

Go to page (83 pages in total): [First page] << < 65 66 67 68 69 70 71 72 73 74 75 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 6901 |
PF04440 (Dysbindin) |
 Estimated TM-score=0.41; very hard target. |
>Dysbindin (Dystrobrevin binding protein 1) (Length=142) TEHAPRRPDMESTQQLKLRERQRFFEEVFQHEVDVYLSSAHLCIKDYRRPPIGSVSSMEVNVDMLEQMDLMDMSDHEALD VFLHSGGEDNSAASPVKGPDVESFTTEISLQVPTQAELRHKLSSLSSTCTDSASQDTEAGED |
| 6902 |
PF14214 (Helitron_like_N) |
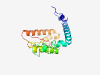 Estimated TM-score=0.41; very hard target. |
>Helitron helicase-like domain at N-terminus (Length=174) YFAFRIHERSNEISTIMYAKRLFQQFLVDAYTMVESSRLHYIRLNKKALRCEAYKGLSDALTRGEVQPSSQGKRIILPSS FTGVARYMIQNYQDAMAICRFVGYPNLFITFTCNPKWPEIERFLEKRNLKAEDRPDIVCRVFKMKLDCLIKEIKNGKLFG RVKKRGLPHAHILL |
| 6903 |
PF16616 (PHC2_SAM_assoc) |
 Estimated TM-score=0.41; very hard target. |
>Unstructured region on Polyhomeotic-like protein 1 and 2 (Length=120) NVGCTKRVGLFHTDRSKLQKAGATTHNRRRASKASLPTLTKDTKKQPTGTVPLSVTAALQLTHSQEDSSRCSDNSSYEEP LSPISASSSTSRRRQGQRDLELPDMHMRDLVGMGHHFLPS |
| 6904 |
PF03338 (Pox_J1) |
 Estimated TM-score=0.41; very hard target. |
>Poxvirus J1 protein (Length=148) MDHNKYLLTIFLEDNESFFKYISEQDDETAMSDIENIVNYLDFLLLLLIKSKDKLESIGYFYEPLSEEYRSLIDFSDMKN FRVLYNRIPINVTPESSIELNKGYLSDFVISLMRLKKEFKGSDSVTKTITYIDPRKDISFSNILSILN |
| 6905 |
PF10930 (DUF2737) |
 Estimated TM-score=0.41; hard target. |
>Protein of unknown function (DUF2737) (Length=53) MSRDQANYLTVTVGGKSDRKHTPMPSREELMKRNSFGSVNNNKYLNRWLGAKK |
| 6906 |
PF15030 (DUF4527) |
 Estimated TM-score=0.41; hard target. |
>Protein of unknown function (DUF4527) (Length=276) LQDPEAEASKEDLRLCVRQLHHQVLTLQCQLRDQGSAHQELQASRDEAARLRCELKDKVDALQKKHHEASLAVTPLKAKL ASLVQKCRERNHLITHLLQELHRHGVENHQLSETASNVVNDVALAEYAATFLAPGVPQTSHHLDVESEKTAVVRAQKYLL NPEMNSVLQRPLHSESWPIPEAEWPAQTAQLDSLKLPWPSGPTPDPGMCQASIAVEPGLPVQRLQEKGGMSCPGLQAEHL LPSPELLSPARILALHQELRQSICSTSQVNKSPLEL |
| 6907 |
PF15334 (AIB) |
 Estimated TM-score=0.41; very hard target. |
>Aurora kinase A and ninein interacting protein (Length=324) QTHLIKSSTKMLTLFPGERKANISFTQRSRPPAGTRQTSIASFFTLQPGKTNGGDQRSVSSHIESQTNKESKEDATQLDH LIQGLGDDCMAPPLATSTPADIQEARLSPQSFQASYHHGIGSPCVTVLSLFQPDTFVCAGESKASLACSFTQDLESSCLL DQKEEEDSSWKREWPYGSKKKNYQNVERHSKTPGHKGHQLLDKTNLEKVSAKRNRQAPILLQTYQDSWSGANKEAVKQSP CPIPVFSWDSEKNDKDSWSQLFTQDSQGQRVIAHNSRAPFRDVTNDRNQGLGQLPNSPWAQCQARTTQLNLQTDLLFTQD SEGN |
