

Go to page (83 pages in total): [First page] << < 6 7 8 9 10 11 12 13 14 15 16 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 1001 |
PF08855 (DUF1825) |
 Estimated TM-score=0.79; hard target. |
>Domain of unknown function (DUF1825) (Length=103) MGFFDSEIVQQEAKQLFEDYQSLIQLGGSYGKFDREGKKIFIDQMEAMMERYRIFMKRFELSDDFMAQMTVEQLNTQLSQ FGITPQQMFDQMHMTLERMKSEL |
| 1002 |
PF07593 (UnbV_ASPIC) |
 Estimated TM-score=0.79; hard target. |
>ASPIC and UnbV (Length=72) ATGARVTLTATINGQQVAQTRHVNTNPGGYGSQSSPLVHFGLGDALSVSGVTINWPSGQTWDTTGVFADQSL |
| 1003 |
PF11171 (DUF2958) |
 Estimated TM-score=0.79; very hard target. |
>Protein of unknown function (DUF2958) (Length=111) LITDEQRARLLANGRAHAEGQELDPLPVVKLFTPDAHATWLLTELDPEDGDTAFGLCDLGLGMPELGCVRLSELENVRGP LNLPIERDLYFIAQRRLSEYAQLARANGSII |
| 1004 |
PF18178 (TPALS) |
 Estimated TM-score=0.79; easy target. |
>TIR- and PNP-associating SLOG family (Length=230) LLGRRIHISGSVVDDLAVAPTADVAAARELVAVLVKELIRRGANFVVPVDAEPLRKADGMPICFDWLIWRTIKENLVLRP ANAPAPLAVAVQHHKNEDQIPPEYEGLWDELRGSQLIKIESAAHWNMNSKRMEAQARFGDILVALGGTEGVLYLANLYHD AGKPVVPLNLALCSESTGARRLYNFGLTSSQSRRLFQVAEDGDSHDWINRIRFPKRQSVADRVAVLIDLL |
| 1005 |
PF04387 (PTPLA) |
 Estimated TM-score=0.79; hard target. |
>Protein tyrosine phosphatase-like protein, PTPLA (Length=162) QTAAIMEILHSIVGLVRSPVSSTLPQITGRLFMIWGILRSFPEIHTHIFVTSLLISWCITEVTRYSFYGMKESFGFTPSW LLWLRYSTFIVCFPVGMVSEVGLVYIVVPFMKASEKYCLRMPNKWNFSIKYFYASVFFMVLYAPVYLHLFHYLIVQRKKA LA |
| 1006 |
PF18819 (MuF_C) |
 Estimated TM-score=0.79; very hard target. |
>Phage MuF-C-terminal domain (Length=101) HKVMQDKHNVTSETLKQLPSQINNPVAVMKSSTRDNSYVVLTELIEQEGNRDKPVIAALHLKKTNQGIELINITSVYGRS ESQIQNGLHNDLLYLNRTKGR |
| 1007 |
PF10145 (PhageMin_Tail) |
 Estimated TM-score=0.79; hard target. |
>Phage-related minor tail protein (Length=201) QQARDIGGSTAFSPTDVARTQDTLARSGYDADAILAATEPTVNLSLASGVDIAEAADIVTNMQSAFNLPLDQIKRVSDVM AKGFTSSNTNLLELGEAMKYVAPIAEAAGASIEDTTALLGVLADNGIKGSMAGTSTSAVFSRLQAPVGKAPEALRELGIT TRDGKGNMLPVEKILKDINRSFKKNKLGTAQQAEYLKVIFG |
| 1008 |
PF18985 (DUF5718) |
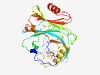 Estimated TM-score=0.79; easy target. |
>Family of unknown function (DUF5718) (Length=254) EAADFMAVKTEEAIQPKAIFPFYVPSETLAPTEQFLSTYPLTHDTIAFPNDADNLQIEPEVALICNIEYQDGKVVTLIPT HFAAYNDCSIRRPNARKICEKKNWGVNSKGISATHIALDKFEQGGNLDQYNIACFHKRDGKLNTYGEDSPAVSYSYFHQK LLNWIIDKMNNQPDQGPMNHIAELIKQANYPAQAIISIGATRYTPFGETNFLQMGDTSIVVVYNATKYTHEQIVEMAEKE EFAEEGISALVQKV |
| 1009 |
PF17277 (DUF5342) |
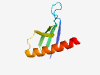 Estimated TM-score=0.79; hard target. |
>Family of unknown function (DUF5342) (Length=69) MLTHFQYKPLFENKELPGWRFSFYFRKERYAGVYHPDGKIEWVTAAPPEKAEDQVTGQIHELMLFHVYD |
| 1010 |
PF09778 (Guanylate_cyc_2) |
 Estimated TM-score=0.79; easy target. |
>Guanylylate cyclase (Length=214) VPIIQQLYHWDCGLACSRMVLRYLGQLHDREFENALQELQLTRSIWTIDLAYLMRHFGVRHRFCTQTLGVDKGYKNQSFY RKHFDTEETRVNQLFAQAKACKVQVEKCTVSVQDIQAHLAQGHVAIVLVNSGVLHCDLCSSPVKYCCFTPSGHRCFCRTP DYQGHFIVLRGYNRATGCIFYNNPAYADRMCSTSISNFEEARTSYGTDEDILFV |
| 1011 |
PF06356 (DUF1064) |
 Estimated TM-score=0.79; easy target. |
>Protein of unknown function (DUF1064) (Length=116) RSKYGAKKIVIDGITFDSKDEGKYYEYLKKLKSQEKILNFELQPKYELQPGFKKNGKTYRAITYAPDFLIYHLDGTEELI DVKGMSTQQGEMRRKMFDYKYPDLKLTWVARSLKYS |
| 1012 |
PF12965 (DUF3854) |
 Estimated TM-score=0.79; easy target. |
>Domain of unknown function (DUF3854) (Length=129) FWQWIINHPSIPICITEGAKKAGALLTAGYAAIALPGINNGYRTPQDEFGNRIGKSRLIPQLEKLAISGRKIYIAFDQDS KPNTIKNVNTAIRKLGYLFAQAGCQVKVITWSPKWGKGVDDFISNKGEK |
| 1013 |
PF13785 (DUF4178) |
 Estimated TM-score=0.79; very hard target. |
>Domain of unknown function (DUF4178) (Length=137) QIGSEGTFRSRHFLVVGRIQLKYEAGLWNEWHILFDDGRSAWLAEAAGEYIVSAQVSVREPIPAFTVLVPEMAVTLDGRQ FTVTDLETARCISGQGELPFRVAAGYDVNTADLRSNDRFLTIDYSETPPLVFVGQST |
