

Go to page (83 pages in total): [First page] << < 4 5 6 7 8 9 10 11 12 13 14 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 801 |
PF04279 (IspA) |
 Estimated TM-score=0.80; hard target. |
>Intracellular septation protein A (Length=174) MKFLFDLFPVILFFVAYKFAGIYVATGVAIVATFAQIGWVHFRHGKVDKMLWVSLVLIVVFGGMTLLLHDPTFIKWKPTI LYWLFALVLLGSAWLFKKNLIRAMMEKQMTLPETIWNRLNLAWAAFFAAMGLLNLYVAYNFSEDAWVNFKLFGGMGLMLA FVVAQGFLLAKYIE |
| 802 |
PF17619 (SCVP) |
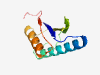 Estimated TM-score=0.80; easy target. |
>Secreted clade V proteins (Length=98) VHVIVTSSEPFDLSKAEENMVTVEKKLKEFADIEGIAFKQLQELPRTAENVGGKFGVHFTVEGAFERCARVLQFIQAACN EIKEVVSGTVKCGAFEPV |
| 803 |
PF15585 (Imm7) |
 Estimated TM-score=0.80; hard target. |
>Immunity protein 7 (Length=129) MFEYHGWVTIQASPSGDDDAALLERIVERVHRAVRDFDDGDLVDLRWAAGVPMLHLGGMDKHGTAIAPELVDLFTRVGDL APGSYGLLHVWDDQDPEHDNEFKVYRMARGLVTERADEHLSPVAPTVLD |
| 804 |
PF02958 (EcKinase) |
 Estimated TM-score=0.80; easy target. |
>Ecdysteroid kinase (Length=284) GENYATVMLKVEIEVQLKDLSTKSLCFMLKVNHEKDQLRELLKGHDVFDAEKSMYDEIIPAFEKLYADVGVEVKFGPKSY DLATKEEYILLENLCVRGFKNANRLEGLDMEHTKCVLKKLAQWHAASAVFVTVKGQFEEKYIKCYFHEEGKESLRTVFEG MGKVFKSCAKNYSNYEEYAADIEALDNKLVDELYKTAIPDPNEFNVLNHGDCWSNNVMFQYDAFGNIKETYLIDLQMPKY GTPAQDLYYFLISSTKYEIKIKQFDYFIKFYHDNLVENLKLLKY |
| 805 |
PF13700 (DUF4158) |
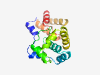 Estimated TM-score=0.80; very hard target. |
>Domain of unknown function (DUF4158) (Length=164) ILSATERDTLLALPESQDDLIRYYTFNDSDLSLIRQRRGDANRLGFAVQLCLLRYPGYALGTDSELPEPVILWVAKQVQA EPASWAKYGERDVTRREHAQELRTYLQLAPFGLSDFRALVRELTELAQQTDKGLLLAGQALESLRQKRRILPALSVIDRA CSEA |
| 806 |
PF14326 (DUF4384) |
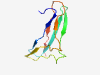 Estimated TM-score=0.80; hard target. |
>Domain of unknown function (DUF4384) (Length=79) TYNIGESITFYFKSNRDCYVHLLDIGTSGKVTLIYPNQFATDNFIKGGQTYKLPAPETFQFVASGPEGTEMVKAIATLK |
| 807 |
PF13564 (DoxX_2) |
 Estimated TM-score=0.80; hard target. |
>DoxX-like family (Length=100) VTTGIIALETGVGSVWDIMKIPFVRTILEQLGYPSYMLTIMGVWKALGVVILLAPRFPRLKEWVYAGLVFVYSGAAASHL VIGHGADAVGPVIFVCLTIA |
| 808 |
PF11172 (DUF2959) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF2959) (Length=190) YYGTMEKFGVHKRDIMVDRVEEARDSQQDAKEQFQSALEQFSQVLNYKGGKLEDKYKTLQAEYDKSEAKADEVTKRIDAV EDVSEALFDEWEDELDQYTNKSLRRDSERKLAQTRKQYEQLIKAMRRAETKIDPVLKAFRDQVLYLKHNLNAQAIASLQS ELVAVESDIADLIKEMEKSIGEANSFIRTM |
| 809 |
PF14192 (DUF4314) |
 Estimated TM-score=0.80; easy target. |
>Domain of unknown function (DUF4314) (Length=68) EKTVKRIKAKYPEGTRVELVHMDDPYTKIPEGTLGTVQVVDDTGTIHVKWDNGSSLGIVYGEDSCRKI |
| 810 |
PF10898 (DUF2716) |
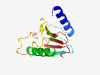 Estimated TM-score=0.80; very hard target. |
>Protein of unknown function (DUF2716) (Length=130) LNDPEYRTLWDTVHERLAFRPHPRCFPGITEPPGAVTWNLGAGEADTLQEVLERGLRSVGEPGERLHWLDWNHIGYGFDP DLCGGEGQPEWPGAVYPDGDYYLYLPSDLRFGTFGHPWEATLCVWGTGLV |
| 811 |
PF04450 (BSP) |
 Estimated TM-score=0.80; easy target. |
>Peptidase of plants and bacteria (Length=198) PKFNLRIEDLSHEGVSIFLNAVNPNVALQEAVVASFTWLYNSETIPTNVKQIVLVLRPMSGVAHTLGSKSHKEIHFSLDH IRNSQKRARDEIMGILVHEVVHCYQFDAKGTCPGGLIEGIADYVRLKDGRAPPHWTKKPGETWDAGYEKTAFFLAWIEDR YGDGTICELNALLNDKKYHRRIFKEVTGRPVRKLWALY |
| 812 |
PF17152 (CHASE8) |
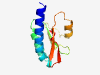 Estimated TM-score=0.80; easy target. |
>Periplasmic sensor domain (Length=101) LRAYADSNQQLIARSISYTVEAAVVFGDAQAAEESLALIASSEEVSSAIVYDRQGQTLASWHRESTGPLHLLEQQLAHWL LSAPTEQPILHDGQKIGSVEV |
| 813 |
PF10080 (DUF2318) |
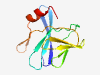 Estimated TM-score=0.80; easy target. |
>Predicted membrane protein (DUF2318) (Length=103) VSDGKAHYYSHEVDGKDVKFFLLKSSDGVIRAAFDACDVCYLEKKGYSQAGDFMVCNNCGQRFHSSRINEVEGGCNPSPL NRTIEGGNVVIKVSDIRDGLRYF |