| 6908 |
PF11088 (RL11D) |
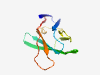 Estimated TM-score=0.41; easy target. |
>Glycoprotein encoding membrane proteins RL5A and RL6 (Length=99) NELKATTGSNFTITHRKDPLTTKWKTVFGNNGDQWLCNVTGIGNATVNGNATICVSSCGHNTLDLCNLKSGDSGFFDLSR WFGENMDEYSGDVWHLEVS |
| 6909 |
PF17665 (DUF5527) |
 Estimated TM-score=0.41; very hard target. |
>Family of unknown function (DUF5527) (Length=139) MNTEEAGNETSPTLSILEFIATATNLVPLSSGPPVEKSDPLPIIIVLICIFLLLATCLIFVTLCKPAALDQSRYGPHECM PYHPEDASEPQLRLWKRLGSLRRSINSFRRSQPVSQNHRTCPRRPPAAQDWDFMESTKM |
| 6910 |
PF15091 (DUF4554) |
 Estimated TM-score=0.41; very hard target. |
>Domain of unknown function (DUF4554) (Length=454) MTDCLIIKHFLRKIIIIHHKIRFNFSVKVNGILSTEIFGVENEPILNLWNGIALVVSYQRYVSRPKFITTESHCSRIHPV LGHPVTLFIPDDLAGMGLLGELILTPAAALCPSPRVFSSQLNRISSVSIFLYGPSGLPLVLSNQEQPSTTVLKDICYFID WKKYQLCMVPNLDLNLDGDFVLPDVSYQTESSEGAQSQNMDPQRQTLLLFLFVDFHSGFLIKQMEIWGVHTLLTAQLSAI LMESHSAVRDSIQAMVDHVLEQHHQAAKAHQKLQASLSVAVNSVLSVVSGSTNSSFRKTCLQAFQAADTQELGTKLHKIF HDVTQHRFLHHCSCDMKQQLTPEKKDLAQNIEDAHENSPELLAEISGQAENKRLKRGSLHQSAEEMEVPHSASAPGTPEI APRRLEPTAASLTVKRAERRKVHVQAPAPDRASPGRVLEDALWLQEVSNLSEWL |
| 6911 |
PF05614 (DUF782) |
 Estimated TM-score=0.41; very hard target. |
>Circovirus protein of unknown function (DUF782) (Length=104) MVTIPPLVSRWFPVCGFRVCKISSPFAFATPRWPHNDVYIRLPITLLHFPAHFQKFSQPAEISDKRYRVLLCNGHQTPAL KQGTHSGRQVTPLSLRSRSSTFHQ |
| 6912 |
PF03875 (Statherin) |
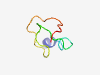 Estimated TM-score=0.41; hard target. |
>Statherin (Length=41) DSSEEKFLRRIGRFGYGYGPYQPVPEQPLYPQPYQPQYQQY |
| 6913 |
PF05680 (ATP-synt_E) |
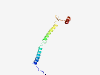 Estimated TM-score=0.41; easy target. |
>ATP synthase E chain (Length=97) MSSNASKAGSNVLRYSALAAGIFYGFTHQRSISAAEKAAAAQREYEHKQELINKAKEAYAKSKQPVSAVSTPSGQTVNQD PMSPNFDIEAYFSALLA |
| 6914 |
PF03034 (PSS) |
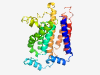 Estimated TM-score=0.41; hard target. |
>Phosphatidyl serine synthase (Length=276) PHPAIWRIVFGLSVLYFLFLVFIIFLNWEQVKNVMYWLDPNLRYATREADIMEYAVNCHVITWERILSHFDIFAFSHFWG WGMKALLIRSYGLCWTISITWELTELFFMHLLPNFAECWWDQLILDILLCNGGGIWMGXXXXLTENDTYHWASIKDIHTT TGKIKRAALQFTPASWTYVRWFDPKSSLQRVTGVYLFMIIWQLTELNTFFLKHIFVFPACHALSWCRILFIGIITAPTVR QYYAYLTDTQCKRVGTQCWVFGAIAFLEALACVKFG |
| 6915 |
PF10814 (CwsA) |
 Estimated TM-score=0.41; hard target. |
