

Go to page (83 pages in total): [First page] << < 1 2 3 4 5 6 7 8 9 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 301 |
PF09512 (ThiW) |
 Estimated TM-score=0.84; easy target. |
>Thiamine-precursor transporter protein (ThiW) (Length=149) LTFSALLVAIGVMAGNIIYIPVGAAKCFPVQHTINVISGVLLGPWYAVANAFLVSLLRNMLGTGSPLAFPGSMIGAFLAG ALYLKLGKKSWAVLGEIFGTGILGGLLAFPIAKLILGKDVAAFYFVIPFLISTIGGSIIGYSILKVMER |
| 302 |
PF10335 (DUF294_C) |
 Estimated TM-score=0.84; easy target. |
>Putative nucleotidyltransferase substrate binding domain (Length=144) IFLACLAANALEHTPPLGFFRQFVLATHGEHEDTLDLKHDGLVPIIDLARLHALAHGVPAVNTRRRLRQLAEQGALNDGD AADLQDAFAFIAQIRLRHQVRQIADGEAPDNYVSPDALSAFDRRHLKDAFRIVKTAQAAVEQRY |
| 303 |
PF10978 (DUF2785) |
 Estimated TM-score=0.84; hard target. |
>Protein of unknown function (DUF2785) (Length=175) LFAHIDEPQNDAVYQRSFAVLLLSVLLYADHAGLKFMTPARLDLIVTQMTIYILLETDTRGFVGTTGWAHAYTHIGNILD ELADESKLARADKLLLLAALLSRYQKLTTPLIFGEPERLSTYLTLITNKDELYCDYLLAALKEWHRQLMMHTRPSSEAEW TQIFNRNRLLEALAL |
| 304 |
PF02677 (QueH) |
 Estimated TM-score=0.84; easy target. |
>Epoxyqueuosine reductase QueH (Length=177) LLLHSCCAPCSSYVLEYLSEYFNITVYYYNPNIHPEKEYRKRVNEQENFIKKFNTKNKVSFIEGEYKPQHFFNMAKGLEK VREGGERCFKCYEMRLRKAAELAKQRGFDYFTTTLSISPHKNAQKINEIGQELEKEYGIKFLYSDFKKKNGYKRSIELSK EYNLYRQDYCGCVFSRR |
| 305 |
PF04843 (Herpes_teg_N) |
 Estimated TM-score=0.84; trivial target. |
>Herpesvirus tegument protein, N-terminal conserved region (Length=174) STHQANCIFGEHAGSQCLSNCVMYLASSYYNSETPLVDRASLDDVLEQGMRLDLLLRKSGMLGFRQYAQLHHIPGFLRTD DWATKIFQSPEFYGLIGQDAAIREPFIESLRSVLSRNYAGTVQYLIIICQSKAGAIVVKDKTYYMFDPHCIPNIPNSPAH VIKTNDVGVLLPYI |
| 306 |
PF08481 (GBS_Bsp-like) |
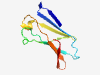 Estimated TM-score=0.84; trivial target. |
>GBS Bsp-like repeat (Length=87) ITNLNNAAGTATLSVNGVSPDDKVRAVSFAVWSKADQSDIYWYHATKVGVGGYSSNFQISNHKYNQGTYYVEGYAEDIYG ISHLLGR |
| 307 |
PF11042 (DUF2750) |
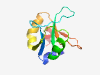 Estimated TM-score=0.84; hard target. |
>Protein of unknown function (DUF2750) (Length=101) NHFVNRVCEQEEIWILTDEHGCMMLTTDDDEDCIPVWPHADYAKDWAVGDWKDCTPEAISLNVWLKRWVPGMTDDELLVA VFPTADETGVVVEPYELKDAL |
| 308 |
PF11823 (DUF3343) |
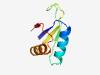 Estimated TM-score=0.84; easy target. |
>Protein of unknown function (DUF3343) (Length=68) YVIVFESTHYAIAAEKMLKEKNYQFDTIPTPREITASCGLSIRFNEESLGYIKKDIEESNIKIKGIYE |
| 309 |
PF10647 (Gmad1) |
 Estimated TM-score=0.84; easy target. |
>Lipoprotein LpqB beta-propeller domain (Length=262) GSMVSVTETGVTPVPGYFGSARNMRSLALSQDGKLVAAVADTGRPAPEPASSLMVGAYEDGAASVLEGGAITRPTWAPDN SAIWAAVNGNTVIRVLREPGTGRTSVVNVDAGAVTALGATITELRLSRDGVRAALIVDGKVYLAIVTQTPGGQYALTNPR AVAIGLGSPALSLDWSTSDTIVVARAASDIPVVQVAVDGSRMDALPSRNLTAPVVAVDASTTTEFVADSRAVFQLNNNDP AGDRYWREVPGLTGVKAIPVLP |
| 310 |
PF09933 (DUF2165) |
 Estimated TM-score=0.84; hard target. |
>Predicted small integral membrane protein (DUF2165) (Length=155) RLSKVALVAAIAFFASLVSFGNITDYGTNYEFVQHVFDMDTIFPNSTIKYRAITSPALHTAGYILIIAAETLTAILCWIG TYAMLTKLGAPAAAFNNAKKWAIAGLTLGFLTWQVGFMSIGGEWFGMWMSHQWNGIDSAFRFFITIIAVLIYVSM |
| 311 |
PF10909 (DUF2682) |
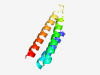 Estimated TM-score=0.84; trivial target. |
>Protein of unknown function (DUF2682) (Length=77) LAVVQDAALDLAAEAQSFLDSGDPAAFPGLESRLTQLLYQTDAVRFGSDQTGLNNLKANVKNCINIFIDLITIKHYT |
| 312 |
PF13826 (DUF4188) |
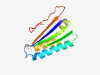 Estimated TM-score=0.84; trivial target. |
>Domain of unknown function (DUF4188) (Length=117) IVVFLIGMRVNKWHAVHKWLPVFMAMPAMIRELYQNKDLGLLSMESYFGLRTTVMIQYWQSTEDLHAYARGQKHLTAWRD FNKKVGDNEAVGIYHETYIVQKGDYETVYGNMPLYGL |
| 313 |
PF14558 (TRP_N) |
 Estimated TM-score=0.84; easy target. |