| 814 |
PF10823 (DUF2568) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF2568) (Length=90) LRFLLELCGLIVFGYWGFQTGDNMMMKFLLGLGVPILFAVVWGTFLAPKSTRRLREPWRSLIELIIFALACWALYSTGAV NLSVAFGGIY |
| 815 |
PF09486 (HrpB7) |
 Estimated TM-score=0.80; trivial target. |
>Bacterial type III secretion protein (HrpB7) (Length=157) RRITALRRAVIRRKRIEDELREMLAQQRDEQATLLHSCEDKGTQIERERDVVRANEERIAQMMTGSSCFSIAEMQASMRY MDLVTDRIRVLQRELAALEQQYREKTDEISRTAQAIASNRGRIDVCERRIGVLRRKLDELASDIADEEAEEAALARA |
| 816 |
PF12530 (DUF3730) |
 Estimated TM-score=0.80; easy target. |
>Protein of unknown function (DUF3730) (Length=219) ILYSNESSSHQLLLELLEMLPSLASHSAMVPFIVQTIVPMLQRDRKPVLHATATRLLCKTWEITDRVFGSLQGILQPKAF QDLVYEKNICISMAASIRDVCQKNPDRGVDLILSVSACIESRDPTVKALGFQSLGHLCEADVVDFYTAWDVIAKHVLDYS VDPIVAHGICILLRWGALDAEAYPEASKNVLQILWEVGTSSHSNEYKWAKARTSAFESL |
| 817 |
PF03350 (UPF0114) |
 Estimated TM-score=0.80; hard target. |
>Uncharacterized protein family, UPF0114 (Length=118) ISDFRFLALLAVAGSLAGSLLYFLSGCVYIKEAYHVYWTSCVRGVHTGQMVLRLVEAIDVYLAGTVMLIFRMGLYGLFIS NDSPDERPRWMRISSLDELKTKLGHIIVMILLVKMFER |
| 818 |
PF19078 (Big_12) |
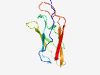 Estimated TM-score=0.80; easy target. |
>Bacterial Ig-like domain (Length=113) DTRAPTLTGTPTIGNRNLIHANQVTTISFVFSEQLDQASFTTADLTIPAGMGRLENLRTTDGGRSWQVDLRPPANLAANT EAKNLQIGINMAGITDIAGNPGTGNVNLVTYNI |
| 819 |
PF14275 (DUF4362) |
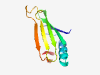 Estimated TM-score=0.80; very hard target. |
>Domain of unknown function (DUF4362) (Length=93) KNDVIVKGVGTSNLDKFEQFVMNVDQGKVDKIRIVQYTDEGDPVFQTLEHSGKDILYVLDSRQDKFAGDHKGLHKDSCKS IVKEQRESESTYR |
| 820 |
PF14332 (DUF4388) |
 Estimated TM-score=0.80; hard target. |
>Domain of unknown function (DUF4388) (Length=99) HGDLTCFKLSDIVQMCCVSGMSGDLQLMHDGIFGDIYLNEGRIVHAQAGLHKGEKALHFLLSFRQGGYEFQKGEMSPEET IQGQWEYLLLEAARLRDEH |
| 821 |
PF05717 (TnpB_IS66) |
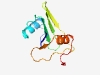 Estimated TM-score=0.80; very hard target. |
>IS66 Orf2 like protein (Length=98) RVWLAAGVTDMRKGFDGLAMLVQQQLAADPFSGHLFVFRGRRGDRIKVLWWDGDGLCLYAKRLERGRFVWPQATEGSVSL TVAQLSMLLEGIDWRRPA |
| 822 |
PF12641 (Flavodoxin_3) |
 Estimated TM-score=0.80; easy target. |
>Flavodoxin domain (Length=159) SIVFSSSTGNTKKLADAVYEILPKDKCDYFGENDSKIPPSDLLYVGFWTDKGSADTKTLELLAKLKNKKIFLFGTAGFGG SDVYFEKILGQVKQSIDSSNAVIGEYMCQGKMPQSVRERYVKMKENPEHPANLDMLIANFDCALSHPDMDDLEKLKKII |
| 823 |
PF14237 (GYF_2) |
 Estimated TM-score=0.80; trivial target. |
>GYF domain 2 (Length=46) WYYADGQNPVGPFTEQEMKDFYTSGKLQDNSLVYQDGTKDWIPFRD |
| 824 |
PF09509 (Hypoth_Ymh) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (Hypoth_ymh) (Length=121) RKVHAEVLNYCRAELLDENYFHAVFESTKGVAERIRSMSGLTMDGAELVSRTFSTQNPILVFGSLATESEKSEQKGFAHL LVGLFGAVRNPLAHAPKTNWPMSEQDALDILTLVSLIHRKL |
| 825 |
PF06516 (NUP) |
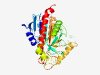 Estimated TM-score=0.80; easy target. |
>Purine nucleoside permease (NUP) (Length=307) VKVLLIGMFAQEAAVWTDKLKLDQAIPVPGLSPTYPAVRCNRDDVCQLTTDMGKANAAASVAALVHSGLFDLRQSYFIVA GVAGVDPAQGTIGTAAWSRYLVDSDLAWEVDARERPAGWSTGYLGIGAKAPGVKPALGYGTEVYRLNEGLLNKALELSRG VTLADAAEAKAYRAQYPSAPANQPPQVTQCDSLTGNNWFHGDLLGQRARDWVALLTDQQGSYCMSSQEDNATYTALMRGA EAGLLDAQRVAVLRAASNFDRPHPGQTALASLTANAGGFPLALQNLYRAAHPLVQDIVQHWPDWKGG |
| 826 |
PF05869 (Dam) |
 Estimated TM-score=0.80; easy target. |
>DNA N-6-adenine-methyltransferase (Dam) (Length=167) SNTPKSFKDRWQTPIEVFRALDAEFNFKLDAAADKSNALCKAFLTEQHDALKSDWNSKGAIFCNPPYSKIMPWVKKAAEQ CKKQNQTIVMLLPSDTSTAWFYEALKTSDEVRFITEGRLSFISAETGKEGKAGNSKGSVLFIWRPWRIPGCWMTYVQRKE LLKKGVN |
| 827 |
PF16156 (DUF4864) |
 Estimated TM-score=0.80; easy target. |