>Cell wall synthesis protein CwsA (Length=133) LTPRERLTRGLTYSAVGPVDVTRGVVGLGVHSAQSTASELRRRYQEGRLARELAAAQETLAQELAAAQEVVANLPQALQD ARRKQRRSKRPYLIAGAAVVVLAGGAVTFSIVRRSMQPEPSPRPPSVDPQPRP |
| 6916 |
PF12346 (HJURP_mid) |
 Estimated TM-score=0.41; very hard target. |
>Holliday junction recognition protein-associated repeat (Length=115) MSRLLSTKPSSIISTKTFITQNWNSRRRHRCKSRMNKTYCKGARRSQRSSKENFVPCSEPVKETGALRDCKNILDVSCHK TGLKLEKAFLEVNKPQIHKLDPSWKELKVTPSKYS |
| 6917 |
PF17195 (DUF5132) |
 Estimated TM-score=0.41; hard target. |
>Protein of unknown function (DUF5132) (Length=46) LVKPLVRGTVKTSMSLVMELKKAAHEAGEELHDMTAEVAAELIGAE |
| 6918 |
PF06593 (RBDV_coat) |
 Estimated TM-score=0.41; very hard target. |
>Raspberry bushy dwarf virus coat protein (Length=274) MSKKAVPPIVKAQYELYNRKLNRAIKVSGSQKKLDASFVGFSESSNPETGKPHADMSMSAKVKRVNTWLKNFDREYWDNQ FASKPIPRPAKQVLKGSSSKSQQRDEGEVVFTRKDSQKSVRTVSYWVCTPEKSMKPLKYKEDENVVEVTFNDLAAQKAGD KLVSILLEINVVGGAVDDKGRVAVLEKDAAVTVDYLLGSPYEAINLVSGLNKINFRSMTDVVDSIPSLLNERKVCVFQND DSSSFYIRKWANFLQEVSAVLPVGTGKSSTIVLT |
| 6919 |
PF12540 (DUF3736) |
 Estimated TM-score=0.41; very hard target. |
>Protein of unknown function (DUF3736) (Length=138) RRRLVSKLDLEERRRREAQEKGYYYDLDDSYDESDEEEVRAHLRCVAEQPPLKLDTSSEKLEFLQLFGLTTQQQKEELLT QKRRKRRRMLRERSPSPPTSKRQTPSPRLALTTRYSPDEMNCSPNFEEKRKFLTIFNL |
| 6920 |
PF15261 (JHY) |
 Estimated TM-score=0.41; hard target. |
>Jhy protein (Length=126) LGGLGPDYQTAQEKAQKVKRQKEYAQQVREQNKKMCSDPFTSLNKTTLSSENKNSVPRQKALEYAKSIRKPKITPKPRST NQEPSKRNFFEHPQYLEDFDLSQIAMLEMLQRRHEEEKQAVAAFKA |
| 6921 |
PF06322 (Phage_NinH) |
 Estimated TM-score=0.41; easy target. |
>Phage NinH protein (Length=60) MTHTVKTIPDMLIETYGNQTEVARRLKCSRGTVRKYVDDKDGKMHAIVNDVLMVHRGWSE |
| 6922 |
PF06072 (Herpes_US9) |
 Estimated TM-score=0.41; hard target. |
>Alphaherpesvirus tegument protein US9 (Length=59) CYYSDSENETADEFLRRIGKYQHKIYHRKKFCYITLIIVFVFAMTGAAFALGYITSQFV |
| 6923 |
PF06708 (DUF1195) |
 Estimated TM-score=0.41; hard target. |
>Protein of unknown function (DUF1195) (Length=135) ANNNNNNHNNNSNKGNYNYKFWVLAAIIVLALWSMFTGSVTLKWSASNLTRFSHDLDHSITLQDLDVLEVEEREKVVRRM WEVYTHSTTAKVPRFWSDAFHAAYEHLVSDVPTIRDAAISEIAKMSIQFLSLQQQ |
| 6924 |
PF14951 (DUF4503) |
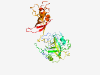 Estimated TM-score=0.41; easy target. |