>ML-like domain (Length=138) LEAKSLNPCMENSKFTATLFSVVFTPDNRTVTFNINGISSISGNVTAELEVIAYGYTAVRQSLNPCKMNLEGMCPMNTGQ INLPGNIPDISPDVVNQVPGIAYTVPDLDGTVKIYINSTDTNTPLACVEAQMSNGKTV |
| 314 |
PF17159 (MASE3) |
 Estimated TM-score=0.84; hard target. |
>Membrane-associated sensor domain (Length=224) SFYSYLLFHSLIELFSIVVAFAIFILAWNSRQFLENNYLLFIGIAYLFIGGIDLIHTLAYKGMGVFPGYDSNLPTQLWIA GRYVESISLLIAALVLGRKLRPYLVFLGYAVGISLLLMSIFYWGVFPQCFIEGVGLTPFKKVSEYIISLILAGSMILLFR SRRQFDKGVLRLLVASIAITIGSELAFTFFVDLYGLSNLIGHLFKVISFYLIYKALIETGFRKP |
| 315 |
PF09983 (DUF2220) |
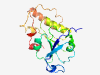 Estimated TM-score=0.83; easy target. |
>Uncharacterized protein conserved in bacteria C-term(DUF2220) (Length=179) GFRDKPLRVRFRILDPKLALLPTNTDQDITLTQATFARLEIPVTKIFITENEINFLAFPEVPEAMVIFGAGYGFENMASV EWMRDRVIHYWGDIDTHGMAILNQLRRFFPQAASLLMDHETLMEHQPLWGAEPSPETGTLTRLTAQEGALYDQLRRNELG SRIRLEQEKIGFEWLVEAL |
| 316 |
PF14430 (Imm1) |
 Estimated TM-score=0.83; easy target. |
>Immunity protein Imm1 (Length=121) IPVTLSTVDEVDALLDRVRVESPPAAPMLMDVHLSGDPYAQGLDVGVAVDRGVIRYSGREWPHGVVSTGDGSADGEPRSY FYMGHGREFPANSEVPIDVIRKAVKEFMESNGARPTCVRWQ |
| 317 |
PF06685 (DUF1186) |
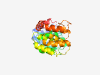 Estimated TM-score=0.83; easy target. |
>Protein of unknown function (DUF1186) (Length=253) QVSNALFELTINSGIFPREALTKAIELREQVTPRLLGYLETTEKYAYEQRDNDNLMLHIYALYLLAQFREQRAYPIIINF FSLPGKISLEITGDVVTEDLHRILASVYDGDDTLLKNLIEDEQVNEYVRDAGIRTFVTLVVCGQKSRDEVIAYLKTLFGK LERKPSFTWNSLVYVCMDLYPEEVDAEIRQVAKEGLTEEVLLTKEINILGLHEEALKKSYALGKEKTLEKLQNDDHYTLI DDTIRELKDWSCF |
| 318 |
PF13699 (DUF4157) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF4157) (Length=78) PLPDPAREFMESRFENDFSGVRLHTDSEAVQLSHNLRAKAFTHENDIYFNAGQYNPNSVGGRRLLAHELTHTIQQTGA |
| 319 |
PF10826 (DUF2551) |
 Estimated TM-score=0.83; easy target. |
>Protein of unknown function (DUF2551) (Length=83) LKGYLSRDRDGIRHELLNLFVKVKSLTIPQIYEKLQKQFSISYHSIASMVGIIASRIGILHVRRNAEGTNTIYELKDQYV DVV |
| 320 |
PF14062 (DUF4253) |
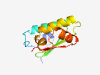 Estimated TM-score=0.83; hard target. |
>Domain of unknown function (DUF4253) (Length=108) LALVPARRSADIPAAIGWTGPLNHENDVARLCAVLRTWEDRFGARVVAIGFATLAVSVAAPPATQEEAEAIAAEHYAFCP DNIEQGSGTIEAYAKDLIGAPSWHFWWD |
| 321 |
PF01810 (LysE) |
 Estimated TM-score=0.83; easy target. |
>LysE type translocator (Length=192) IFIVLVPGPNTLFVLKNSVSSGMKGGYLAACGVFIGDAVLMFLAWAGVATLIKTTPILFNIVRYLGAFYLLYLGIKILYA TLKGKNSEAKSDEPQYGAIFKRALILSLTNPKAILFYVSFFVQFIDVNAPHTGISFFILAATLELVSFCYLSFLIISGAF VTQYIRTKKKLAKVGNSLIGLMFVGFAARLAT |
| 322 |
PF10134 (RPA) |
 Estimated TM-score=0.83; easy target. |
>Replication initiator protein A (Length=228) KNDLASMEHPLFSLGTRPDRRILTYEHNGAEITVTPSVKGLATIHDKDILIFCVSQLMAALNAGRQVSRTLHLTAHDLLV ACNRETSGDAYRRLREAFERLAGTRITTNITTGGQEVTSGFGLIDSWEIVRKARGGRMVSVAVTLSDWLYRAVLSKSVLT LSRDYFRLRKPLERRIYELARKHCGRQDSWQISVETLLKKSGSASPRRVFRKMIRDMIEAQPLPDYEL |
| 323 |
PF07456 (Hpre_diP_synt_I) |
 Estimated TM-score=0.83; trivial target. |
>Heptaprenyl diphosphate synthase component I (Length=146) LVAQALVLHVIERAIPVPFITPGAKLGLANLVTVIALYTFGFKETFAIVVLRIIMAALLGGSLSSFLYSIAGGILSFFVM FVVMQLGEENVSAIGVSVAGAVFHNIGQVLVASMIVQNIRIVSYLPVLMVAGLGTGIFIGLTARYL |
| 324 |
PF08444 (Gly_acyl_tr_C) |
 Estimated TM-score=0.83; easy target. |
>Aralkyl acyl-CoA:amino acid N-acyltransferase, C-terminal region (Length=81) FCLLGPKGTPVSWCLMDHTGELRMGATVPEYRGQHLIYHSSLYQIQSLVKLGFPLYAHTDKANHIIKKTSVRLRYVPMPS E |
| 325 |
PF12679 (ABC2_membrane_2) |
 Estimated TM-score=0.83; easy target. |
>ABC-2 family transporter protein (Length=303) WNLALKELYVSVKSRRFIVIVVLYFIIFGLAVYSIKDYLVQMGVPSVDSNELGLWGVSAEIYTTPLAMLFTVNMTIITVL GAVLGVALGADAINREVESGTVRVLLGHPVYRDEIINGKFLGMGLLIAVTYMVSYIVMIAVMLILGIPLDGESLFRSFLA ILVTMLYTMVFLSLGILLSTLSKKPETSMLAGVGLAIFLTVFYGIVVGIVAPKLAGPEPPIGTSAYQIWADELHVWMNRL HSINPAHHYVQLVSYIFAGDRFFNYYIPLSDSLIYGFNNLAALLVMLLLPFALAYARFLTSDL |
