

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MANLDKYTETFKMGSNSTSTAEIYCNVTNVKFQYSLYATTYILIFIPGLLANSAALWVLCRFISKKNKAIIFMINLSVADLAHVLSLPLRIYYYISHHWPFQRALCLLCFYLKYLNMYASICFLTCISLQRCFFLLKPFRARDWKRRYDVGISAAIWIVVGTACLPFPILRSTDLNNNKSCFADLGYKQMNAVALVGMITVAELAGFVIPVIIIAWCTWKTTISLRQPPMAFQGISERQKALRMVFMCAAVFFICFTPYHINFIFYTMVKETIISSCPVVRIALYFHPFCLCLASLCCLLDPILYYFMASEFRDQLSRHGSSVTRSRLMSKESGSSMIG | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCC | |
| 987556765566788888887777776652288975999999999999999999889888467778699999999999999999989999999846999867078999999999999999999999999825775241621232102778554999999999998898804578579808998478741237999999999999999999999999999999998378887653044525999999999999998179999999999998457898089999999999999999999999899999339889999999830650577778899888789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MANLDKYTETFKMGSNSTSTAEIYCNVTNVKFQYSLYATTYILIFIPGLLANSAALWVLCRFISKKNKAIIFMINLSVADLAHVLSLPLRIYYYISHHWPFQRALCLLCFYLKYLNMYASICFLTCISLQRCFFLLKPFRARDWKRRYDVGISAAIWIVVGTACLPFPILRSTDLNNNKSCFADLGYKQMNAVALVGMITVAELAGFVIPVIIIAWCTWKTTISLRQPPMAFQGISERQKALRMVFMCAAVFFICFTPYHINFIFYTMVKETIISSCPVVRIALYFHPFCLCLASLCCLLDPILYYFMASEFRDQLSRHGSSVTRSRLMSKESGSSMIG | |
| 654234223314133333344434043343402100002201200231332322001000122343310000000001000000000001000003633100100110012112311100000000003100100010020242332200000010111010011010001214556331000203273242111001111212301331221012000100010132446465454110000000000000120323111000010023132244141230020011001010130002100000000460142025003422444434454444448 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCC MANLDKYTETFKMGSNSTSTAEIYCNVTNVKFQYSLYATTYILIFIPGLLANSAALWVLCRFISKKNKAIIFMINLSVADLAHVLSLPLRIYYYISHHWPFQRALCLLCFYLKYLNMYASICFLTCISLQRCFFLLKPFRARDWKRRYDVGISAAIWIVVGTACLPFPILRSTDLNNNKSCFADLGYKQMNAVALVGMITVAELAGFVIPVIIIAWCTWKTTISLRQPPMAFQGISERQKALRMVFMCAAVFFICFTPYHINFIFYTMVKETIISSCPVVRIALYFHPFCLCLASLCCLLDPILYYFMASEFRDQLSRHGSSVTRSRLMSKESGSSMIG | |||||||||||||||||||||||||
| 1 | 5unfA | 0.25 | 0.25 | 0.93 | 2.90 | Download | VGQIDDALKLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGPKKVSSIYIFNLAVADLLLLATLPLWATYYSYYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPF------PWQASYIVPLVWCMACLSSLPTFYFRDVRTIGVNACIMAFP-PEKYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSYGKNRITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVINSCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR------------------ | |||||||||||||||||||