| 1014 |
PF08410 (DUF1737) |
 Estimated TM-score=0.79; hard target. |
>Domain of unknown function (DUF1737) (Length=53) MKAYRFLTADDTSAFCHKVTEALNKGWELHGSPSYAFDAANGVMRCGQAVVKD |
| 1015 |
PF16402 (DUF5010) |
 Estimated TM-score=0.79; trivial target. |
>Domain of unknown function (DUF5010) (Length=338) SDVGVTMGFTTTTLNGGTPEYAGNTIYNLPLYMSSSDTSQWWDNIVEEIAYSGADFVAPTIRGYLDSVPDYNNNGDPRKL ADMVAAMNRRGVADKFKISFLDDTAASMADHKNRDNGGGGYDPKFDIGDTGNYKYIWDYNMRAFFNAIPDNMRYKIDGRP AIWEWGIGDYAFTNYGNGNTKALLMYIREKCQAEFGFNPYIIVDQSWVKCDPSVNDPSVIDGIHGWFILGPGYSTENFNG NSFGALCPAFRVVNGSASMFLDSNHGKTFDTNLSLTAGKKCLSTLVEGFSDWEENCSVWRSKDTTYYDYPNQRINTLRKY TKNPFPDDLKVEAEACDS |
| 1016 |
PF11972 (HTH_13) |
 Estimated TM-score=0.79; easy target. |
>HTH DNA binding domain (Length=54) LPKLIELVLSRPLVSAGMIAKELAITPRAALRIVEELGLREMTGRGRFRAWGVL |
| 1017 |
PF18737 (HEPN_MAE_28990) |
 Estimated TM-score=0.79; hard target. |
>MAE_28990/MAE_18760-like HEPN (Length=202) SDRLSNDLAWRKKELSEVKSLVETKSFSDSKHKALIRSAICLLYAHWEGFVKLAANSYLEYVRMQKLCYQELATNFLALA MKERLKEAKETNKPSLYIPVCNFFLDELNQRCKLPEDAISSASNLSSEIFKEIIYILGIDFSPYSTKSVLIDTKLLKTRN EIAHGEYSLFDKEEYIELHKEVIAMLDIFRNQIENAAINKDY |
| 1018 |
PF08379 (Bact_transglu_N) |
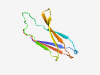 Estimated TM-score=0.79; trivial target. |
>Bacterial transglutaminase-like N-terminal region (Length=77) IYHLTHYKYDRPVTLGPQVIRLRPAPHSRTRVISHSLRVSPAGHFVNHQQDPYGNWLARFVFPEPVTELKIEVDLVA |
| 1019 |
PF05805 (L6_membrane) |
 Estimated TM-score=0.79; easy target. |
>L6 membrane protein (Length=186) MCTGKCSLFIAISLYPLVLLSILCNILLFFPDWSTKSVKEGHITDEVKYMGGFIGGGVLVLIPALYIHLTGEQGCCANRL GMFFSVIFAAVGAAGAVYSFIVALLGLYNGPLCRTSSYKWERPFMDKNLEYVKNSNSWNDCLEPKNVVQFNMGLFITLMV ASVLQGLLCVSQVINGFVGCICGTCN |
| 1020 |
PF07423 (DUF1510) |
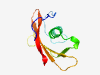 Estimated TM-score=0.79; hard target. |
>Domain of unknown function (DUF1510) (Length=93) IVNPDWKPVGTTQTGEHAAVYDESSVDWQEMLQAISYASGLDKNNMTVWRLGNNGGPNSAIGTVSQKGSDQKYRVAIEWV DGQGWKPVKVEEL |
| 1021 |
PF14704 (DERM) |
 Estimated TM-score=0.79; easy target. |
>Dermatopontin (Length=152) NCWDCPFKFKCPNRQGLAALTSIHSNRREDRRWNFACASAGVGTTGQTSRSGGLDFNQFDRPLSFTCPEDYYLTTLESAH SNNKEDRVWRFECRKIANAYLRGCSTTGYLNGFDQPLNYIASNDRIITGLHSYHNNRKEDRRWKVSTCKVEC |
| 1022 |
PF11026 (DUF2721) |
 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF2721) (Length=117) TTPALLFPAISLLLLAYTNRFLAIATLIRNLHEKHENEPTKNLIEQIKNLRKRVNLIRNMQFLGVSSLLLCVVCMFVLFW GQIELGHWLFGISLILLMLSLSFSVIEIQISVKALNI |
| 1023 |
PF06578 (YscK) |
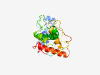 Estimated TM-score=0.79; easy target. |
>YOP proteins translocation protein K (YscK) (Length=205) ALTPYQFRFCPASYIHSDHLSPEWLTVLSSLPEWRHSPRLNGLLLTQFDLNVDYELPTGLGNIALLEQSCLEQLLTWLGA LLHGQAIRHCLMATELRHLHDSLGKEGHRFCLKYLDIIIGNWPTGWQRSLPPEINANYFRTSALQFWLTAMESPPIDFAK RLSLRLPSYENLAAWPVSQAERPLAQALCLKLAKQVNTECYHLLK |
| 1024 |
PF02076 (STE3) |
 Estimated TM-score=0.79; easy target. |
>Pheromone A receptor (Length=286) AAFAGFLMCTIPLPWHLEAWNTGTCLYMIWTGLACLNQFINSVVWNRNAINWSPTWCDISSRFIVGSAVAIPAASLCINR RLYHISSVRSVTITKAEKRRDIIIDLAIGLGLPLLEMVLQYIPQGHRFNIFEDVGCYPFTYNTWPAFVLVYCPPIAIGLV SAVYAIMSIIQFNKSRSQFNDLLSGHNNLTSSRYVRLMCLAGIEVLCTVPLGSYALYLNIKAGINPWISWENTHFGFSRV DQIPSVLWRANRVNEISLELTRWLVIICACIFFGFFGFADEARKHY |
| 1025 |
PF08309 (LVIVD) |
 Estimated TM-score=0.79; easy target. |
>LVIVD repeat (Length=41) GGAHNDIKIVENYAYVAQTQGLYIYDISNVVNPTLVGSYKT |
| 1026 |
PF16734 (Pilin_GH) |
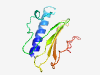 Estimated TM-score=0.79; easy target. |
>Type IV pilin-like G and H, putative (Length=107) ESQGIQYIWAMNIAQQGYYAENGDFTDTIPNLGSSIPTQTANYNYSISKGNQAVFNYATAREPNLKSFVGGVFLVGDEIQ TILCQANVADTTKPANPINDNGVLLCG |