| 326 |
PF06902 (Fer4_19) |
 Estimated TM-score=0.83; easy target. |
>Divergent 4Fe-4S mono-cluster (Length=64) YTGENIDVYFNTAMCQHSGNCVRGNANIFKLSRKPWIMPDNASVEEVKRVIDTCPSGALQYREK |
| 327 |
PF14082 (DUF4263) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF4263) (Length=165) YNESQWQNEILQIILLLYPKYIYVIEEAKIRDTYNQKTRRLDFLLIDSSGNTDIVEIKQPFDQCIVSRNTYRDNYIPVKE LSGSVMQVEKYIFYLNKWGKKGEQALTEKYKKELPDEFQIKITNPSGIIIMGREHNLSLEQRQDFEVIKRKYKNVIDIIT YDDLL |
| 328 |
PF13550 (Phage-tail_3) |
 Estimated TM-score=0.83; trivial target. |
>Putative phage tail protein (Length=157) TTQRKAWDVLSDFCSAMRCMPVWNGQTLTFVQDRPSDKVWTYNRSNVVMPDDGAPFRYSFSALKDRHNAVEVNWIDPNNG WETATELVEDTQAIARYGRNVTKMDAFGCTSRGQAHRAGLWLIKTELLETQTVDFSVGAEGLRHVPGDVIEICDDDY |
| 329 |
PF05272 (VirE) |
 Estimated TM-score=0.83; easy target. |
>Virulence-associated protein E (Length=203) YLNTLVWDGTERIRFCLRHFLGADADDYTYEALKLFLLGAISRAFQPGCKFEIMLCLVGGQGAGKSTFFRLLAVRDEWFS DDLRKLDDDNVYRKLQGHWIIEMSEMMATANAKSIEEIKSFLSRQKEVYKIPYETHPADRPRQCVFGGTSNALDFLPLDR SGNRRFIPVMVYPEQAEVHILEDEAASRAYIEQMWAEAMEIYR |
| 330 |
PF14771 (DUF4476) |
 Estimated TM-score=0.83; hard target. |
>Domain of unknown function (DUF4476) (Length=91) ITDANFNTLVTNVKNKWSQSLKGEAIREALVNSNYYYTSTQVRQLMGLVSSETDRMDLIKLAYPVVSDTNNFPGLYDVFA STANRTEFNEF |
| 331 |
PF13349 (DUF4097) |
 Estimated TM-score=0.83; easy target. |
>Putative adhesin (Length=243) IDSMVIESDSKDVRIIAEERSDISAGISGDSGKLFVTENNRKLELTVKEKEFQFLNGFNRSTLIVRLPYDYKGDLAVRTS SGDVSVAGNDHLALSRLNTVSASGNASVTDVRLQDLKVKASSGDVTISNTVSKTSGIDLASGDANLVHVSGSLDVKMTSG DFNAVLTKITGPVSVTLTSGDANLSLPKNGSFTVKAKSASGDVSSPYSFADKAHKEQHHITGTQGSGRHPIDIKTDSGDL AIR |
| 332 |
PF08862 (DUF1829) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function DUF1829 (Length=87) GKSGYVHNFDFVIPASKKSPERIIKTINNPNKNNTTSVLFSWSDTRENRTLSSKMYVFLNDTEKEVKPEIINAYIQYQIK PVLWSKR |
| 333 |
PF12264 (Waikav_capsid_1) |
 Estimated TM-score=0.83; trivial target. |
>Waikavirus capsid protein 1 (Length=197) EQAFQDEEKRTVDPNISDMYNAIKSEYLVKSFSWKVSDGQDKVLSNINIPEDLWNTNSRLNDIMSYFQYYKATGLTFRIS TTCIPMHGGTLFAAWDACGCATRQGIATAVQLTGLPGIMIEAHSSSLTTFSVEDPLTQSTVCLSGSEHSFGRIGILKICC LNVLNAPQAATQSVSVNVWVKFDGVKFHFYSLKKQPV |
| 334 |
PF10354 (DUF2431) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF2431) (Length=169) LLVGEADFSFAASLAAAFGSSVSNLTATSLEYKRELEKVYSKAVDNIQELKSKGCNVFHAVDATVMASHPSFKNSVFDRI VFNFPFAVISEEEWGNLHLPAVLEKQHSLVREFLENAKKLISENGEIHITHRKDGIFGAWRIKFLASEQGFQVVEAVDFN ICDYPGYNP |
| 335 |
PF07788 (PDDEXK_10) |
 Estimated TM-score=0.83; trivial target. |
>PD-(D/E)XK nuclease superfamily (Length=74) SDVEYDIVIKDGSVILVEITSSLKRGDLPTIKRKKEFYEKVKNIKVNLVYVVTPFIHDRYPERVKAMAKDMGIE |
| 336 |
PF09792 (But2) |
 Estimated TM-score=0.83; hard target. |
>Ubiquitin 3 binding protein But2 C-terminal domain (Length=141) EYPHLIVPINSSTPDTAYGTQYFGTVSSTVSTIFNFDIPSSDSGKTCSLIFLFPTKDQLETSDYTFSGDGKIDFSQLESA ASESTTYNNAPGVKQDYGDFTVSPGNSYLISTFSCPAGQTVSYEMKEAGSTYLNFFEDYNP |
| 337 |
PF17747 (VID27_PH) |
 Estimated TM-score=0.83; trivial target. |
>VID27 PH-like domain (Length=108) EILAKETAELHLFDFPSGTFVLQDQEVTATVSEIGTWQYWLQISGKDKEWLGQPVVADINPVFNFEYLSFIFNHYTEDGS AYSWLLRFKDQPTEERFQEGLMQALWEQ |
| 338 |
PF02988 (PLA2_inh) |
 Estimated TM-score=0.83; easy target. |
>Phospholipase A2 inhibitor (Length=82) DCEHCVVWGQNCTGWKETCGENEDTCVTYQTEVIRPPLSITFTAKTCGTSDTCHLDYVEANPHNELTLRAKRACCTGDEC QT |
| 339 |
PF14321 (DUF4382) |
 Estimated TM-score=0.83; hard target. |
>Domain of unknown function (DUF4382) (Length=142) TGTVNMYISDQQNAIDQFDHLTVTVTSVAAHRVDDPDTEANESGWVEQEIDNVTVDLTELQGENASKLGAIEAPNGTYNK VFVHVDSDINATLADGSSQTVKLPSNKLRLNTEFTVGNGEEIDFVFDMTVFERGNGDYILKP |
| 340 |
PF09938 (DUF2170) |