>Domain of unknown function (DUF4503) (Length=386) QDAYGMFGEVYLEGAMLKGRQLEGKSCRLAGMKVLQKATRGRTPGLFSLIDTIWPPAIPLKAPGHSQPCEEVKSHLPPPS FCYVLSVHPNLGQIDMIEGDSISKLHQPPILHCLRDILQTDNLGARCSFYARVIYQRPQLKSLLLLAQKEIWLLVTDITL QMKDGNDHHLPKILPVCVTPSCVLGPEILEALSTAAPHSILFRDALRDQGQIVCVERTVLLLQKPFWGMASGAGSCELTG PVILDELDSLTPVNSICSVQGTVVGVDENTAFSWPTCDRCGNGRLEQRPEDGGTFFCGDCCELVTSPVFKRHLQVFLDCP SRPQCSVKVKLLQGSISSLLRFAACEDGTYEVKSVLGKEVGLLSCFVQSITTHPTNFIGLEEIELL |
| 6925 |
PF07131 (DUF1382) |
 Estimated TM-score=0.41; hard target. |
>Protein of unknown function (DUF1382) (Length=60) MHKASPVELRTSIEMAHSLAQIGIRFVPIPVETDEEFHTLAASLSQKLEMMVAKAEADER |
| 6926 |
PF15792 (LAS2) |
 Estimated TM-score=0.41; very hard target. |
>Lung adenoma susceptibility protein 2 (Length=75) SLDIEKNPNFQGPYTPVGNDNFVTPVIRTNINGKQCGRLKNPKLIHRTNNCISESSLSFPKKSSFKDSSEHSLEK |
| 6927 |
PF15094 (DUF4556) |
 Estimated TM-score=0.41; very hard target. |
>Domain of unknown function (DUF4556) (Length=215) KCPMLRSRLGQESVHCGPMFIQVSRPLPLWRDNRQTPWLLSLRGELVASLEDASLMGLYVDMNATTVTVQSPRQGLLQRW EVLNTSAELLPLWLVSGHHAYSLEAACPPVSFQPESEVLVHIPKQRLGLVKRGSYIEETLSLRFLRVHQSNIFMVTENKD FVVVSIPAAGVLQVQRCQEVGGTPGTQAFYRVDLSLEFAEMAAPVLWTVESFFQC |
| 6928 |
PF15330 (SIT) |
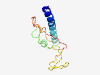 Estimated TM-score=0.41; hard target. |
>SHP2-interacting transmembrane adaptor protein, SIT (Length=107) ILLPLILAVVLLLLMVASLVVWRMVKQQKKAAGISPEQVLQPPEGDICYANLSLQKTENSGSSQKKAPTRSSSCTQANAG EVEYVTMTPFPREDIAYAALSLETSDQ |
| 6929 |
PF06589 (CRA) |
 Estimated TM-score=0.41; very hard target. |
>Circumsporozoite-related antigen (CRA) (Length=121) NTKLYSLFVFICLIIVYCATVQGSKDNGKNGVYELKMIKKKHKKNNKDNKHVQRAGEDEDEDDNIDYTNLVKEEEKRNEE VKRLSKMHKAYTLGMSTIIFLSTLFLIRDCVLESISRPDSY |
| 6930 |
PF04168 (Alpha-E) |
 Estimated TM-score=0.41; hard target. |
>A predicted alpha-helical domain with a conserved ER motif. (Length=311) MLSRVADSIYWLNRYVERAENIARFADVNFNLMLDSPTGITQQWEPLVKITGDLPLFQARYGEATAENVIQFLTFDTEYP NSIISCLRSARENARSIQEIISSEMWQQVNQFYMMVQEAANQQTMSQLLEFLAEVKMAGHLFAGVMDATMTHNEGWHFGQ MGRFLERADKTSRILDVKYFILLPSVQYVGTALDEIQWMALLKSASAYEMYRKRKQHRITPTGVAEFLIMDREFPRSIHF CLLQAESSLHEITGTPSNTWKTPVERALGRSRAELDYLTIEEIIKKGLHEFMDDLQQQINDVGHKIFESFF |
| 6931 |
PF06567 (Neural_ProG_Cyt) |
 Estimated TM-score=0.41; very hard target. |
>Neural chondroitin sulphate proteoglycan cytoplasmic domain (Length=119) LYLLKTENTKLRRTNKFRTPSELHNDNFSLSTIAEGSHPNVRKFCDTPRVSSPHARALAHYDNIVCQDDPSAPHKIQDPL KSRLKEEESFNIQNSMSPKLEGGKGDQDDLGVNCLQNNL |
| 6932 |
PF17220 (DUF5137) |