>Domain of unknown function (DUF4864) (Length=100) IRSVIERQLAAFQKDDAQTAFALASPGIREQFQNADNFMRMVSISYPAVYRPRSVFFEKITTIQDNITQPVLLLAPDGKP LRALYFMEKQQDESWKINGC |
| 828 |
PF18905 (DUF5661) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF5661) (Length=72) AKEVGEILRIDWRKIDFEQFYMGINVELEHGSRFGCTTNVSSDSAVISGRIALAHLFEIPDYYTRLDAMEEK |
| 829 |
PF16927 (HisKA_7TM) |
 Estimated TM-score=0.80; easy target. |
>N-terminal 7TM region of histidine kinase (Length=224) LAIAAITSASLALLGWRRRDVSGAQAFLGLMGAVTVWSLGYALELGSTKEAVKLLWAKTEYLGIVIVPAAWLIFALQYTG RGRVVTRRLLALLAIEPLVTLALVWTNEGHGLIWREVRLVTWGAFSLLDPTHGPAFWVHTAYSYLLLLAGTALLFRAFLR APTLYRRQVGAILIGALVPWAQNALYLSGLNPFPYLDLTPFTFTITGLAVAWALFRFRLFDIVP |
| 830 |
PF09990 (DUF2231) |
 Estimated TM-score=0.80; easy target. |
>Predicted membrane protein (DUF2231) (Length=103) YEVSWWNMLFATISIFIAIIFGQTEAGLAQPYDAVKPILNLHTLIGWSLSAILATITGWRFILRSRDPQKVPVSYLAVSL VLVGIVCFQVYLGDELVWVYGLH |
| 831 |
PF18847 (LPD29) |
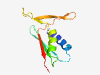 Estimated TM-score=0.80; hard target. |
>Large polyvalent protein associated domain 29 (Length=93) TCAETAKLIRKALKEAFPGVKFSVKSHTYSGGASINVSWTDGPNADQVEAVAGHFKAAYFDGSIDYKGSIYHMMEGQSVH FGADFIFCERQYS |
| 832 |
PF12976 (DUF3860) |
 Estimated TM-score=0.80; trivial target. |
>Domain of Unknown Function with PDB structure (DUF3860) (Length=92) MDSQNLRDLIRNYLSERPRNTIEISAWLASQMDPNSCPEDVTNILEADESIVRIGTVRKSGMRLTDLPISEWASSSWVRR HERARYNERKES |
| 833 |
PF08734 (GYD) |
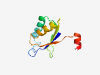 Estimated TM-score=0.80; easy target. |
>GYD domain (Length=90) YISLTKYTQQGIQNIKDSPSRLDAAKKAFQSMGAEIKQFYLVMGQYDTVIVFDAPDEETVAKLMLATGSLGNISTQTMRA FTEDEFRKIV |
| 834 |
PF03715 (Noc2) |
 Estimated TM-score=0.80; easy target. |
>Noc2p family (Length=294) FLSTVLKVMYLSFVRNARFVSPTTLPGINFMRRSLAEMFTLDTECSYQHAFLYIRQLAIHLRNAVTLKRKESYQAVYNWQ FVNALKLWGDVLGATVNSELSALIYPLVTIIQGVIKLIPTAQWFPLRFHCIRVLINLTKSTRIFIPVLPFILEILQSATF NKKHTQISLNAMPFTCILRCTRPQLEELSSRTQVTENIFSTALEYLAAESYSLTFPDLVVPAIVNLKQYLKTCQVAATSR KLKQLLDLIIDNSRIIEAERMKINFSLRDAALIQSWETQMENNGTPLLTFYTNW |
| 835 |
PF07279 (DUF1442) |
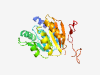 Estimated TM-score=0.80; easy target. |
>Protein of unknown function (DUF1442) (Length=224) MKLIWSPETASKAYIDTVKSCENLGTPGAAELVAAMAAGWNANLIVETWSEGETIAISVGLNIASRHTNGRHICIVPNAR SQTAYLQAMAEQSCSNLPETIIMNEEGEELEHTMQTLQGIDFLVVDWDQKDFAANVLRNAVFGSRGAVVVCRSGYRRSTS CFSWTKAFSDRNVVRTVTLPVSGGLEIAHVAAARSSGKSDNNSNKRKWIKHFDQRSGEEHVIRK |
| 836 |
PF13808 (DDE_Tnp_1_assoc) |
 Estimated TM-score=0.80; hard target. |
>DDE_Tnp_1-associated (Length=86) HISIIPDYRQTWKVEHKLSDILLLTICAVISGAEGWEDIEDFGETHLDFLKQYGDFENGIPVHDTIARVVSCISPAKFHE CFINWM |
| 837 |
PF07308 (DUF1456) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF1456) (Length=68) NNDILRRVRYALNLKDSAMLEIFKSVDCVITHEELLNLLKKEDEEGFEKCNNKVLEAFLDGLIILKRG |
| 838 |
PF04240 (Caroten_synth) |
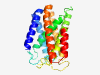 Estimated TM-score=0.80; hard target. |
>Carotenoid biosynthesis protein (Length=214) GGVVYMLLATAAVAVYAYRTLGLWHWLGFMAPAIALSLCSELLGTSTGFPFGHYRYLTGLGYKVAGLVPFTIPLSWFYLG FSAYLLARAGLESRQIPNWVQYVGAIALGATMLTSWDFVLDPAMSQTTMPFWIWEQPGAFFGMPYQNFAGWLGTGTVFMT VATLLWKAKPVTLPRTQLGLPLAIYLSNFAFATIMSVAAGIYIPVFLGVILGVV |
| 839 |
PF08865 (DUF1830) |
 Estimated TM-score=0.80; hard target. |
>Domain of unknown function (DUF1830) (Length=65) CCYVNATSKIQIARITNIPNWYFERVVFPGQRLLFEALPDAQLEIHTGMMASAILSDTIPCTRLQ |
| 840 |
PF08892 (YqcI_YcgG) |
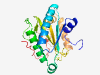 Estimated TM-score=0.80; hard target. |
>YqcI/YcgG family (Length=210) ADDADTFPCIPGRQAFLTDQLRIAFAGDPRENRTAEELAPLLAEYGKISRDTGKYASLVVLFDTPEDLAEHYSVEAYEEL FWRFLNRLSEQDEKEWPEDIPADPEHYKWEFCFDGEPYFILCATPGHEARKSRSFPFFMITFQPRWVFEELNGSTAFGRN MSRLIRSRLEAYDEAPIHPQLGWYGEKDNREWKQYFLRDDEKQVSKCPFS |