 Estimated TM-score=0.83; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2170) (Length=137) WTAQALHEALAATELFQSGQAGIELIQGADASLHITMREYGDLPLFIAVFGEQIIVEALLWPASDVKDAAAFNEEVLRTH KLFPLSSIGLEKMGDGRDCYTMFGALSSSSTLSDVVFEIELLADNVIKATEAYEGFL |
| 341 |
PF09970 (DUF2204) |
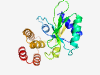 Estimated TM-score=0.83; easy target. |
>Nucleotidyl transferase of unknown function (DUF2204) (Length=181) LNDLSWLFRKLQEHGIKGVVIGSTVIELALRRKSFEDDVDVFALEPSPLIEEDFYRSVAEREDWQVSYTALGTPKFIVKL PSGEEVIVEFYENIHDFYVPPEILEQAPKKSIGGVEIRVIRPEDYIVLKAKAAREPDIEDLRIIREYIDEGKLKIDERIV KRDIELLPEEEQGFVVNKLRE |
| 342 |
PF12386 (Peptidase_C71) |
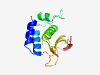 Estimated TM-score=0.83; easy target. |
>Pseudomurein endo-isopeptidase Pei (Length=141) LKGHWTTKVIEKIGTFHDATSLYERVKKTCKYKYYYNDQVPNHVAVMRMTTSGINCTDACQLFSKVLEEMGYEVKIEHVR VKCNDGKWYGHYLLRVGGFELKDGTIWDYVSATKTGRPLGVPCCTAGFQHLGWGIVGPVYD |
| 343 |
PF07209 (DUF1415) |
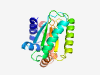 Estimated TM-score=0.83; very hard target. |
>Protein of unknown function (DUF1415) (Length=169) VIAATEEWLEKAVIGLNLCPFAKSVHVKKQVRYVVSEATTPEELLEQLMDELQLLADTEPEKVDTTLLIHPHVLTDFEDY NEFLDVADAALEDMELVGELQVASFHPDYQFADTEKDDIGNYTNRAPYPTLHLLREDSIDRAVEAFPDAEDIFEKNIETM EKLGHEGWA |
| 344 |
PF13576 (Pentapeptide_3) |
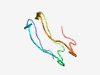 Estimated TM-score=0.83; trivial target. |
>Pentapeptide repeats (9 copies) (Length=44) EGADLTGADLSGASFAGASLARVTLTRAKLTGVTLDGVSFNGST |
| 345 |
PF13464 (DUF4115) |
 Estimated TM-score=0.83; hard target. |
>Domain of unknown function (DUF4115) (Length=72) VVLRARAESWVMLREQATGRVVFDRVLQPGESHAVPDRAGMLLTTGNAPGLEVLVDGEPLPALAGAGRVRRD |
| 346 |
PF14273 (DUF4360) |
 Estimated TM-score=0.83; very hard target. |
>Domain of unknown function (DUF4360) (Length=179) VQVKSINAAGTGCPLGTIQKNISDDKKAFTMTFSEYFAEIYDGSFPGDSRRNCNLTIDLDFPEGWTYSLVSFDYRGYVFL DDQVEATAKAAYYFQSESEGVEFQSQITGFADEDYHFRDELGIDSVVWSPGCSGARRPLNLQTQIRVNNEENAGGEGFIT VDSIDGEFTQKYGLVWRRC |
| 347 |
PF06059 (DUF930) |
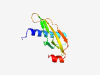 Estimated TM-score=0.83; hard target. |
>Domain of Unknown Function (DUF930) (Length=98) EKLDPQTRLEQRCDTEAMERIGADKNRFKPDKVIAYTFADPVMKGDKMKATGAVFRSKGDWYKLAFRCKTDSEHLEVLSF EYKIGERVPRENWDQLYL |
| 348 |
PF15970 (HicB-like_2) |
 Estimated TM-score=0.83; easy target. |
>HicB_like antitoxin of bacterial toxin-antitoxin system (Length=81) NYPATSHFDEESETYEITYRDFDNIHAVAYTEDDIELEASDILHVGLEEFISSKMPIPMPSEPQSGDFIVYLPPISCLKI A |
| 349 |
PF05150 (Legionella_OMP) |
 Estimated TM-score=0.83; easy target. |
>Legionella pneumophila major outer membrane protein precursor (Length=304) TVPCERTAWDFGVQALYLQPTLSAGNGYDFYNTDFNGVRRYHDFDPDWGWGFKLEGSYHFHTGNDLTVSWYHMNKSTDNH FGDFVLPNLNIVTANGIQLGNSLRTSFEPKWDAVNVELGQHVDFSEWKVIRFHGGVQYARIEHDYDANLTTPVSFPRYGV VAGVPFRLASAETKFNGFGPRVGADMSYNLGNGFAVYANGATALLIGQGKFRSSFGYVPSLTTPLFGIPGHGSKTTMVPE MEAKLGAKYTYAMAQGDLTLDAGYMWVNYWNANHILVGHSDGTYSSRESNFALSGPYVGLKWVG |
| 350 |
PF17263 (DUF5329) |
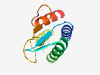 Estimated TM-score=0.83; hard target. |
>Family of unknown function (DUF5329) (Length=93) KATAEIDALLNFVEHSDCQFIRNGSEYSGSKARAHLQQKLDYLENKDLVNSAEDFIERAASKSSMSGKPYQVSCPGGKQE ASTWLTTELKRLR |
| 351 |
PF14871 (GHL6) |
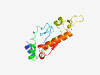 Estimated TM-score=0.83; easy target. |
>Hypothetical glycosyl hydrolase 6 (Length=139) DKEEFAKTLEEAHVDSITCFARCHHGWLYYPSKKFPELIHPGLKNKNFLLEQIEACHKRGIKVPVYTTAQWDGRIMREHP EWLAVDENGEFIDTQGVPAPHFYHTICLNSGYRQFFKDQLQDMIEVIGVENLDGIFMDI |
| 352 |
PF18451 (CdiA_C) |
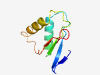 Estimated TM-score=0.83; easy target. |
>Contact-dependent growth inhibition CdiA C-terminal domain (Length=83) NPDFLIDGKPFDCYAPDVNTSINNICRTITKKTKKQAERIVLNLDDYPIEKVDELIENISRKANPNGDLKHLQELFIVKD GKI |
| 353 |
PF07014 (Hs1pro-1_C) |