| 1027 |
PF07040 (DUF1326) |
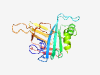 Estimated TM-score=0.79; very hard target. |
>Protein of unknown function (DUF1326) (Length=184) ENCNCDVVCPCLLSTNAQLTSKPTQGVCDVALVFHIDQGNYGDVRLDDLNVAMIAHTPGPMAEGNWTAAAYIDERADDRQ MEALGAIFTGAAGGPMAAFAPMIGNNLGAKKTPIRYQVDGKKRSAEIPGVMHMAVEPLPTMREDGEMWAATGHPVNPDRL TMAMGVQGSTFSDHGMNWDNSGRN |
| 1028 |
PF06115 (DUF956) |
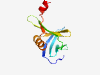 Estimated TM-score=0.79; easy target. |
>Domain of unknown function (DUF956) (Length=116) MVQSLNTKVDLTIKGTSYMGMSDYGQIMVGDNAFEFYNDRDANKFIQIPWEEVDYVVASVMFKGKWIPRYAIQTKRNGTY SFASKEPKKVLRAIREYVDPDRMVQSLSFFDVVKRG |
| 1029 |
PF13289 (SIR2_2) |
 Estimated TM-score=0.79; easy target. |
>SIR2-like domain (Length=135) AVITTNYDKLIEQAYALTGRNIPPTYTFDSSPDLISAVSHNRFFVLKVHGDIDRKGTIVLSERDYRDLIYRSPGYRAALN TLFITRTVLFVGASLTDPDITLVLESVSESFAGKGPRHYALLPRREAGDAESDHW |
| 1030 |
PF11161 (DUF2944) |
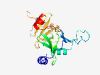 Estimated TM-score=0.79; easy target. |
>Protein of unknown function (DUF2946) (Length=184) MDDIVRQALAKWPNVPHCTGWLLLDRRGQWRLRDDAAQAAGEPGSPVRHAALNAFIARNYECDAHGQWFFQNGPQRVYVE LAYTPWIVRLAERDSRLALTDHTGAPFEPAQAWLDDAGGVLFRDTGTPPRIAALHDHDLGLFADHADFDAAPPVLRWRDG CALPLDSIARADVPARFGFVASPA |
| 1031 |
PF14424 (Toxin-deaminase) |
 Estimated TM-score=0.79; easy target. |
>The BURPS668_1122 family of deaminases (Length=140) EFRQELPKEIQKTGNFGLAEVKIEGLRTEFKSHSRVNELKDAGNAEGYSVLKPDPKDWNFTPQEVNTKDIINGEGAYSRK WDTEFKIIEDITDQLKALKPEQRKGSIDIYTDREPCASCGPILEQFKARFPDIQVNVYFA |
| 1032 |
PF05231 (MASE1) |
 Estimated TM-score=0.79; hard target. |
>MASE1 (Length=281) LGLFVVAYILGCGFAQSLAIVPGTGISIWPPSGLFIATLVLASGYSWPWWVLGGCLAELFSNFLWFHSPLPAAVLIYIGN ALEAAVGAWLVNRTCRRPVRLETLQEVLAFVVLAAGVAPTIGATVGSATLAWFGIQSQTFAAAWPLWWIGDATGILIVAP LALVVFQNWRGKSQLSAARWMEACVLGLIFLGVAALSLSGYLPFAYIIMPPLLWAAARFEFRGAAVALTLLALITAVFTI SGASQFAGDPESQKHKQIMLQLFLAISAFSALIVAAISRQH |
| 1033 |
PF06197 (DUF998) |
 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF998) (Length=171) APAVIPVGSTVAAAAQPPGYDPLRQSLSTLARIGATDRWIMTGTLVLLGLGYVAGALVLTRIPWPGRIAVGLGGAGTLFA GLLPQPETGSSPWHMGAATVGWIAFVGFPLAVSRHPSASPLLGRRAAWVVTGTLLALLGWFFVQLQTGGPYIGLSERVLI IAQTLWPAVVV |
| 1034 |
PF11968 (Bmt2) |
 Estimated TM-score=0.79; easy target. |
>25S rRNA (adenine(2142)-N(1))-methyltransferase, Bmt2 (Length=225) AIEAEIEALGGIEKYQEASLQGQRHDRGGDSSNVLMEWVGPYLSSEEYKEGRLRMLEVGALSTKNACAQSNHFDITRIDL NSQEPGILEQDFMERPLPKDESEKFDIISLSLVLNFVPDHKDRGEMLVRTTQFLTSPKRFSGTTAAFFPSLFLVLPAPCV TNSRYLDQERLAAIMESIGYEMVESKTTQRLVYYLWRRTSDQVGNDRPFRKEELRSGSSRNNFAI |
| 1035 |
PF09858 (DUF2085) |
 Estimated TM-score=0.79; hard target. |
>Predicted membrane protein (DUF2085) (Length=90) CHRMPERSFFIKGHQFPVCARCTGFYTGLMIYLIVNLFYKHPYDWNMLFISMVLMVPVAIDGFTQYFGPRESTNTLRFIT GFIGGIGLII |
| 1036 |
PF07608 (DUF1571) |
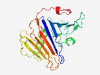 Estimated TM-score=0.79; easy target. |
>Protein of unknown function (DUF1571) (Length=230) YPGHRAQFKRLERIHGQLGPWQTIELKVLHNPLSVYMKFLEPDDGKEVLYSQGRFDNHLIAHPGGMARLLTPRLKLDPNG PLAMAENRHPITSAGLLNLVRRLIHFRRLDLNHPTSHLRLDRIETPDGQMVFRSCHRHLKRTDQHPYSRSEIWYDSATCL PIRVENHDWPAGIILGHTISSPADLGDPFAIFDAPDNGETLVEIYEYAHLDFETPLTPLDFDPANPAYGF |
| 1037 |
PF06949 (DUF1292) |
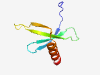 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF1292) (Length=70) EDGGEQQCRVVFTFDSEEHSYVLFSLIGDEEAGISALRYVLDENGEMSGFSDIETDDEWAMVEEVMNTVL |
| 1038 |
PF10128 (OpcA_G6PD_assem) |
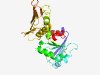 Estimated TM-score=0.79; very hard target. |
>Glucose-6-phosphate dehydrogenase subunit (Length=246) ANDASREHPCRVIVVARGARKAAPRLDAQIRVGGDAGASEVIVLRLYGELANEGASCVVPLLLPDTPVVAWWPNEAPAVP AQDPIGQLAQRRITDAAAEKNPIKALEQRRAGYTPGDTDLAWTRLTLWRAMLASAFDLPPYEKVTEAEVSGESDSPSTDL LAAWLAGCLRVPVKRAKAASGEGIVDVRLERRSGVVELSRPDGRVGTLTQPGQPERRVALQRRQVRDCLAEELRRLDPDE VYEAAL |