| 841 |
PF09979 (DUF2213) |
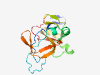 Estimated TM-score=0.80; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2213) (Length=165) RRKTPEGYLIVPARFARTGIQHYAAHELGVSDADPQRVIRVYRPPEEVFAAEAIASFDGRPITDEHPDEEVTAENWRAHA VGFARNPRREGEYLVADLTITDEATIEKIEAGKQELSGGYSAEYDWTPGLTAEGEPYDVKQIRIRGNHIATVAAGRAGPQ CRVAD |
| 842 |
PF06730 (FAM92) |
 Estimated TM-score=0.80; easy target. |
>FAM92 protein (Length=222) MFRRGKLSFLNTKDDRVKIINERINITERHLMEMCSSFALVTRKMAKYRDSFDELAKSVKSYADDEEINESLCQGLKSFT NAVTIMGDYMDINVHRLEHKIVNELAQFEQLCKSTRDNLRLAVIARDKEVLRQRQMLELKSKFSANNSAADSELFKAKME VQRTNKEIDDIIGNFEQRKLRDLKQIISDFILIAMKQHTKALEILSASYYDIGTIDERDDFI |
| 843 |
PF11756 (YgbA_NO) |
 Estimated TM-score=0.80; hard target. |
>Nitrous oxide-stimulated promoter (Length=103) KREREQYVVSQMIELYCKKQHKSKNGALCKDCQELKDYAWQRSEKCPFMETKTFCSNCKVHCYKPAMREKIREVMRFSGP RMLFYHPMLAIRHLIESKWEKKN |
| 844 |
PF09664 (DUF2399) |
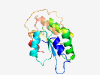 Estimated TM-score=0.80; easy target. |
>Protein of unknown function C-terminus (DUF2399) (Length=154) VLTLRQIRDSMAAFDTADTGRYIVSVCENAAVVAAAADRLGPDCPPLVCTAGQPSAAVIGLLDLLTAAGGRLRYHGDFDW PGLTIAGGLYRRYRWQPWRFTAADYASCEPPVATAPLTGIPVQAEWDTELTVAMSRRGIQIEEELTLPQLLADL |
| 845 |
PF07087 (DUF1353) |
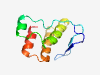 Estimated TM-score=0.80; easy target. |
>Protein of unknown function (DUF1353) (Length=89) RLTQPFRVRTGAGRVIEVPAGFETDFASVPRLFWRMVPPWGRYSPAAVVHDYLYHTGKVSRLAADRIFLELMAALGVSLW KRQVMYWAV |
| 846 |
PF10321 (7TM_GPCR_Srt) |
 Estimated TM-score=0.80; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Srt (Length=312) MTLFYMLTNSFELWPDAYECSENTKYPETRWPRFGIYLISSGSILIVLYSLCFIAILKVKKLTPAYQLMLLISVFDIKSL SIGSVMSGFFTYYGIFFCQYPRLFYISGCIVMLTWLTNCVACIILAIERCVEVNPRFFLYFLFGKRVFKLVILCLAVYGI NSLMFSKPVIFSPEYSSYVFDPMIGKDPTLYINSWLALNNLMIAVSTTTLYFYLCYYLIFQFGYSTSMWLYKTKRQRILR ISDCNARSDNVSVPFRGSLYLRVHDSPLYLIVTGQTVWQLACGCLSVVYLTLNRTIRNSVLKMVIPKAIRQK |
| 847 |
PF07344 (Amastin) |
 Estimated TM-score=0.80; easy target. |
>Amastin surface glycoprotein (Length=158) FVAFMFVLVATPIDMFRIRSRDSQVLPDRCYTLWGLKTNCDSFSYQFLSDDLWESCPPRRDRFRVAQAFAVMSIFVYLAA VVLGVIMLFCCRYLRWVCLALNCFGAVTVCIVWACMAVTYNRNEGLNCLPLKRTHIYGAGFVLLVLAWLLDILNIPVL |
| 848 |
PF08861 (DUF1828) |
 Estimated TM-score=0.80; hard target. |
>Domain of unknown function DUF1828 (Length=89) TPYLDRHNDYLQIYVVKEDGSYRITDDGYTINDLEDSGCALESQKRQALLKTALDGFGVRHEGEELFVLAKADNFAQKKH NLVQAILAV |
| 849 |
PF04165 (DUF401) |
 Estimated TM-score=0.80; trivial target. |
>Protein of unknown function (DUF401) (Length=389) ASSSLALLFSIAVVLILIRKVNIGLAIFVGSAILAYLTLGADGLLVMLKSLANPQTIEIVVIVILAFTLGYSMEYFGMLE KVTGGLSESFGVYSFLLIPLLIGLLPMPGGALISAVMLGPLTKMYRISPEKATLLNYWFRHVWVTFWPLYPSVIIGAAVV NVDYFHFAAATFPIGVFALTAGLTLTRGMERRFRVSRNLFDSLINLYPIAILLILSVLLRLDLLLALVVAIAAVSIHNRA KLEDFREIFRRTVDKKIILLVFAVMVYKSVIQNSGAAESLFTDISLGFPAPAAAFLLTLLVGFATGIEMSYSSIALPLLT SFTGSGIVVAKNLMLVIAAGYIGVMLSPMHLCYILTAEYFGTEIGKTYRQLLAIAGITAALVWIAYLVI |
| 850 |
PF03756 (AfsA) |
 Estimated TM-score=0.80; easy target. |
>A-factor biosynthesis hotdog domain (Length=136) LVHRSSIAEVFVTDGVRTGENAFSVGAQWPRDHALYHPDENGLNDPLLFAETLRQAHFYGAHTYFGVPVGSRFIGQDVSF EITDPTALRVGAAPLAVVLNGTWTEERDRRGRPAGARLDVTLTVDGRPCGRGHTRG |
| 851 |
PF14995 (TMEM107) |
 Estimated TM-score=0.80; hard target. |
>Transmembrane protein (Length=117) LVPARFLTLTAHLVIVITIFWSRDNNVRACLPLDFSQDEYRSQDTQLVVALSVTLGLFAIELAGFFSGVSMFNSTQSLLS IAAHASASVALLFFLFEQWSCAIYWWIFGFCRLVSLT |
| 852 |
PF08578 (DUF1765) |
 Estimated TM-score=0.80; easy target. |