| 2 | 4mbsA | 0.20 | 0.21 | 0.87 | 4.30 | Download | -------------------------KINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA-AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 3 | 4mbsA | 0.20 | 0.21 | 0.87 | 3.87 | Download | -----------------------PCQKIVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ-WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK---EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 4 | 4djh | 0.25 | 0.27 | 0.83 | 1.53 | Download | -----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFTPLKAKIINICIWLLSSSVGISAIVLGGTKVVDVIECSLQFPDDDY-SWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------- | |||||||||||||||||||
| 5 | 4xnv | 0.26 | 0.27 | 0.86 | 1.16 | Download | ---------------------SFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKNAICISVLVWLIVVVAISPILFYSGTGVRKTITCYDTTS--DEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYTEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------------------ | |||||||||||||||||||
| 6 | 4mbsA | 0.20 | 0.21 | 0.86 | 3.36 | Download | -------------------------KINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK---EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 7 | 4djh | 0.26 | 0.27 | 0.82 | 1.70 | Download | -------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVIECSLQFPDDDYS-WWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-----A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------ | |||||||||||||||||||
| 8 | 4mbsA | 0.21 | 0.21 | 0.86 | 4.79 | Download | -----------------------PCQKINVQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK-------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 9 | 4mbsA | 0.21 | 0.21 | 0.87 | 3.56 | Download | -----------------------PCQKINVQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA-AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK---KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 10 | 4n6hA | 0.22 | 0.26 | 0.94 | 3.98 | Download | LANEGKVKETTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
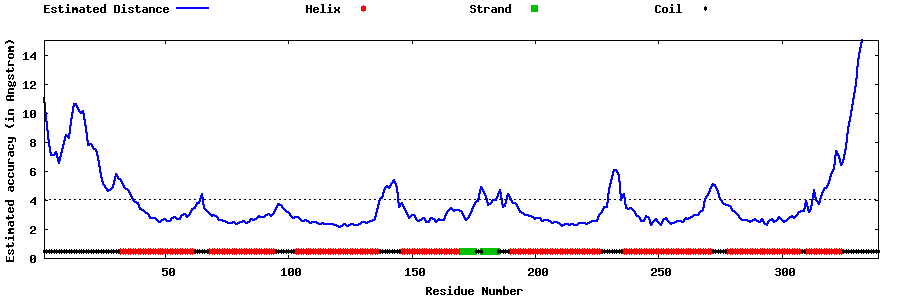
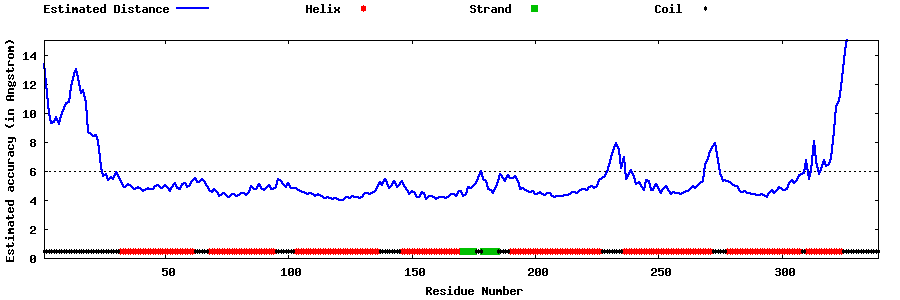
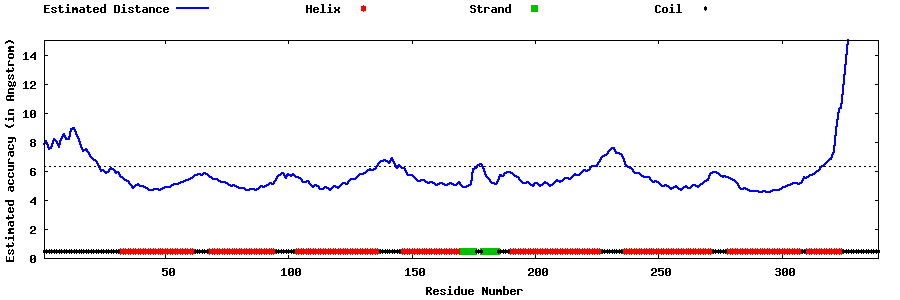
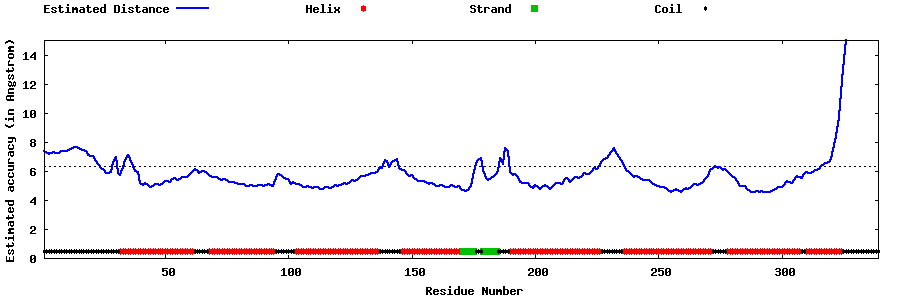
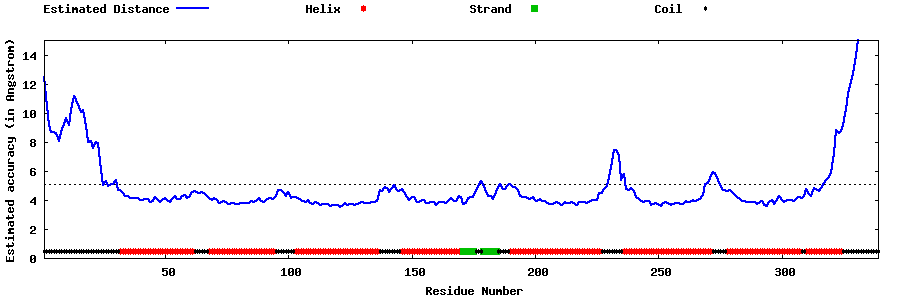
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||