| 1039 |
PF04827 (Plant_tran) |
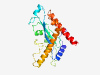 Estimated TM-score=0.79; hard target. |
>Plant transposon protein (Length=204) EYLRKPNSNDVRRLLEMGEARGFPGMMGSIDCMHWQWKNCPKAWKGMFMSGHKGVPTLLLEAVASSDLWIWHAFFGVAGS NNDINVLDRSPLFDEVLEGRAPEISYTLNGNNYNFGYYLADGIYPEWATFDKTIPRPQGEKRKLFSKYQEGQRKDVERAF GVLQSRFAIVRGPARFWDKGDLARIMRACIILHNMIVEDERDTY |
| 1040 |
PF04339 (FemAB_like) |
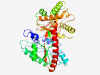 Estimated TM-score=0.79; easy target. |
>Peptidogalycan biosysnthesis/recognition (Length=384) VGAIAAADWDACARAALTPPAACASEAAAAAYNPFLSHAFFSALEQSKSASPRTGWGPRHLVAKQGDRVIGIVPCYLKSH SQGEYVFDRGWADAYERAGGNYYPKLQVSVPFTPATGPRLLIREGEDSATVAAALTTGLTALCQMSEASSVHVTFARESE WRFLAEQGFLQRTDQQFHWHNAGYASFDDFLGSLNSRHRKAIKRERRDAVANGITIHHLTGADITEDAWDAFFEFYMETG SRKWGRPYLTREFYSLIGQSMSQDVLLVMAKRDGRWIAGAINFIGGDTLFGRHWGAVEQHPFLHFEVCYYQAIDFAISRG LKVVEAGAQGEHKIARGYLPQTTYSAHYIADPALRRAIADYLKRERMYVDEMGRELTEAGPFRK |
| 1041 |
PF10994 (DUF2817) |
 Estimated TM-score=0.79; easy target. |
>Protein of unknown function (DUF2817) (Length=353) FTETYYQARALFRERARGAHATLHTLRLDHLQDLDLTIDVAVMEGSKKNVVVHVSGTHGAEGFAGSAIQSAALQRYSTWM THSDADPTVIFVHALNPYGFAQFRRNNEHNVDLNRNWLSDDEIQALTKRDANAFGYLDVYDLLNPSVTDVESWACPLMLR QIWHFATKGWSSFKRAVVTGNYHFPASLFFGGFKKQPSIVLLEEFLQRNIDFQSLERFAVVDVHTGLGKSGEETLFLTGK GSSEVMSRVFAHEIKENVANSGANSTNAVTSGYDAVAGVVCDGIAQLVPVNRRGHVVSVAQEFGTVPPPFVLKAMVEENA MYHQAPTRRLPYAERARDVFYVHRSVSWKQSVL |
| 1042 |
PF18800 (Atthog) |
 Estimated TM-score=0.79; easy target. |
>Attenuator of Hedgehog (Length=133) GGGETSLGLMWTCTTLYNRPKVCFSPDLTPEWMMALVCIFIGCVCITTTVVLLISSHWDYNVVPYARWVGFAAMVLFCLA AVIFPLGFSMSEIGGEPYQLPNSFQVGLSYIFFVLALWITVISELFAGKVCLP |
| 1043 |
PF12860 (PAS_7) |
 Estimated TM-score=0.79; easy target. |
>PAS fold (Length=109) LPLGVWVVGADGRLAFANAATRQVTGIDPAETPPGLPAAELVRRFAYRGIYGEGDPAALAAEHLAIDRSRPFRRQFRRTD GASFEQAGVPLPGGGYVAAVIELTGHLAR |
| 1044 |
PF14342 (DUF4396) |
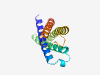 Estimated TM-score=0.79; easy target. |
>Domain of unknown function (DUF4396) (Length=143) WQSVGVGASHCGSGCTLGDIIAEWFVFFVPIVIFGSHVVGTWVLDYIVAFVFGIAFQYFTIKPMRELSVGEGLKAAVKAD TLSLTAWQIGMYGWMAISFFLLFSPETLRPNNPVFWFMMQIGMVCGFLTSYPVNWWLLSKKIK |
| 1045 |
PF14087 (DUF4267) |
 Estimated TM-score=0.79; hard target. |
>Domain of unknown function (DUF4267) (Length=107) ATIFIGFGVNAILRPDHALTFFEFQPPTSVVDKQMVDSLMAVYGVRDIFMGIAIYVASYFGTRSTLGWILLAASSVAFAD GIVCWTHGQGQWNHWGYAPMIAVAGSM |
| 1046 |
PF08616 (SPA) |
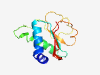 Estimated TM-score=0.79; easy target. |
>Stabilization of polarity axis (Length=113) QHSHLHSNGSLTHPIIILFNAMATQKRVIFLGHGQPANDVATFVLSACALGSGCGAVFRGFTARAFPYANLTSLDEMEKV PGYIAGVTNPRFEDLPAWDVICNIETGRITISK |
| 1047 |
PF14626 (RNase_Zc3h12a_2) |
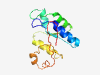 Estimated TM-score=0.79; easy target. |
>Zc3h12a-like Ribonuclease NYN domain (Length=124) YEPELNLPVKPLTDIILLFLIRGHKTTVYLPKYYQEFLSDCGVSKVDDLVAFQKLIDLGFIKFLEITNPFNFKWFNVVAD LADRCGAVFVSSEDYRQRKMKINYSKPSGRIITPCFLNAEDRLL |
| 1048 |
PF19174 (DUF5856) |
 Estimated TM-score=0.79; easy target. |
>Family of unknown function (DUF5856) (Length=93) HFDTKSYARHKAYEVFFNEIPDLIDAFGEQWLGFSGKSYTPALPSQKELPKDTIEMLDLILAKADGIYKSVPAALQSVLD DITGLCYKTKYLL |
| 1049 |
PF19147 (DUF5829) |
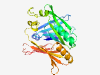 Estimated TM-score=0.79; easy target. |
>Family of unknown function (DUF5829) (Length=265) YNHSYGVFDRETADAIEHSDYLRDFANFQVRTTTGTGGQTWTGRYLMGRETYLELFGVGDIAGQDGTLGSAGLGLSTEQD GDLATVTERLKSEGVADPIEFLQTRDFGDGVPVPWFDAILTATEYDAFGAWAMEYRPEYFADPRSKTEPASFPGDVGRER VLSDDYRTHLMRDVTSVRIAVTEGDLADTVPLLRAGGFAVKNVAGGGVVAQGGGTTIRFDSVPREQAGFQRVEMSLNRPV QDRHVEQIGHSTLTVGPGSRAVWTF |