>Protein of unknown function (DUF1765) (Length=123) FEKMMQAAAKKTSIFDHNACFILCDFMEEALNIYVHFQEASKFIDWRFWLDVCQKILESENTMSEIRLFALIYCVWDIIT ADDRRKEVLCLDWLLTEKFFDKYFNHWCPMIRAYYMRLLCWRV |
| 853 |
PF12612 (TFCD_C) |
 Estimated TM-score=0.80; easy target. |
>Tubulin folding cofactor D C terminal (Length=189) SLIRGICKQAVEKMDKLREAAANVLPRILYNQMIYIPYIPFREKLEEIIPKEAEPKWAVPSYSYPCFVQLLQFGCYSRDV LSGLVISIGGLQDSLKKISLSALLEYLEGVESEEHNTRMSREYMLSVDILWVLQQYEKCDRVIIPSLKTIEALFSKKVFL NMEANTPTFCAGVLDCLAIELKGSKDFSK |
| 854 |
PF08874 (DUF1835) |
 Estimated TM-score=0.80; hard target. |
>Domain of unknown function (DUF1835) (Length=123) IHILFGESPAGSLKNAFRDMGIYEQERVVFFRDIFSVGPVWMLHEESGRENRIEWMKDLISDDYEEYPEYRNKFEKTIRQ IQDIPDGSHIIIWHGDNAHEQTGLQYVVYLLKGKNIDISAINT |
| 855 |
PF14017 (DUF4233) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF4233) (Length=104) RTLCSSTLIGEFLVIGFAALVAMKDDDLSTSTVWTVSGIAMFLCLVLCGMVTRPGGIALGWVLQVALIASGFAVPTMFFL GAVFAALWWASVHYGRKIDEAKAR |
| 856 |
PF06258 (Mito_fiss_Elm1) |
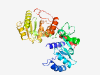 Estimated TM-score=0.80; easy target. |
>Mitochondrial fission ELM1 (Length=321) PGSENQSLGLVRALGLSDNHILYRVTRPRGGINEWLHWLPVSFHKKDGPLLVVASGRDTISIASSIKRLASQNVFVVQIQ HPRSNLNRFDLVITPHHDYYPLTPHAQEQVPRFLRKWITPHEPPDRHVVLTVGALHQIDSAALRSAASAWHDEFAPLPKP LLVVNIGGPTRHCRYGADLAKQLTTYLFSVLESCGSVRISFSDTTPEKVSNIVAKELGNNPKVYIWDGEEPNPQMGHLAW ADAFVVTADSISLISEACSTGKPVYVMGAERCRWKLSEFHKSLSERGVVRPFTGSEDISESWSYPPLNDTAQAARRVHEA L |
| 857 |
PF08547 (CIA30) |
 Estimated TM-score=0.80; easy target. |
>Complex I intermediate-associated protein 30 (CIA30) (Length=174) WEFRSLEDLDKWTLSSDQEIGGKSQVSLKLGRNNQTALFYGTLNTEVPRDGETRYSGYCTMKSKTPLGAFNRKLHYDWSN FNTLYLRVRGDGRPWMVNIKSDSYFSQQRDDLYNYFIYTQGGPYWQDIKIPFSKFFLSSRGRIQDNQHPLWTDKITAVGF TLGDKANGPFQLEI |
| 858 |
PF06848 (Disaggr_repeat) |
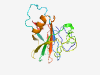 Estimated TM-score=0.80; easy target. |
>Disaggregatase related repeat (Length=180) DNGLRTSSPTSAFPTINYIDIGRNSASCRDLMMFDLSNYKTTDVISKATLSLYWYYPAGKTRTSDTVVQIYRPAADWDPR YVSWTNRASSTKWTTAGGNWFDKNNVAQGATPYASLTFAGSKVPDNKYYDFDVTQLVQEYVSGKYKNTGFFLKAKTESGN YIAFYSSEWTNAAQRPKLTV |
| 859 |
PF11735 (CAP59_mtransfer) |
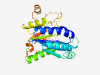 Estimated TM-score=0.80; easy target. |
>Cryptococcal mannosyltransferase 1 (Length=251) RIFIASTHWNNEPILRSHWNNAVVELAKIFGAENVFVSVYESGSWDNSKGALRDLDAQLEQLGVRRKFTLSETTHQDEIS APPASEGWIDTPRGKKELRRIPYLARLRNLTLKPLEELAEQGITFDKVLFLNDVVFTVDDVIALLNTNQGRYAAACSMDF SKPPLYYDTFALRDSNGDAHVMQKWPYFRSTASRHALFTMAPVPVKSCWNGMVAMPTEPFMSNTPLRFRGISDSLGESHL EGSECCLIHAD |
| 860 |
PF04019 (DUF359) |
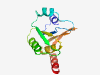 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF359) (Length=123) TTFHLLEAGIIPDICIVDNRTKRKPVSRDVSARNRDKVYEEVSVDNPAGIITDELIKTLCEAFASEKLLRIFVRGEEDLA TLPVILMAPLGSVVLYGQPDEGVVFVEVTEEKKEEIRALFEKL |
| 861 |
PF03155 (Alg6_Alg8) |
 Estimated TM-score=0.80; easy target. |
>ALG6, ALG8 glycosyltransferase family (Length=478) ATAFKVLLFPAYKSTDFEVHRNWLAITNSLPVRDWYYEKTSEWTLDYPPFFAYFEWILSQFAKLVDPEMLRVYNLQYDSW QTVYFQRATVIITELVLLYALHLFVQSAPPALKRPSHAAALSILLSPGLLIIDHIHFQYNGFLYGILILSLVLARKKSTT LASGLLFAALLCCKHIYLYLAPAYFVYLLRAYCLGPKSIFHIKFGNCLKLGTGIIAVFGAAFGPFVYWEQIPQLLSRLFP FSRGLCHAYWAPNIWAMYSFTDRILIYVAPRLGLAVNQNAINSVTRGLVGDTSFAVLPEITPRTTFILTLFFQILPLIKL FFDPTWDTFVGAVTLCGYASFLFGWHVHEKAILLVIIPFSLLALKDRRYLGAFRPLAVAGHVSLFPLLFTAAEFPIKTVY TIFWLVFFLMAFDRLAPASKRPRIFLLDRFSLLYIAVSIPLILYCSLVHGVVFGDKLEFLPLMFMSSYSAVGVVGSWV |
| 862 |
PF09612 (HtrL_YibB) |
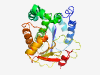 Estimated TM-score=0.80; easy target. |
>Bacterial protein of unknown function (HtrL_YibB) (Length=265) STTIITAYFDIGRGDWTANKGFREKLARSVDVYFSYFERLAALENEMIIFTSPDLKPRVEAIRNGKPTTVIVIDIKKKFR YIRSRIEKIQKDESFTNRLEPRQLKNPEYWSPEYVLVCNLKAYFVNKAINMGLVKTPLVAWIDFGYCHKPNVTRGLKIWD FPFDESKMHLFTIKKGLTVTSQQQVFDFMIGNHVYIIGGAIVGSQHKWKEFYKLVLESQKITLNNNIVDDDQGIFVMCYY KRPDLFNLNYLGRGKWFDLFRCFRS |
