.0 | Yes |
| 503383 | 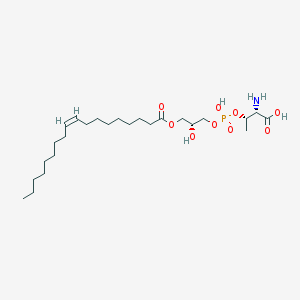 CHEMBL3577162 CHEMBL3577162 | C25H48NO9P | 537.631 | 10 / 4 | 2.6 | No |
| 503384 | 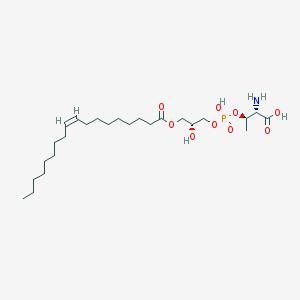 CHEMBL3577161 CHEMBL3577161 | C25H48NO9P | 537.631 | 10 / 4 | 2.6 | No |
| 504463 | 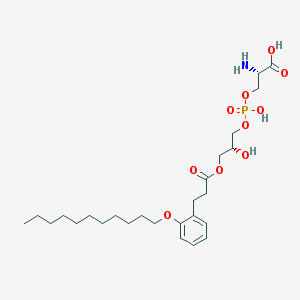 CHEMBL3577171 CHEMBL3577171 | C26H44NO10P | 561.609 | 11 / 4 | 1.6 | No |
| 509778 | 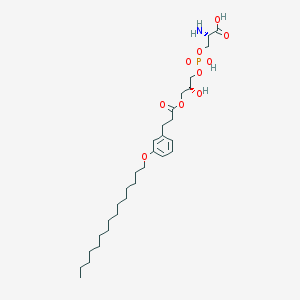 CHEMBL3577175 CHEMBL3577175 | C30H52NO10P | 617.717 | 11 / 4 | 3.7 | No |
| 555184 | 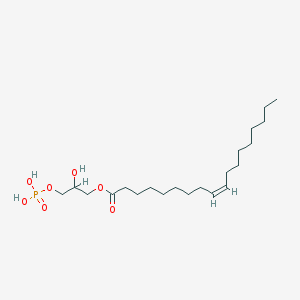 lysophosphatidic acid lysophosphatidic acid | C21H41O7P | 436.526 | 7 / 3 | 5.2 | No |
| 509935 | 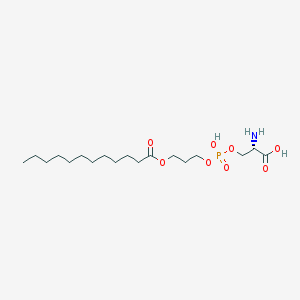 CHEMBL3577152 CHEMBL3577152 | C18H36NO8P | 425.459 | 9 / 3 | 0.8 | Yes |
| 511416 | 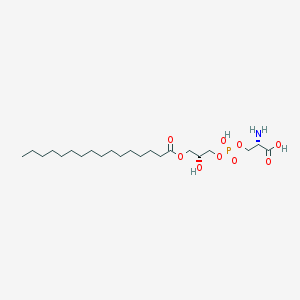 CHEMBL3577145 CHEMBL3577145 | C22H44NO9P | 497.566 | 10 / 4 | 2.0 | Yes |
| 511639 | 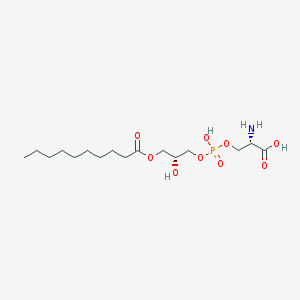 CHEMBL3577148 CHEMBL3577148 | C16H32NO9P | 413.404 | 10 / 4 | -1.3 | Yes |
| 512214 | 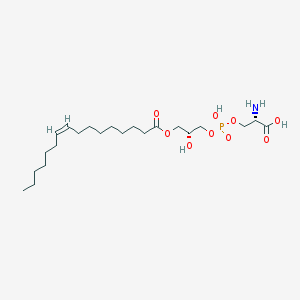 CHEMBL3577144 CHEMBL3577144 | C22H42NO9P | 495.55 | 10 / 4 | 1.1 | Yes |
| 513052 | 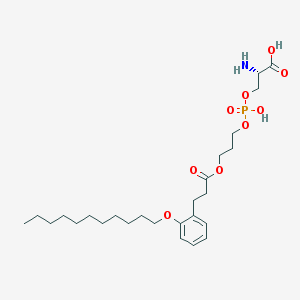 CHEMBL3577179 CHEMBL3577179 | C26H44NO9P | 545.61 | 10 / 3 | 2.5 | No |
| 515909 | 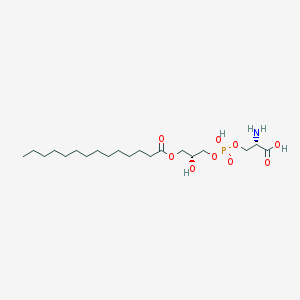 CHEMBL3577146 CHEMBL3577146 | C20H40NO9P | 469.512 | 10 / 4 | 0.9 | Yes |
| 515974 | 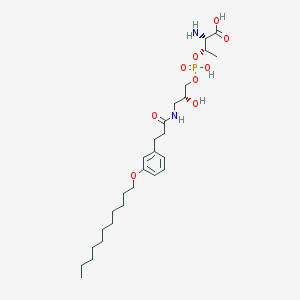 CHEMBL3577265 CHEMBL3577265 | C27H47N2O9P | 574.652 | 10 / 5 | 1.4 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417