| 1050 |
PF14168 (YjzC) |
 Estimated TM-score=0.79; very hard target. |
>YjzC-like protein (Length=55) MGQQRHFQPGDKAPNNGVYIEIGETGDNVKNPKKIKLKAGDTFPETSNHNRHWTY |
| 1051 |
PF17074 (Darcynin) |
 Estimated TM-score=0.79; easy target. |
>Darcynin, domain of unknown function (Length=124) MQRTYTVTIPSFEPALTVFMLVKTSPEWLGFPVERRFALLEEQFTPILRKHAAHVTLRFFDVEFYATRVTDLWMWDARDH HSYQLLVEDLRETAFWDRYFEIVEILPGVENAYARNYGRPAINA |
| 1052 |
PF12588 (PSDC) |
 Estimated TM-score=0.79; hard target. |
>Phophatidylserine decarboxylase (Length=140) HPVIQEFKDLIEKNTRIYILVNSMFEEIPTKKPYKRDPGGHKQVRDYHHMLELFNHILTTAPEWSDHEYSVGVVGTPFNA ILDWPMGTPSGFAFFLDPEVNKMIKKVLNAWAEFLASPDSAYVLDNNKIGWFSEHGTHDL |
| 1053 |
PF18726 (HEPN_SAV_6107) |
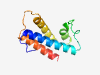 Estimated TM-score=0.79; easy target. |
>SAV_6107-like HEPN (Length=101) ATALETPNERYATAHLAALRTAAAVLAARGRPEPTPRRRARIRSAWEVLPEIAPELTEWSALFASGADRRARAEAGIRGA VGTRDADDLIRDVAMFLRLVE |
| 1054 |
PF05069 (Phage_tail_S) |
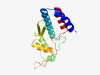 Estimated TM-score=0.79; very hard target. |
>Phage virion morphogenesis family (Length=147) DDLQALEDWAAPLLAQLEAKERRRLARAIARDLRRSQRQRIRSQTNPDGTPYAPRKPQKWRARQGSIRRSAMFDKLSTAK WMKATAHGDTAVVSFMGNVARIARTHQYGLRDRVDRDGPTIEYPQRELLGFTDADRELVINALFTHL |
| 1055 |
PF07603 (DUF1566) |
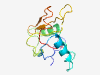 Estimated TM-score=0.79; easy target. |
>Protein of unknown function (DUF1566) (Length=118) GTVTDKNTGLMWGNDSVVTGTWESALASCEASGANGYDDWRLPSAAELLSIVDYGRKDPAVDDTVFTNTVSEFYWAATLI FDGSDRAWIVNFLNGWASASTYPRNLVESDTHYMRLVR |
| 1056 |
PF04754 (Transposase_31) |
 Estimated TM-score=0.79; hard target. |
>Putative transposase, YhgA-like (Length=202) TPHDAVFKQFLYHPDTARDFLDIYLPSTLRELCDLQTLKLESGSFIEDSLRASYSDVLWSLKTNEGDGYIYVVIEHQSSP DAHMAFRLMRYAMAAMQRHLDAGHKTLPLVIPMLFYHGALSPYPFSLCWLDEFDDPALARQLYSATFPLVDITVIPDDEI MQHRRIALLELMQKHIRKRDLMGLVEQLVSLLATGYANDSQL |
| 1057 |
PF14173 (ComGG) |
 Estimated TM-score=0.79; hard target. |
>ComG operon protein 7 (Length=97) YHTEIQFFEEEKKWYEADYLMKQALTYVKQTLASTPENVSSLTQTVSFVHGQATYRATRIEPDVWSVTVSCITNAQFRYD VQFQFDASTNNISVWRE |
| 1058 |
PF02654 (CobS) |
 Estimated TM-score=0.79; hard target. |
>Cobalamin-5-phosphate synthase (Length=224) AVQFLTRLPTPQVRDFKPELLAGAVGWFPAVGLLIGVLLTIPVALLAPLQPWLAALVGLAGWAWITGGLHLDGLADLADG LGAAHRDRERFLAVVKDPHVGSFGVLVLVLQCAAKLVLLMLVCRLGHWWLLVLAPAWARWGVFRWQTLPPLAPGMAEQFG WDRSNAVWFWTGLLVTMSLLLAPALLAAPFIVWAYRRWLKRRLGGVSGDCLGAGVEITETALLL |
| 1059 |
PF10474 (DUF2451) |
 Estimated TM-score=0.79; easy target. |
>Protein of unknown function C-terminus (DUF2451) (Length=236) MFGLSERIIATESLVFLAGQFESLHAHLEAMIPANKRAFLSQFYSQTVSTAQELRRPVYRSVASRVIDYDQTLQLMTGVR WDIRDIMSQHNAYVDILVRELQVFSMRLTQLANKQIPVPQETRAVLWDHCIRLINRTFVEGFASAKKCSNEGRALMQLDF QQYLMKLEKLTDIRPIPDREFVETYVKAFYLTENEMERWIREHKEYTAKQLTSLVMCGVGSHVTKKGKQKLLAIIE |
| 1060 |
PF15650 (Tox-REase-9) |
 Estimated TM-score=0.79; easy target. |
>Restriction endonuclease fold toxin 9 (Length=88) YTKSNLKLGRQMHKSYKANKVLKGVREKEFRLPSGKRIDFIDFEKKIIYELKPNNPRAIKKGLKQLKAYKKEVEKIYGKG WRTVLEVY |
| 1061 |
PF17202 (sCache_3_3) |
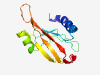 Estimated TM-score=0.79; easy target. |
>Single cache domain 3 (Length=100) LVNNNFEVVDEIKEKTGSLATIYLGNERVATNELDKDGERTVGTMVSNEVAEIVLKKGVPYERAETVLGEKYSVKYVPIK DNAGKVIGMWSVGIPKSSIS |
| 1062 |
PF06793 (UPF0262) |
 Estimated TM-score=0.79; very hard target. |
>Uncharacterised protein family (UPF0262) (Length=151) RLIEITLDESTVVRRSPDIEHERKVAIYDLLEGNSFRPEGSEVGPYRLHLSVEENRLVFDIRTGEDEQHAKVMLSLTPFR KIVKDYFLICESYYDAIKTAAPAQIEAIDMGRRGLHNEGSELLKARLDGKIAVDFDTARRLFTLICVLHIK |
| 1063 |
PF18475 (PIN7) |
 Estimated TM-score=0.79; easy target. |