| 863 |
PF12422 (Condensin2nSMC) |
 Estimated TM-score=0.80; hard target. |
>Condensin II non structural maintenance of chromosomes subunit (Length=151) MMYCSPALMACKEGIKLLALILDKIGVRNGLTLLEKQILKSDLTDTDCQRYGDVFATTWLNAHSDANFDLTEELETDVYH VFFQHALFGQKPLCAKFQKVLSAFAAQRKENPDFAIMISGMLNGCIYRALDSKNSAVQTSGMEVFFMFFPF |
| 864 |
PF13704 (Glyco_tranf_2_4) |
 Estimated TM-score=0.80; easy target. |
>Glycosyl transferase family 2 (Length=106) KNEGPFIVDWVAHHRAIGVTDFIVYTNDCTDGTDQLLDLLVEKGLVTARYDNPFQPDTPGMSNPQFTAFEDAEKRPEMAQ MDWLLPMDVDEFIDVKVGDGTLGALM |
| 865 |
PF13929 (mRNA_stabil) |
 Estimated TM-score=0.80; easy target. |
>mRNA stabilisation (Length=297) LLQNLQSQDYPSAFRLINNNKDQSLQLMQTLVEWVSNLPIEEEAAVPWSKWVTAFESCWPLVLPEEKASQYWSLRLKFFK LVNMADMKHYNLSNFFNDYLLAKKATGAGLVPQDLVEFLKLAIVNTRYCDKHRGYWDLVHSNDIVVKALRLFEDVRGADA IVKDSSVVTMLLRTMVSGQEIGERATRLHAVYEVSDYLVDTFGPELTPPTIAGILEVLAAAKDWNKYLQFWEAGIGGLVP GADQRPWAHFIRLVVESGDVEMMRKLLSEGHLLWLDRLQVEIDDELKAQLGLLFAKA |
| 866 |
PF11866 (DUF3386) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF3386) (Length=209) QTSARELFQAAYENRYTWDKNFPGYTADITFKQDEQVFTGKVRVNPDLKAEVFEVDDEDAKQAIHSQLWETAIHRIRRTF EETHGKNSFSFGETDETGAIEILMGGKAEGDRYKIRDNEVCHVHRHIHGVVVTIDTFSSHKTGEGYLSHRYDSVYHDPKT GEQKGGKSVFEDSYEKVGNYFILTRRLIHTETEGKTSVQEFIFSNVQLL |
| 867 |
PF12990 (DUF3874) |
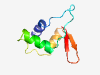 Estimated TM-score=0.80; easy target. |
>Domain of unknonw function from B. Theta Gene description (DUF3874) (Length=70) CPAEDVFNCCFRVALPEEECLHLSAAEIYQELKKMNSAALRGFNPNQFAQVLTKMGVGRKHTEYGNVYSV |
| 868 |
PF18929 (DUF5678) |
 Estimated TM-score=0.80; hard target. |
>Family of unknown function (DUF5678) (Length=48) ELKKYRGKHVAIIGDKVVASGDNAIEVYKKAKEKYPDKNPVLAFVPRE |
| 869 |
PF09954 (DUF2188) |
 Estimated TM-score=0.80; trivial target. |
>Uncharacterized protein conserved in bacteria (DUF2188) (Length=57) YHITFNKDDRWQVKKVGALKILKTFKTQKEAISFADKLAKNQKAGVVIHRTSGQIRS |
| 870 |
PF14974 (P_C10) |
 Estimated TM-score=0.80; hard target. |
>Protein C10 (Length=103) FTLETAKAILTDILTALNTPENLQKLAEAKENSGNEMLKMMQFVFPLVTQIQMDVIKNYGFPEGREGTVQFAQLIRTLER EDAEVAQLHSQVRSYFLPPVRIN |
| 871 |
PF04505 (CD225) |
 Estimated TM-score=0.80; hard target. |
>Interferon-induced transmembrane protein (Length=69) PPRDHLVWSLFTTLYGNFCCLGLLAFVFSVKSRDRKVLGDYSGALSYGSTAKYLNITALVINILIVIIV |
| 872 |
PF11667 (DUF3267) |
 Estimated TM-score=0.80; easy target. |
>Putative zincin peptidase (Length=103) LGLLVIHELIHGLFFKLYQPEKKVEFGFKNGMAYATSSDSYYPRNQYIWIAAAPFVLITLLLLALFIAEALSKVLFVMLA SIHAAGCVGDFYYLYLLSKQPKD |
| 873 |
PF14568 (SUKH_6) |
 Estimated TM-score=0.80; easy target. |
>SMI1-KNR4 cell-wall (Length=120) EEEIHKVEEELGISFPGDYKEYVKKCNGGFPNRDEFYFDDGFYSVSLRNLISLHHDMLATYEDVKSYIPEKVIPFMETRS GDFLCFDFRESLTSPPIVYMNHEEEGDESLSALCDTFTDL |
| 874 |
PF13430 (DUF4112) |
 Estimated TM-score=0.80; hard target. |
>Domain of unknown function (DUF4112) (Length=105) LKKVRRRAYRLDQSISCCCCSFRVGWAAIIGIIPVIGDVIDALLAMRIISTAKSIDGGLPESVVAQMYANVVVDFGIGLI PFVGDIADMIYKCNTRNLQILEKYL |
| 875 |
PF06250 (YhcG_C) |
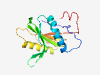 Estimated TM-score=0.80; easy target. |
>YhcG PDDEXK nuclease domain (Length=155) QETLKDPYIFDFLTIEEPFHERELETGLVGHVEKFLLELGAGFAFVGRQYHLEVSEQDFYIDLLFYHLKLRCYVVIELKK GKFQPEYAGKMNFYCSVVDDKLRHEQDNPTIGLILCQTKDKIVAEYALRDVNKPIGISEYELTRALPDNLKSSLP |
| 876 |
PF16127 (DUF4839) |
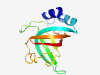 Estimated TM-score=0.80; easy target. |