 Estimated TM-score=0.83; trivial target. |
>Hs1pro-1 protein C-terminus (Length=259) MRRCPYTLGLGEPNLNGKPNLEYDAVCKPSELHSLKRNPYDHIHNHENQTVYTAHQILESWIHVSQQLLKRITERIESQK FEKAASDCYLIERIWKLLSEIEDLHLLMDPEDFLRLKNQLNVKSFNETKPFCFRSKALVEIAKQCKDLKHKVPYILGVEV DPMGGPRIQEAAMKLYSEKREFEKIHLLQALQAIESAMKRFFFAYKQLLVVVMGSLEANANRVVVSSDSCDSLSQIFLEP TYFPSLDAAKTFLGDFWNH |
| 354 |
PF03444 (HrcA_DNA-bdg) |
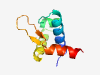 Estimated TM-score=0.83; trivial target. |
>Winged helix-turn-helix transcription repressor, HrcA DNA-binding (Length=78) ELTPSQKNILQELVNLYRESESAVKGEDIADKVDRNPGTIRNQMQSLKALQLVEGVPGPKGGYKPTATAYDALQIQEM |
| 355 |
PF08889 (WbqC) |
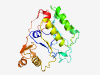 Estimated TM-score=0.83; very hard target. |
>WbqC-like protein family (Length=215) QPSYLPWLGFFEQMTRSDKFVLLDDVQYTRRDWRNRNKVRVKEGWVWLTVPVQQKSRFSQSLLETRIDNSVSWRRKHLET LRQHYCKAPFFEKYFPRCQQIYEKDWEFLFDLCLETIQFLKEEMGIETPLLRSSEMKLSGEKTERLVSICRELGATHYLS GESGSDYISQEDFSNQGIELEYQNYEHPVYPQRYPGFVPHLSTIDLLFNCGEQSL |
| 356 |
PF09335 (SNARE_assoc) |
 Estimated TM-score=0.83; hard target. |
>SNARE associated Golgi protein (Length=119) IPAIPLTMSAGLLFGSLTGTILVSISGTVAASVAFLIARYFARDRILKMVEGNKKFLAIDKAIGENGFRVVTLLRLSPLL PFSLGNYLYGLTSVKFVPYVLGSWLGMLPGTWAYVSAGA |
| 357 |
PF11695 (DUF3291) |
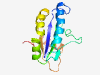 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF3291) (Length=137) LAQLNIAWMKAPLESPEMADFVANLERINALAEGSPGYVWRLQDEAGDATAIRPFGDEVLVNMSVWEDVQSLSDYVYKSA HTEMLKRRREWFERVEQAHQVLWWVPAGHRPSVTEAAERLAHLREHGATAQAFTFRH |
| 358 |
PF11454 (DUF3016) |
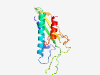 Estimated TM-score=0.83; hard target. |
>Protein of unknown function (DUF3016) (Length=139) GPVQVRWTDPAQFSDIRYSGNRWEAQRGDWVTQLATYFQRSAERQLPDGQRLSVTITDIRRAGQYEPWHGPRMQDVRVVK DIYPPRMSFEYTLTDASGAVIDQGERKLVDSAFLMSGPRMNDTDPLRFEKAMIDDWVRK |
| 359 |
PF06940 (DUF1287) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF1287) (Length=162) VNDARKQINVTVSYDSTYRKMAFPMGDVPQHTGVCTDVVIRAYRQQQIDLQKLVHQDMKKAWVSYPKIWGLKTTDTNIDH RRVPNLETFFKRHGKSLSLTDLSSFKAGDIVTWRLPRNLPHIGIVSDKKNKEGIPLIIHNIGQGTQEEDILFKYKMIGHY RY |
| 360 |
PF17656 (ChapFlgA_N) |
 Estimated TM-score=0.83; easy target. |
>FlgA N-terminal domain (Length=73) HSVEQFLRTQTTGLPGQVKISVGNLDSRLNLADCDALEPFIPTGSRIWGKTTVGVRCNAPANWTIYVQADIRV |
| 361 |
PF16238 (DUF4897) |
 Estimated TM-score=0.83; very hard target. |
>Domain of unknown function (DUF4897) (Length=158) FEIVYYRTKMEYDYNGSAVFTTTAGLFFKDKNKQASYVQQYQQVSLDTFRKYFEDVSKKIGREIEVVSMESSINERAGIL EIVEVAKLNNAAVVTDGVVDTQMKDITLSASGDSEIVIVIPKDAVVVSVEPTPTKIVENQIYWQPIGSRMAFPRVVFK |
| 362 |
PF07485 (DUF1529) |
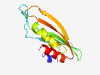 Estimated TM-score=0.83; hard target. |
>Domain of Unknown Function (DUF1259) (Length=119) AVGQALGKEGSMQPGDVYKVSLPRSDLQVTVGGVAVKPALALGSWVAFKKMGDTAMVMGDLVLTEDEVTPVMTKLQEGGV MQTALHNHVLHESPRVMYMHINATGDAVKIAKAIHDALS |
| 363 |
PF14385 (DUF4416) |
 Estimated TM-score=0.83; hard target. |
>Domain of unknown function (DUF4416) (Length=160) PAKLVISLFMNDQSIVQRVLALLEEKFGSIDMISPWFDFDYTNYYHREMGSPLFRRVVVFKALVAQDSLAEIKISTNNIE AMWKENGKRAVNIDPGYLVKSRFILATGKDFAHRVYIGQGIYADLTLMYQKGRFIDLDWTYPDYRGNAIKSFLEIVRAKY |
| 364 |
PF14252 (DUF4347) |
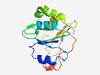 Estimated TM-score=0.83; hard target. |
>Domain of unknown function (DUF4347) (Length=160) VVFIEDNVADYQTLIDGIGAGIEVVLLDSTQDGLAQMALWAQSNSDYDAIHIISHGAEGQVNLGGFSLDNFTVNTRSADL AQLGAALNEDGDLLLYGCEVASGEGQDFIIALAQATQADVAASDDLTGSAVLGGDWQLEFQQGQISETQFLDDSVEDIYH |
| 365 |
PF00609 (DAGK_acc) |
 Estimated TM-score=0.83; easy target. |
>Diacylglycerol kinase accessory domain (Length=156) VMNNYFGIGIDADLCLDFHNAREENPNKFNSRLRNKGVYVKMGLRKMVGRKMCKDLHKAIRLEVDGKLVELPQLEGIIIL NILSWGSGANPWGPEKDDQFSKPNHWDGMLEIVGVTGVVHLGQIQSGLRGAMRIAQGGHIKINLKSEIPVQVDGEP |