>PIN domain (Length=101) LIDYENVQPKNLEILAHHNFRVYLFVGANQTKLSFELAKAMQELGDKARYIKISGNGPNALDFHIAYYIGEFIKDDPNAY FHIISKDTGFDPLIRHLKARG |
| 1064 |
PF11143 (DUF2919) |
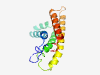 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF2919) (Length=144) YAIEQYDTHGFLKAPIWLWLGWLFLARAWIVFVVAGVSRDSGSQILSFVYPGNTALYLGLAAGVPSIALMWLISLRHPER RWVNALCHIGRWVTLLTAVAQFLQTGYHVYLQHGAFHWTNALTLLILLWLILYVYRSRSVRDCF |
| 1065 |
PF15639 (Tox-MPTase3) |
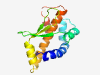 Estimated TM-score=0.79; easy target. |
>Metallopeptidase toxin 3 (Length=132) RKYPKLAYYVKVNLPQVVHVPAIVQALKKIANLNDVKLRDALIWGKGPQIKVTYLVDAYGEFSPNQQSNELRIDTSLVTD FEKGRGKRMARAGNVYLVGITLLHELTHWGDDQNGIDRPGEEGEEFEKLVYG |
| 1066 |
PF12276 (DUF3617) |
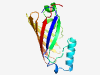 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF3617) (Length=143) IRPGLWEVTTTSMLLALVPQIPPDQMQQLTNLAKQYGLDMPQIQNGAATSKICITQQMADEKIPSYFHQNQTGCSFKNAV RTENSYTMDLVCTNPQLKGNGRAEGTFMTPESFSGWTIFKGTVQDRPVNEQADTSGRWISASC |
| 1067 |
PF09365 (DUF2461) |
 Estimated TM-score=0.79; easy target. |
>Conserved hypothetical protein (DUF2461) (Length=226) NTLLFLKDLKAHNQRSWLKSHDGEYRRSLKDWNTFVETITPKITEVDFTIPELPAKDLVFRIHRDIRFSKDPTPYKAHFS AAWSRTGKKGPYACYYIHLQPGSCFVGGGLWCPEASHLQKLRESIDERPRRWRRVLNDERFKKTFLPKSLQKGGEEAALK GFAETNKGNALKTKPKGYLVDHRDIELLKLRNFTISKKVDDKVFTEDDAQEQICEIIAAMHPFITF |
| 1068 |
PF04955 (HupE_UreJ) |
 Estimated TM-score=0.79; easy target. |
>HupE / UreJ protein (Length=179) AALLLPSVACAHTGTGDVGGFMHGLAHPLGGLDHLCAMLAVGLWAAQTGGRALWAVPLTFVGVMTLGGMLPLLDIGLPFV EPGIVLSVLLLGVLIAAAVRLPLWLGSSMVGLFALWHGHAHGAEMPENASGIGYALGFMLATASLHLAGIGFGLGMQRLA RERLVRFAGAGIALCGVYL |
| 1069 |
PF07457 (DUF1516) |
 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF1516) (Length=107) THAHITTWVVAIILFFVAHSLYKSGKEKPSKIVHMVLRLFYILIIVTGGLLVSAIYKIDGEYIAKMILGVWTIGAMEMVL VRLKKGKPTRVFWIQFVIVVILTILLG |
| 1070 |
PF10319 (7TM_GPCR_Srj) |
 Estimated TM-score=0.79; easy target. |
>Serpentine type 7TM GPCR chemoreceptor Srj (Length=305) HWFHHYTPQIFGFLAFIVNPYFVYLIFTEKVNLGNYRYLFSYFALFNIFHSLMDIFVPIGVHGFRYCFLIFLSDGWLFKQ SELGMFLLALRCSLIGCTYAVLISHFVYRYLTVKGSSLTQKYFPIYMIASFFLCAFFSMFWIVIAYIVAVPNLETRKYIQ EDFLELYEANSLKLNFFALMYKEAPLEILIKSWFGAIAGTTVSVGSISIFMIFSYLIMKNLKQKQLGMSEKTAKLQRELF RALIVQTVVPICLSFAPCMLSWYTPMFNMKLGKWLNYTGAVPLSAFPFVDPLAVILCLPGLRRRI |
| 1071 |
PF15405 (PH_5) |
 Estimated TM-score=0.79; easy target. |
>Pleckstrin homology domain (Length=128) IVFKSQLRKTPHDSSPANEITSYLFDHAVLLVRVKQAGKAEEIKAYRRPIPLELLAIREMDEVIPQQGVKRTSSSLLPLG GNNAKDGKKSEGWPITFRHLGKAGYELTLYAANQAGRDKWLQFIDKAQ |
| 1072 |
PF08349 (DUF1722) |
 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF1722) (Length=117) KLLVMAHSPKHVGDLGKLVANGKRSHLDEVCAEYHRTLMEALKRKATVKKNTNVLQHLMGYFKKQLSSDEKQELSELIEH YHQGYLPLIVPVTVIRHYVRKYDQPYLKEQYYLNPHP |
| 1073 |
PF03812 (KdgT) |
 Estimated TM-score=0.79; easy target. |
>2-keto-3-deoxygluconate permease (Length=304) ILKAIERIPGGLMVVPLLLGAIINTFFPQALQIGGFTTALFKQGAMPLIGLFLLCNGAQITLREAGVSLSKGVALTAVKF FIGAALGWVVGKFMGPTGLLGLSSLAIIAAVTNSNGGLYTALASQYGDPTDVGAIAILSINDGPFLTMVAMGATGLANIP FIALVAVVIPILVGMILGNLDEEWKKFLAPGQKLLIPFFAFPLGAALDFRTILKAGFPGILLGVLTVVLTGLGGYLTIKY IFRENKGVGAAIGTTAGNAVATPAAVAAVDPTLEPLVAAATAQVAASVIITAILCPLLVSYLDK |
| 1074 |
PF14452 (Multi_ubiq) |
 Estimated TM-score=0.79; easy target. |
>Multiubiquitin (Length=63) LNFLSRQVSDPVPLGRQLLEAAALDPQDGYSLFAVLPSGDFEDVRLDEPFDLREHGTERFVTF |
| 1075 |
PF13547 (GTA_TIM) |
 Estimated TM-score=0.79; easy target. |
>GTA TIM-barrel-like domain (Length=299) WGLRRMVLHYAHLCAAAGGVDAFLIGSEMRGLTTIRSGASTYPAVQAYRDLLADVRSILGSGTRIGYAADWSEYFGHQPS DGSGDVFFHLDPLWADPAIDFIGIDNYMPLSDWRDGFDHADAQEGWPAIYDRAYLQGNISGGEGFEWFYASAADRTAQVR TPITDGTAGKPWVFRNKDLRAWWSNPHYDRPGGVESGAPTAWVPQSKPIWFTELGCPAIDRGTNQPNVFFDPKSSESFTP HFSRGWRDDAIQRAYLEATYLFWSTQANNPVSSVYGGRMVHVSECAAWTWDARPYPYFP |