>Domain of unknown function (DUF4839) (Length=123) ENNADFAALLLTEDTCHEANLDFATKHEGRTVAFNGSIVHMVPHGDYETHYDFLLGPGDKGPQTTIGPAFKYEDVNVFNL NLTGEEIPATVGTGDKFRFIAKVGEFNPVQCLFHLDPVSTEIR |
| 877 |
PF06510 (DUF1102) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF1102) (Length=144) ITSDDNELIDLDPVQPYAYLNNGKLTIEISPNNPNYPGYGDGMSHNTTYVFEEMFNVSNDLWENENQNFPICVQISVPQN SSVKVFAGNYNDDGTATIANPGTQIQFTVEHGQPVSVGFIFDNTCEELGTHSVNMDIQAWAGAC |
| 878 |
PF11761 (CbiG_mid) |
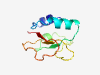 Estimated TM-score=0.80; easy target. |
>Cobalamin biosynthesis central region (Length=87) PGIDMLGVPFGWQRGEGDWTGVSAAVARGEAVEVIQDAGSTLWQDHLPQGHPFQFLATDVAKARIWISPTQRRFSSASDF PKIQWHP |
| 879 |
PF12959 (DUF3848) |
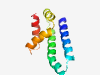 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF3848) (Length=93) NALLYDKMKAEQDKYRDWLLSQPPEEILNHTYEYTMKEDIVMCMEELELSPKQAKALLRSPCPLDDVYKEFKDREVEHMD TIRDSIETRANDV |
| 880 |
PF14742 (GDE_N_bis) |
 Estimated TM-score=0.80; easy target. |
>N-terminal domain of (some) glycogen debranching enzymes (Length=200) VKDDDLFLVTDTMGNISSCSPDESNSSMGLFCRDTRFLSRLELQINGRSPVLLSSTTDKGFALSVLCTNPKITEELKPDT IGIHREIVLNGALFEEIEIANYSTTTINFELSLSFDADFVDLFEVRGHSREKRGQILRLIEKNTEHQKEESLTLAYQGLD NSIMESRIQFQHRQPDDFQGYTAIWRLELGSHETQKLGYR |
| 881 |
PF11731 (Cdd1) |
 Estimated TM-score=0.80; easy target. |
>Pathogenicity locus (Length=84) RAALKALQSMPNIGPAMARDLVSMGFRTPEELRGQEPMELYRRLEQITGSRQDPCVLDTFMSAVHYAETGERRPWWSFTA ERKE |
| 882 |
PF17278 (DUF5343) |
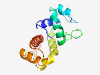 Estimated TM-score=0.80; hard target. |
>Family of unknown function (DUF5343) (Length=137) PYTTVPGKIKPFFQKIQEAGLPSKVTQKWINSIGFTSSNDPSIIPLLKKLGFIDRSGVPTELWARYRDKDLAPKVMAEAI KNAYKELFETYPDAYRKDKEALRNFFASQTGKAENTIEKMIQTFKALCELADFESNS |
| 883 |
PF17099 (TrpP) |
 Estimated TM-score=0.80; easy target. |
>Tryptophan transporter TrpP (Length=172) NTKLITINALLLAIGVILHQITPVLGLPMQPDFALTMLFIIIILNDNFKITMVSGTLMGIFTAMTTKFPGGQLPNVVDKI FTAFIVFLFIALIKKSNILGRLSVEKRNNILMAIVLPLGTLVSGILFLGSAKIIVGLPASFSILFLTVVVPSVALNTVIG LVLYRIVEKSLR |
| 884 |
PF06055 (ExoD) |
 Estimated TM-score=0.80; hard target. |
>Exopolysaccharide synthesis, ExoD (Length=189) LSQELARFSRLPESPEQPEPSPEGLIRIEDILKLAGERSFGLFLAVLSFPSALPLPAPGYSTPFGVVILLLAAQMLAGRT TPWLPKWVLQTSIERKHFQKWVQAGIPWLQRIEKIARPRWKQLCTTRLGQVWLGTWIGLMGISMIIPVPGTNTLPAMGVF VTAFGLLEEDGILCSLGSLICVAGGLLTT |
| 885 |
PF10382 (DUF2439) |
 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF2439) (Length=75) VKKWSVTYTKHLKQKRKVYHDGFLELQSSNHKAMLYDDCEKLLGIKILKNDDDVKTGETLPFDSYLVDIGDPHGD |
| 886 |
PF09842 (DUF2069) |
 Estimated TM-score=0.80; hard target. |
>Predicted membrane protein (DUF2069) (Length=101) ASFSLIALILLCLAWEGWLAPLRPEGSMLILKAVPLLLPLFGILRGKRYTYQWASMFILLYFTEGVVRAWSDEGLSAQLA LLEVVLTVTFFICAIFYAKLT |
| 887 |
PF06813 (Nodulin-like) |
 Estimated TM-score=0.80; easy target. |
>Nodulin-like (Length=247) HWFMLFASLLIMAASGAGYMFGMYSNEIKTSLGYDQTTLNLLSFFKEVGATVGIISGLVNEITPPWVVLSIGIIMNFLGY FMIWLAVTARIAKPQVWQMCLYICIGSNSQTFSNTGALVTSVKNFPGSRGSLLGILKGYVGLSGAIIAQLYHAFYGDHNP QALILLIAWLPAAVSFLFLPTIRILNTVHQTNDNKVFYHLLYISLGLAGFLMVLIIVQNKLSFTRSEYIADGVVVFFFLL LPLVVVV |
| 888 |
PF01531 (Glyco_transf_11) |
 Estimated TM-score=0.80; easy target. |
>Glycosyl transferase family 11 (Length=311) LSLLCPDRSRVTSPVAILCLNPNTTFSCPKHPASVSGTWTIDPKGRFGNQMGQYATLLALAQLNGRQAFIQPSMHAVLAP VFRITLPVLAPEVDRHAPWQELELHDWMSEEYTHLKEPWLKLTGFPCSWTFFHHLRGQIRSEFTLHQHLRQEAQRSLSGL RLPRTGDRPSTFVGVHVRRGDYLEVMPLHWKGVVGDRAYLQQAMDWFRARHEAPVFVVTSNGMEWCRENIDTSRGDVIFA GDGQEDAPVKDFALLTQCNHTIMTIGTFGFWAAYLAGGDTVYLANFTLPDSSFLKIFKPEAAFLPEWVGIN |
| 889 |
PF15494 (SRCR_2) |
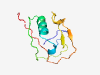 Estimated TM-score=0.80; trivial target. |