| 366 |
PF11265 (Med25_VWA) |
 Estimated TM-score=0.83; easy target. |
>Mediator complex subunit 25 von Willebrand factor type A (Length=214) DHTVQADIVFVIEGTAINGAYLNDLKTNYLLPTLEYFSQGNLEDRDYVCEGTSTLYGLVVYHAADILPKSSCDCYGPYSS PQKLLMTLDKLELIGGKGESHANIAEGLATALQAFEDLQQKREGNNIITQKHCILVCNSPPYFMPVMEASTYSGHTIDQL AAILNEQNINLSILSPRKIPAIFKLFEKAGGDLTQSQNKNYAKDPRHLVLLKGF |
| 367 |
PF14280 (DUF4365) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF4365) (Length=133) ERIGVSAVSKCIAQMGLIFREQPTDDYGIDAQIETFDRGYASGRLIAVQIKSGDSYFDEVTNGNIIFRGDIKHYNYWINH SLPVILVLYNPQDDKCYWKEVNEETAVLTDKSWKIEIPMTNVLDRSAKSQLVR |
| 368 |
PF03594 (BenE) |
 Estimated TM-score=0.83; trivial target. |
>Benzoate membrane transport protein (Length=378) QDWSLSATIAGFLAVLISYSGPLIIFFQAAQKAHVSTEMMISWIWAISIGAAVTGIFLSIRFKTPVVTAWSAPGTALLVT LFPNISLNEAVAAYITAAIVIFLVGITGYFDKLLKWIPQGIAAGMMAGILFQFGLGLFLATDTLPLIVFSMLLCYLISRR FSPRYCMLWVLICGVAFSFFLGKMNPVPLDFQIARPQFIAPEWSWFSTLNLALPLILVSLTGQFLPGMAILKLSGYNTPA KPIITAASLASLFAAFAGGITIVLASITATLCMGKDAHELKDKRYIAGIANGLFYILGGVFAGSIVALFSLLPKELVAAL AGLALLGAIASNIKIAMQEDQQRDPALITFLATVSGMHFLGLSSVFWGMCIGMLAYFI |
| 369 |
PF03222 (Trp_Tyr_perm) |
 Estimated TM-score=0.83; trivial target. |
>Tryptophan/tyrosine permease family (Length=400) KSPSLLGGAMIIAGTAIGAGMLANPTSTAGVWFIGSILALIYTWFCMTTSGLMILEANLHYPTGSSFDTIVKDLLGKSWN IINGLSVAFVLYILTYAYITSGGGITQNLLNQAFSSAESAVDIGRTSGSLIFCLILAAFVWLSTKAVDRFTTVLIVGMVV AFFLSTTGLLSSVKTAVLFNTVAESEQTYLPYLLTALPVCLVSFGFHGNVPSLVKYYDRDGRRVMKSIFIGTGLALVIYI LWQLAVQGNLPRTEFAPVIEKGGDVSALLEALHKYIEVEYLSVALNFFAYMAISTSFLGVTLGLFDYIADLFKFDDSLLG RTKTTLVTFLPPLLLSLQFPYGFVIAIGYAGLAATIWAAIVPALLAKASRQKFPQATYKVYGGNFMIGFVMLFGILNIVA |
| 370 |
PF08821 (CGGC) |
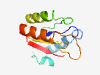 Estimated TM-score=0.83; hard target. |
>CGGC domain (Length=104) KIAVVRCETVSEVCPGTGCFKAFNEKKVHFDRYDKKDEIIGFFTCGGCPGRRVYRLIKALKKHGASAVHLSSCMKTKKYP PCPHIDSIIKIIEDAGMEVVEGTH |
| 371 |
PF12371 (TMEM131_like) |
 Estimated TM-score=0.83; easy target. |
>Transmembrane protein 131-like (Length=81) TLSPDELNFNGVPVGSSRTIPVTALNTGTAALEISAITIEGADFALGAVVAPVTLQPNDTLTFDVIYQPSSPGEASGVVR V |
| 372 |
PF13761 (DUF4166) |
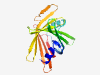 Estimated TM-score=0.83; very hard target. |
>Domain of unknown function (DUF4166) (Length=186) LHPELRRRFSVGLASGELCVGRGVMDRIWHGRAFVKPFLALGGLRNILLPRTGRDVPFTIENVPYLDSYGRETVTFVRTF DLPGGPRRFDATMVHSPERDCVLDYLGTHQHLASDLHPTVEPDGSLLITSGEHRFREGPVDVRVPELLAGRAEVRESFDE RTGRFRIDVRVVNRHFGPLFGYQGSF |
| 373 |
PF06904 (Extensin-like_C) |
 Estimated TM-score=0.83; easy target. |
>Extensin-like protein C-terminus (Length=172) RCAETLATSGLGYTVQADSPASATCPLSNVVRVQDAPVRLSSSFLATCPVAVAFALFERHGLQPAAQRVFGQPVVQVDHL GSYACRNMYNRAEGRRSQHASANALDIAGFRLKDGQRIELQKDWNGEGPKAQFLRQVHAGACESFNTVLGPDYNAAHHNH FHLDMGGWQICR |
| 374 |
PF18180 (LD_cluster3) |
 Estimated TM-score=0.83; easy target. |
>SLOG cluster3 family (Length=170) EYAATADVVAITAAVSALVHVTLGRRQLVWGGHPAISPMIWVVAQEMGIEYGRWVRLYQSRFFQDEFPEDNDRFQNVIYT PNVPEDREKSLLLMREQMFSENQFKAGVFIGGMAGIVDEFELFSRLQPNAAVVPVVSAGGAALQVAERIGVIAPDLRDDL DYVALFHRYL |
| 375 |
PF18299 (R2K_2) |
 Estimated TM-score=0.83; easy target. |
>ATP-grasp domain, R2K clade family 2 (Length=146) FLGRKIWKSRINYIAKHPETWNVFIKPTTLTKKFTGRVVRSPKDLISCGDEFEDTEIWCSEPVKFLAEWRCFVRHGEILG VKSYYGDWRVHFDPQVIEQAIAAYTSAPAGYAADFGVTDKGETLLIEVNDGYSIGAYGLFPPDYAR |
| 376 |
PF10223 (DUF2181) |
 Estimated TM-score=0.83; easy target. |
>Uncharacterized conserved protein (DUF2181) (Length=236) DGLHIKWYHGANSLSEMHEAIAGDHMMLEGDIILRWWGLPNQTEEPVMAHPPAVNSDNTLDNWLTQILNREDKGIKLDFK GLQVVRPSLELLKEKENLVNHPIWANADVVAGPGSSADPIDPKQFLSIVNEVWPNMTLSLGWTTGCCEPFTRQMMEDMLE VCHGVTQPVTFPARAASFRASWEHFKWLLEQNPESYTITVWTPSGAEDWEGTDIYDLLYVRNDCDKTLIYYDLPGE |