| 1076 |
PF12224 (Amidoligase_2) |
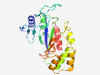 Estimated TM-score=0.79; easy target. |
>Putative amidoligase enzyme (Length=234) QDFGIEIEMTGLTRSRAAEVIAEYFGTTKEYEGSFYDAYFARDTTGRKWKVMSDGSLDCQRKEGRRKVSADRNYSVELVS PICQYKDIETVQELIRKLREAGAFVNSSCGIHIHINAAPFDAPHLRNLVNIMAAKEDMIYKALKVSRGRESHYCRKIDPA FLERLNRQKPATLDRLKSLWYNGRDGSREHYHDSRYHCLNLHSVFQKGTVEFRAFNGELHAGKIKAYIQFCLAM |
| 1077 |
PF11255 (DUF3054) |
 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF3054) (Length=104) LAGDIVCVVVFCAIGRRSHAEGLTLAGIAETAWPFLSGTVAGWLISRAWRAPTAIAPTGVIVWLCTVVVGMLLRKASSQG VAVSFIIVASVVTAVLLLGWRGVA |
| 1078 |
PF01610 (DDE_Tnp_ISL3) |
 Estimated TM-score=0.79; easy target. |
>Transposase (Length=244) DEYSFKKGKMSFIAQDFDSLTILAILDGRTQTTIRNHFLRYSRQVRNRVKVITMDMFSPYYDIAKKLFPNAKIVLDRFHI VQHMSRAMNHLRIQIMKQFDRKSHEYKALKSYWKLIQQDSRKLSDKRFYRPTFRMHLTNKEILKKLLSYSQELREHYELY QLLLFHFQKKQAEHFFDLIEELLPSVNPIFQTIFKTFLKDKDKIINALELPYSNAKLDATNNLIKVIKRNAFGFRNFDNF KLRI |
| 1079 |
PF02498 (Bro-N) |
 Estimated TM-score=0.79; easy target. |
>BRO family, N-terminal domain (Length=88) VRVQLDEHKRRWFNANDICAALELLNPRAALAQHVDAENVSKRAAIDTIGRTKHVNYLNESGVYALLIGSTKEAAKRFRQ WLISEALP |
| 1080 |
PF14301 (DUF4376) |
 Estimated TM-score=0.79; hard target. |
>Domain of unknown function (DUF4376) (Length=97) AEINTWRDQQEQTPVEHDGHHWDTDAASRARIISVLMAGVMPLDYWTDADNNDQPMTLDQLRELYAAIVQQGGRIHDRQR TMKADVSALTTIEEVQA |
| 1081 |
PF17157 (GAPES4) |
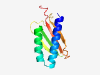 Estimated TM-score=0.79; easy target. |
>Gammaproteobacterial periplasmic sensor domain (Length=97) IQYKFSHRVQAVATAIDTHLVSNDFSALRPQITELMMSADIVRVDLLHGDKQVYTLARNGSYRPVGTSDLFRELSVPLIK HPGMSLRLVYQDPMGNY |
| 1082 |
PF11459 (AbiEi_3) |
 Estimated TM-score=0.79; easy target. |
>Transcriptional regulator, AbiEi antitoxin, Type IV TA system (Length=149) RVMLFGYKDEKLPAWFLKHQWDVKTEYHSSSFLPAETGLVDLEFKTFSMQISGAARAMLECLYLVPKKQQLLECYQIMEG LNNLRPKIVQTLLEECRSIKVKRLFLYLANKAGHQWVQYLNLDNIDLGTGKRSIVKNGVYIHKYQITVP |
| 1083 |
PF17441 (DUF5419) |
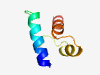 Estimated TM-score=0.79; hard target. |
>Family of unknown function (DUF5419) (Length=54) NFTRWLRRLDIELDKITGGIGLTRNDFADWRYAVAFTNGIAPRQAAIDMLAEDH |
| 1084 |
PF06853 (DUF1249) |
 Estimated TM-score=0.79; hard target. |
>Protein of unknown function (DUF1249) (Length=119) RLMRLLPDMRSGCETRRIAISQGDQLLGVLVLEVLESCPYTTTVRVSQEHCLSWLPVPKMEVRVYHDARMAEVVRAENAR RFRGIYHYPNAQMHQPDEKNQLNLFLGEWLGHCLACGHE |
| 1085 |
PF11205 (DUF2987) |
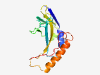 Estimated TM-score=0.79; very hard target. |
>Protein of unknown function (DUF2987) (Length=144) AQQYMFTYSKLFSQMKSNTKEDHPDVKMGIFFVDAQSKQNCVIEKAWMEKEEHYEELPIGPSNELVIPLDNNLRQANPLV FVNTPQDKRCDYSMVVMAKKSFEGKVTYQEVEVLLPQMQAMLEQLGGMFASWFTPAVEGVTLEF |
| 1086 |
PF14208 (DUF4320) |
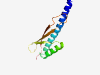 Estimated TM-score=0.79; hard target. |
>Domain of unknown function (DUF4320) (Length=112) VLCAMLVIALAVNVFPVYIQKQQIDTFAAELIREAEISGQVGSETERREQYLREKTGLSPTVAWSKTGRIQLNEEVTVTV TYQTNIGLFGGFGSFPITLRADAAGKSEVYWK |
| 1087 |
PF04586 (Peptidase_S78) |
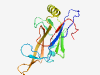 Estimated TM-score=0.79; easy target. |
>Caudovirus prohead serine protease (Length=159) NLKTRSEEDSQTQIVTGYAAVFNSPTELWEGLNEVIKPGAFSRALSNSDVRCLFDHDWGKVLGRTRSGTLKLEEDDKGLR FEVELPNTTVANDLIQSMSRGDINQCSFGFYPTEETWDYRSDPVLRTIHEVELYEVSIVSLPAYEDTEAALSRNKQEMK |
| 1088 |
PF12147 (Methyltransf_20) |
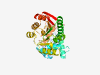 Estimated TM-score=0.79; easy target. |