>Scavenger receptor cysteine-rich domain (Length=89) EVLSPATKLWLPVCQDSLTSFTANLICRELGYDQGHHQCCSAFGPVSSTNFSLVECTGQESDLSHCRLKTEASCFSGEYI SLACSSAGR |
| 890 |
PF10184 (DUF2358) |
 Estimated TM-score=0.80; easy target. |
>Uncharacterized conserved protein (DUF2358) (Length=112) RVIQTLKRDLPTLFEKDISYDIYTQDIYFQDPVNKFKGKLNYRIIFWTLRFHAQLFFTKIYFDLHDVNQSAENTILAKWT VRGVLRVPWKAKIFFNGYSTYKLNKDGLIYEH |
| 891 |
PF14219 (DUF4328) |
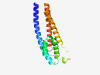 Estimated TM-score=0.80; hard target. |
>Domain of unknown function (DUF4328) (Length=159) ARDAEHADKLYSAAGGVQLVALVATCVVFLVWFHRVRVNAEAFDPHIHSKSRGWTVGSWFVPVVNLWFPRRIAIDIWDAS GDRSKSLEAPAERETPWLINAWWTLWLVSLLAGRYAGRRYSAAEEPEEINSALTGMMVSDALDIAAAVLAILFVHRLTR |
| 892 |
PF08401 (DUF1738) |
 Estimated TM-score=0.80; trivial target. |
>Domain of unknown function (DUF1738) (Length=122) DVYARLTAEIVAAIEAGAGEYVMPWHHNGSSTALPVNVASGKAYRGVNTLALWVAAQSAAYPMGLWGTYRQWQALGAQVR KGERSTTVVFWKQIGGDCDEDDSGSDDGAERRRFFARGYSVF |
| 893 |
PF16976 (RcpC) |
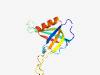 Estimated TM-score=0.80; hard target. |
>Flp pilus assembly protein RcpC/CpaB (Length=118) GFLSGVLSPGMRAITVGVDPETGGGGFVLPGDRVDLIVIFDVPDEDEVTGETTNLVVSETILQDIRILAVGQEVAIEGGG GDETLTEVAETVTLEVTPNQAQVVSVADQMGRLRLVLR |
| 894 |
PF07774 (EMC1_C) |
 Estimated TM-score=0.80; easy target. |
>ER membrane protein complex subunit 1, C-terminal (Length=236) AENWYACTFFGQYALKDAQGHALSGQSLKGYQIVVTDLYESNESNDRGPLGSAANFSSIETVDEPTGAPTPFLVSQAWVL SAPIVALAVTQTRQGITNRQLLGYQPETHGIAGLPRQVLEPRRTVGRDPTAQEVEAEGLIRYTPVIEVDPRQVITHQRDV IGVKDIIATPALLESTTLVFAYGIDIFGTRLAPSLSFDILGKGFDKVTLIGTVLALVAGVAALKPIVRRKQTDLRW |
| 895 |
PF17201 (Cache_3-Cache_2) |
 Estimated TM-score=0.80; easy target. |
>Cache 3/Cache 2 fusion domain (Length=293) TSRSATKEAERTLEQQAQALVESMASYHGALADSAGKLAAVFRTHFPNSFTIDTSKTVTVGDKQTPLLASGSMAINLNTD IVDRFTSVTKAVATVFVRTGDDFVRVSTSLKKEDGSRAIGTALDRSHPAYGGLLKGDEFVGKATLFGKDYMTKYLPVKDA QGKVIAVLFIGLDFTDGLKALKEKVKAIKVGKTGYIYVFDAKEGKDQGKLVIHPAKEGTSVIDSKDADGREFIREMIKNK NGIIRYPWMNKEVGDRFQRDKVAAYRHLKEWNWIIGVGSYQDEFNGLARLVRN |
| 896 |
PF13251 (DUF4042) |
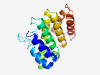 Estimated TM-score=0.80; easy target. |
>Domain of unknown function (DUF4042) (Length=182) RVRQTALGCFNAVIKGTDRRVMFGYWTSFIPDALGSGSSQSHTLFTVILKDPVPKTRTAAIAALGSLLEGSKQYLAAADD SEQHKAAFTSYSVVLGLMIKEIHRCLLVALVAENSPLAITQIIKCLATLVPNVPYQKLKPGLLSRISKQIRPFISHRDPN VRVACLTCLGAIVTVQAPLMEV |
| 897 |
PF18924 (DUF5674) |
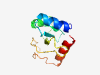 Estimated TM-score=0.80; hard target. |
>Protein of unknown function (DUF5674) (Length=109) IILEHPISRAELKKYAANTFGDMIKCVADVEKGLLAIDADLHADLERMMLENGSEQTSLWGFNLYPDEMGDDFIEYDSLI NIRSWQGNPSRDVLDKDVRERIASIVGKF |
| 898 |
PF09562 (RE_LlaMI) |
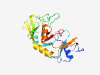 Estimated TM-score=0.80; trivial target. |
>LlaMI restriction endonuclease (Length=241) VRGRKPDTSNINQDHDGKAGHWLEHQMGIKINNKTEADLFGYEMKNQTTSGKITFGDWSADYYIFKDYKYFTSGNTGVNR DCFMQIFGTYKLDKGRYSWSGEVTPKIKNFNRYGQKLIITENKDITAIYSYDQDSRQNKKDIIPVNMQINDLILARWTSN SMKKRVESKFNDKGWFKCLQNSLGVYTNIQFGKPITFESWIDLVAEGVVFFDSGMYQGNNRPYSQWRALNNFWDSLIIET Y |
| 899 |
PF15915 (BAT) |
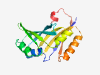 Estimated TM-score=0.80; hard target. |
>GAF and HTH_10 associated domain (Length=156) DTVVELELSLGDPALFFVDLSDRCGCLLEYSGSAPQPDGTLHMFFTVSGADPDAVVEAAADCPDVAGASLLTEHEDGALV EFALSESSLVSYLAEQGGRTKEITASDGGARLRVDLPQEANVRSVVETVQDRYPGTELLSYHERERPEKTRQEFVA |
| 900 |
PF08670 (MEKHLA) |
 Estimated TM-score=0.80; easy target. |
>MEKHLA domain (Length=139) TLAHWICNSYRCYLGVELLKSNNEGNESLLKSLWHHSDAILCCTLKALPVFTFSNQAGLDMLETTLVALQDTPLEKIFDD HGRKILFSEFPQIIQQGFVCLQGGICLSSMGRPVSYERVVAWKVLNEEENAHCQKLRFF |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417