| 377 |
PF14269 (Arylsulfotran_2) |
 Estimated TM-score=0.83; trivial target. |
>Arylsulfotransferase (ASST) (Length=303) QAAKLHGEDVLFSFEGDHNPSYGHGHGHVTLLNNNYETIRELRAGNRKLSDKHEFQILNEKTAVIQIHQPIPYDLGPFGG TKKQQWIVNSIFQELDIETGEVLFEWSSLDHVSPHETVFSVEDGSVGSGHNSSDAWNYFHINSIDKDSDGNYLLSARICA SIYKINGKTGEVIWKLGGLPGITSSTITSVGNFTFSFQHHARYLETSEDKSRQLISLFDNSAHGSENKDGKVKLNGISSA KIIEINTKTWEATLLYNAEAPYNFSVKSQGSAQVLPNKNVLVNWGSLGAVTEYNANEEIVFHA |
| 378 |
PF11210 (DUF2996) |
 Estimated TM-score=0.83; trivial target. |
>Protein of unknown function (DUF2996) (Length=121) AVEDKPFAEFIQQDYLPALQTALTKQGVEDLDVSFAKQKIPVAGMSSAGDCWQVVGKFQSGQRQFNLYFPQENIQSQRAF SCAENNTKTSTLEPFLIDERKITLDLMVFGVVQRLNAQKWL |
| 379 |
PF14119 (DUF4288) |
 Estimated TM-score=0.83; hard target. |
>Domain of unknown function (DUF4288) (Length=82) WYVAKIIFQIVSGEGNHQPQFDEQLRLVKADDEKQALEKACDLGRKNQDRFVNSQQQTVQWHFIDVAELNKLSELADGTE LY |
| 380 |
PF12857 (TOBE_3) |
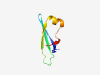 Estimated TM-score=0.83; easy target. |
>TOBE-like domain (Length=58) GEGGIAAHVSHILSVGPIVRLELMRNDGKDKELIEVEISRERFRELQLTRGDQVFIKP |
| 381 |
PF05506 (DUF756) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF756) (Length=83) PEVRVCYDIANGNVYLQMRNDGKAPCKFTVRAKAYRDDGPWNATVNGGAQAEQHWELAASGQWYDFAVTCDSDPAFYRRF AGR |
| 382 |
PF12616 (DUF3775) |
 Estimated TM-score=0.83; hard target. |
>Protein of unknown function (DUF3775) (Length=72) EELKQFIEALNEEEQIELVALAWLGRGSFSIEEWEDAKAEAQRAHNDRTADYLLGMPMLADYLAEGLAAFGI |
| 383 |
PF14305 (ATPgrasp_TupA) |
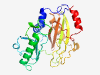 Estimated TM-score=0.83; easy target. |
>TupA-like ATPgrasp (Length=241) EKLQWLKLYDRHPEYTQMVDKATVKEYVASIIGEKYIIPTLGVWENFDDIDFENIPSSFVLKCTHDSGGVVICENKNLFD VEKARSKLLKSMKKNYYYVGREYPYKDIKPRIIAEKYMVDESHIELKDYKIHLFHGIPNFIQVDFDRFTSHHRNLYTIDW KPLGKAIKYPAVYEREIDKPIVLDEMLELAMKLAFNIPYVRADFYCIGNKVYFGELTFYHGSGFETFSPKSFDEELGALI K |
| 384 |
PF10230 (LIDHydrolase) |
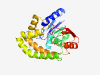 Estimated TM-score=0.83; easy target. |
>Lipid-droplet associated hydrolase (Length=257) DVVIVITGNPGVPDFYKDFAEQLQAKLPSEVPIWIIGHTGHTKPPDNLPNSYPDTKSARHLYDLKGNLKHKIEFIKQYVP ADARIHIIAHSIGSWFTINLLRETEIADRVVKCYLLFPTIERMAESPTGRFLTGIVLRIAAVIIFLSWIFTLLPYLLQAV LIRVFGLFYGIPAHQTGAVLQLVHPIPLKRIFLLAKDEMILVKELDDNLISQHKGKLFLYYGANDGWTPIRYYEDLKAKF PDIDAHLCRRGFQHSFV |
| 385 |
PF09478 (CBM49) |
 Estimated TM-score=0.83; easy target. |
>Carbohydrate binding domain CBM49 (Length=78) VNITQTIVNSWNDGNQDFIQVQVVITNIGPTTIRSFSFKLNNIISIWEVETLSSNSYQLPNWVGSISSGSSHSFGYIQ |
| 386 |
PF18154 (pPIWI_RE_REase) |
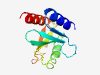 Estimated TM-score=0.83; easy target. |
>REase associating with pPIWI_RE (Length=115) GLAEVELEDRLTDLGLDVTMWPGFDAYDLLVVFPDGHRWAVDVKDWAHPAFLGRAARPVRPEPPYDEAFWVVPVYRVRER PGYQQAFQRARPADAAVRLLTDDELTRAARQRLAE |
| 387 |
PF12281 (NTP_transf_8) |
 Estimated TM-score=0.83; easy target. |
>Nucleotidyltransferase (Length=214) LTGDVVEAMATAGIFRLRGVLVGTVAFQTYAGHLGVRLPGASLQTGDADFAQHYSISSSVEDSLPPILEILRSVDPTFRE IPHRSDPLRVTQFQNASRYKVEFLTPNRGSDDYTDNASPMPALGGASAQPLRFLDFLIHEPIRTVMLHRSGVPVTVPSPQ RYAVHKLIVASRRQSDPNGIAKREKDIYQASLLVEALHATRRQDDLAEVFAEAW |
| 388 |
PF13875 (DUF4202) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF4202) (Length=183) EQALAGIDDAHRGDPNIVTVDGSEIPYELHYANKMTKYLEKHNPSASEALKLAVRAQHLRRWEIPRSSYPMTKPGYFSWR TALKNRQADIADEICRNCGYDEDAAGRVAALVRKENMKRDEECQVLEDVACLVFLDDQFEKFEKEHDEEKIVGILKKTWA KMSERGHELALQIDMSDRAKGLV |
| 389 |
PF11231 (DUF3034) |
 Estimated TM-score=0.83; easy target. |
>Protein of unknown function (DUF3034) (Length=255) TGGVSQLEGAAGGGLTPWAVIGGYGTRDEIGANAFYTRVNLPDYHVDTAGAMVGLYNRVELSFARQRFDTEKVGAALGLG EGFTFRQDVVGVKVRLFGDVVLDQGSWLPQVSVGAQYKKNDQDGVVRFVGAKRSQGTDYYISATKLFLGQSLLVDGTVRF TKANQIGILGFGGDLNNSYKPEFEASVAYLLSRHWAIGAEYRQKPNNLGIAREDDWYDAFVAWAPSKHVSLTVAYADLGN IVIKDRQRGLYASVQ |