>Putative methyltransferase (Length=309) IRRFVGQCFQQPAERPSLLDADRQGASCAEAESLAAPLPKYSPRNLYWQATRASLRLGKQLSEGVKLGFDTGFDSGSTLD YVYRNQPNGKGKLGQLVDRNYLDAIGWRGIRQRKLHAEELLRNAIERLRQQGRPVHIVDIAAGHGRYILEALQGLEQRPD SILLRDYSELNVTQGNALISEKGLGDIARFVQGDAFDRASLAALDPKPTLAVVSGLYELFPDNDLVRTSLAGLAEAIEDG GYLVYTGQPWHPQLEMIARALTSHRGGEAWVMRRRSQAEMDQLVQAAGFRKLEQRIDEWGIFSVSLAQR |
| 1089 |
PF11236 (DUF3037) |
 Estimated TM-score=0.79; very hard target. |
>Protein of unknown function (DUF3037) (Length=118) YAVIRVVPRVEREEFMNVGVILYCAGKGFLQTRCTLNEARVQAFSPELDMDELQERLQAFDRICKGRSEGGTIGQLPIAS RFRWLTATRSTIVQTSPVHPGLCEDPEEKINQLFQQLV |
| 1090 |
PF10049 (DUF2283) |
 Estimated TM-score=0.79; easy target. |
>Protein of unknown function (DUF2283) (Length=48) VTYDPEVDAAYIRFSTGKVEESEEVAAGIVLDYDEEGRIVGMEVLNAR |
| 1091 |
PF09318 (Glyco_trans_A_1) |
 Estimated TM-score=0.79; trivial target. |
>Glycosyl transferase 1 domain A (Length=199) YFISGGLPSNYGGLTKSLLLRSKLFGEECNQNTFFLTFRFDLELSSKIDELYSNGKIDKKFTSVINLFDDFLSVRTNGKR SYEERIGLDQIKKQVGMGKFAKTLLRLFGKKNNEMSVVYYGDGETIRYVDYWNDKNQLIKREEYTKNGNLVLVTHYDVQL NKMYLQEYINDQNQVYLDKLYVWNNEEKDVQLSHIIWYS |
| 1092 |
PF14907 (NTP_transf_5) |
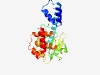 Estimated TM-score=0.79; easy target. |
>Uncharacterised nucleotidyltransferase (Length=251) LDWARLLRLAEHHRVVPAVYLQLKRINHPAIPAELLRNLQAQYQRNTLRMLRLQAEAGRVTRLLMNHGIRVLMLKGPALA QQLYGDVSLRTSKDVDLLIAPDDMDQAEQWMQEAGYVSKSGEARVLGSWKWKDHHASFLHPHKRVEIELHWRLHPDAGGE PSFEDLWKHRQIGEVTATGVPCVYTLGSEHQFLYLSAHGARHGWFRLRWLVDIDRLAVRAVDWEALLPMMRRYGGLPAGG QAWALAAALLG |
| 1093 |
PF13933 (HRXXH) |
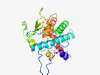 Estimated TM-score=0.79; easy target. |
>Putative peptidase family (Length=238) ILSSAAIAAALPTPETSEGNQSQPFSGLAPWNAGAVNQFPIHSSCNATQRRQIELGLNETITLASHAKDHILRWRNESEI YKKYFGDRPSMEAIGAFDIVVNADKKDILFRCDNPDGNCDNESWAGHWRGENGTDETVICDLSYQTRRSLSTMCGLGYTV SESETNTFWAADLLHRLYHVSAIGQNWIDHFADGYEEVIDQAKENATLSTRDSETLQYFALEVYAYDIAVPGIGCPGE |
| 1094 |
PF15474 (MU117) |
 Estimated TM-score=0.79; easy target. |
>Meiotically up-regulated gene family (Length=107) NNKGSSQCGSIDDACDRALNDGFDDETIYKDYVSRYARIQSGMVMVATFGQAGCTAQFKCDDYGLGMKGKDIKDAVQHMK DNDGVSKCGTAYLSNTCQITLNYCTNC |
| 1095 |
PF17641 (ASPRs) |
 Estimated TM-score=0.79; trivial target. |
>Ancylostoma-associated secreted protein related (Length=121) CKELGRYKMRDNIKKIVIGQVSTAEPLRGHLISTTIIYDCVLEGLAVLEMKRRGPANACTSAGQFFTEKFEDTKRAGYKP KQFIEQAVKSFYPTIGKLGVMGKYGCSYATDEIKYKLVCVF |
| 1096 |
PF13446 (RPT) |
 Estimated TM-score=0.79; easy target. |
>A repeated domain in UCH-protein (Length=59) TVEQALVFLGVDEQTSDDFIITMYTAKISDSPSSKDLARRAVELIAEARKSDTLKHFLK |
| 1097 |
PF07564 (DUF1542) |
 Estimated TM-score=0.79; easy target. |
>Domain of Unknown Function (DUF1542) (Length=73) EEAKTAVNNAANAKNAAIDNNNNLTAEEKAAEKAKVEAAKNAALAGIDQAKTTAARNAAQNKGTSDINAVNPV |
| 1098 |
PF14216 (DUF4326) |
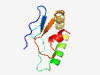 Estimated TM-score=0.79; hard target. |
>Domain of unknown function (DUF4326) (Length=76) VVHCKRDEYDVYIGRPSKWGNPFVVGRDGDRETVIQKYREWIMEQDELLADLHELRGKRLGCWCAPKACHGDVLAE |
| 1099 |
PF01955 (CbiZ) |
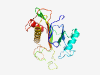 Estimated TM-score=0.79; hard target. |
>Adenosylcobinamide amidohydrolase (Length=208) FPAPRRVVSTARLNGGYREDIKAVFNHHVPPCSHGHDPKTLPGRSWEGYLAYTAQELGLSPKHSVGLLTAARMENASCKS ERYRNLEVTAIVTAGVDVNGGRAGDPARYYEEEGKWIFVGGTINIILLINGNLPPHTLVRSVVTATEAKAAALQELMVPS RYSRGLATGSGTDQIVAVADQESPYRFSDAGKHSKLGELIGAVVKEAV |
| 1100 |
PF14089 (KbaA) |
 Estimated TM-score=0.79; hard target. |
>KinB-signalling pathway activation in sporulation (Length=179) GGITTAIIGIILNWSEYEKLFLRSDVLEIIAVLVWHIGVGFIFSVISQMGFFAYLTIHRFGLGIFRSDSLWNSVQVVLIL FVLFDLVYFRYQVFAVEGESIVSYILVALFILIVGLIVAYIKSAQTNKGAFVPALFFMVVVTVIEWFPALRINNEDWLYL MLFPLLVCNAYQLLILHKL |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218