| 390 |
PF03653 (UPF0093) |
 Estimated TM-score=0.83; hard target. |
>Uncharacterised protein family (UPF0093) (Length=143) GNLYDWVKALHVISIIAWMAGLLYLPRLFVYHCEAAAGSATSETLKVMERRLLRAIMNPAMIASYVFGIWMLALNPGWFQ SGWIHAKLALVIGLTGVHMMMAGWRRKFAEDRNQRPQRFFRVMNEVPTLLMIGIVILVIVKPF |
| 391 |
PF14315 (DUF4380) |
 Estimated TM-score=0.83; easy target. |
>Domain of unknown function (DUF4380) (Length=295) IQRLPLNTSAISVEVTPDIGGRILAIALPGKENFLRQGEAVISEPEPYVAPDADNVGYLAQETWLGPQSQWWTQQLVNPA RAQAKAVWPPDPYLILAKNQVVQADKRRVVMRTPASPVSGVVVEKHYALVEGKPDQLHLMARATNIRDTTVAWDIWFNTR VPPSSQVYVPVASQEDVRVEHMENEQRGSLGYRLVDGFFTFVSKAHPDKPGVRGKAFIQPSAGWMAAFREGQVFIIHFPL QPTAVIHPEQGQVELYLDSIPEYPSQGLLEMEVHAPYRKLQPGAHMEAEQTWRLL |
| 392 |
PF17680 (FlgO) |
 Estimated TM-score=0.83; trivial target. |
>FlgO protein (Length=126) VTDQLIKKIGVKNISKVSVLMTTIVPVDNILKPTSFGRMCAEQLMTEMSMGGFNVIESRMTEGYFIRNNVGEFMLSRDVK LVGKQHNARLALVGTYSESGGQVLINVRLVDTENAGVVATANTMMN |
| 393 |
PF06687 (SUR7) |
 Estimated TM-score=0.83; easy target. |
>SUR7/PalI family (Length=211) FPCALTIGSLICLVIVGLGCTRAHKAENLYFLRFIQALDEVNNTTGLNDFYSIGLWSYCTGNITDNGTYKTTYCSKPRGG FWFDPIDTWGLNGTNTNDFLPGKLKKSLNMYRNVSLWMAIIYLIAVIATVLEIMTGLMAVGGNRLVSCFAWLLAGVSLLF TTAMSITSTTIFATLAATVKTVLKPHGITGHMGRHIYAAAWLAVALSFLAS |
| 394 |
PF10012 (DUF2255) |
 Estimated TM-score=0.83; easy target. |
>Uncharacterized protein conserved in bacteria (DUF2255) (Length=115) ELDTIGAAEELEIAPRRRDGTLRTPVTIWVVHVGDSLYVRSYRGRGGAWFRAAQVRHEGRIRAGGVEKDVTFVEETDPGI NDQIDAAYRTKYRRHAARYVDPMVASEARATTIKL |
| 395 |
PF12183 (NotI) |
 Estimated TM-score=0.83; trivial target. |
>Restriction endonuclease NotI (Length=231) EVFGHPVTDASSRADRYRSQRLCPFNNKVPNCTKDKAKSPLGVCSIQHDGSPVITCPIRFREDWLITDDAASFFFPEGTK WSSLTEIRLNDGNGKSAGNIDIVLVAYDDNGKVKDFGALEIQAVYISGNVRDPFEYFMEEPKGRAFMDWSNQPNYPRPDY LSSSRKRLVPQLFFKGGILHSWKKKSAVALNKSFFDTLPPLTTVSRKKADIAWLIYDIELCGSGAEKRYRL |
| 396 |
PF10216 (ChpXY) |
 Estimated TM-score=0.83; trivial target. |
>CO2 hydration protein (ChpXY) (Length=351) YIYRLERGEALLKDSPQNVTEVVGILKSYGVVLDAYSRNLIYISENQFLVLFPFFKYFNGEVKLSQLLRHWWHDRINFEY AEYCMKAMFWHGGGGLDAYLDTPKFEAAAEKAIQAKFKGNILMLGLHKIFPNFLPEQVRQLAYYSGLGQFWRVMADIFLE LSDRYDRGEIKTIPQVVQHILDGLVKDANRPITYQVKINNKVYEILPKSVGLTFLMDTAVPYVEAVFFRGTPFLGTVSYN AQAYQIPYDQAMFTYGALYADPLPVGGAGIPPTLLMQDMRHFLPEYLHNIYRQSFRQEDDLLVQICESFQKSMFCVTTAA IKGLAPYPLDTTDPKEKKANRSYLEKWMNRF |
| 397 |
PF13650 (Asp_protease_2) |
 Estimated TM-score=0.83; trivial target. |
>Aspartyl protease (Length=89) IDIKVNGKPTRAYIDSGAEPVLVKQSVARELGLMWQPSLHLIRGYGQGITKVHGEVEVELEVDLVKANVKALIVEDSAQR VPVIVGQTF |
| 398 |
PF16760 (CBM53) |
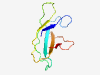 Estimated TM-score=0.83; easy target. |
>Starch/carbohydrate-binding module (family 53) (Length=87) VTVFYNPRDTPLNGRQQIYLMGGWNRWTHKRHFGPIAMHRPVDGGEHWQATLTVPKDAFKMDFVFADVPGGDGTYDNRGG FDYHLPV |
| 399 |
PF03659 (Glyco_hydro_71) |
 Estimated TM-score=0.83; easy target. |
>Glycosyl hydrolase family 71 (Length=383) VFAHVVVGNTAAHTQSTWQNDITLAHNAGIDAFALNIATPDSNTAPQVANAFAAAKALNNGFKLFFSFDYLGGGQPWPAT GSNSVVSYLTQYKNSGVYFYYNGLPFASTFEGTNNINDWAPGGPIRSGAGDVYFVPDWTSLGPSGVASHSANIQGAFSWD MWPAGANDMTTTPDQQWVSALAGKSYMMGVSPWFFHSASGGTDWVWRGDDLWAQRWQQVLQINPAFVEIVTWNDFAESHY IGPVYTTSEIAAGSSTYVNNMPHESWRDFLPYYIAMYKKSTFNISRDQMQYWYRTAPAAGGSTCGVVGNNPGWQTTMSPN SIVQDKVFFSALLMSPATVTVQIGNYAAVSYNGVQGINHWSQPFNGQTGSVKFSVVRNGATVK |
| 400 |
PF10741 (T2SSM_b) |
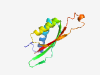 Estimated TM-score=0.83; easy target. |
>Type II secretion system (T2SS), protein M subtype b (Length=109) GTPFLEGPTVTVAGANLLQRVAAAVGNQGGSVQSSQVDVSGAQTKDGFVGLVISCELEQPALQKVLYDLEAGMPFLFVDQ LDVQVPQTTALNDAPSGRVRVILGVSGQW |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218