 Estimated TM-score=0.41; very hard target. |
>Protein of unknown function (DUF5137) (Length=85) MEQENIDPYEEFYLNSISAEQNKRKDELSQVQTEHLYYRSLEPEKFPALQSIDISESSKGITRKRGVDDNLKSSCPSEND GKKKR |
| 6933 |
PF17462 (DUF5424) |
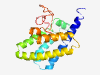 Estimated TM-score=0.41; very hard target. |
>Family of unknown function (DUF5424) (Length=174) YNQEGYEFKITQAFNCVNDAIALTCSSVNHFLDIPKVINKYGVLATVALSKTEQLNLTDTKLESIVKNGFIIYEVVETII KASNAITKDQQLFSKAIDDGINAVNKLQEAAIHLSSVGEPIVEYVTSFFGSHDQHEPLQHESLLFETYTPNSTSAVIEDY KLIGSNAQLIAVEI |
| 6934 |
PF06540 (GMAP) |
 Estimated TM-score=0.41; hard target. |
>Galanin message associated peptide (GMAP) (Length=61) SKRELQPEDDMKPGSFDRSIPENNIMRTIIEFLSFLHLKEAGALDRLPDLPAAASSEDIER |
| 6935 |
PF16951 (MaAIMP_sms) |
 Estimated TM-score=0.41; hard target. |
>Putative methionine and alanine importer, small subunit (Length=60) MGTTAIIMMVLFMVIIWGGLVYATIALRREPDEKVGLFGTSPYATDTVLIEQESERPATA |
| 6936 |
PF04899 (MbeD_MobD) |
 Estimated TM-score=0.41; easy target. |
>MbeD/MobD like (Length=70) MTELETQLLSALEQLQQDYSQRLDEWESAFAEWQRMCGLMQRENAALSERVTNLSQQVDRLSGQLSRLSR |
| 6937 |
PF11126 (Phage_DsbA) |
 Estimated TM-score=0.41; easy target. |
>Transcriptional regulator DsbA (Length=67) IKEASDHKLKISGYNELIKDIRIRAKDELGVDGKMFNRLLALYHKDNRDVFEAETEEVVELYDTVFS |
| 6938 |
PF05755 (REF) |
 Estimated TM-score=0.41; hard target. |
>Rubber elongation factor protein (REF) (Length=215) NDEKGLKYLEFVQVAAIYTIVCFSSIYEYAKENSGPLKAGVQTVEGTVKTVIGPVYEKFHDVPFDLLKFVDRKVDNALSE LDGHVPSLVKHVSSQALAAAHKAPDVARSVAAEVQRSGVVDSAKNITKALYDNYEPTAKELYHKYEPVAEQYAVSAWRSL NRLPLFPQVAQMAVPTAAYWSEKYNQAVRYSAESGYAVALYLPLIPIDRIAKVFE |
| 6939 |
PF17567 (DUF5473) |
 Estimated TM-score=0.41; very hard target. |
>Family of unknown function (DUF5473) (Length=106) MERRVWFLSRSARSLAAMFSCTYSRATHRHSGKTVVRSSGTRCTRQPRLCRVTRSTLVATSPRRRSLVQQRRPPLREQNG GSGSSCVSSGGSASTVKTPGSRRASK |
| 6940 |
PF13322 (DUF4092) |
 Estimated TM-score=0.41; hard target. |
>Domain of unknown function (DUF4092) (Length=176) AEEIDKELGRQKLAFGEQYRKPKQIKAVDSDAQNVQRDVERLWGATQQAQREGWKPVERFHIFHDSTNFYGSTGSARAQA AVNISNKAFPVVMARNDKNYWLDFDKPQAWDENGLAYITEAPSKVKPKKVDASNATFNLPFISIGDLGKGKVMVMGNARY NSVLVCPNGFSWNGGV |
| 6941 |
PF15700 (DUF4667) |
 Estimated TM-score=0.41; very hard target. |
>Domain of unknown function (DUF4667) (Length=305) SSNRPISAHLTIHYKAIQEEEGEDMRSGAGSGGHHDDYFLESNRSPTPNKKHEFIKTVLNINDNDSEFSESCSPREKLHN ERACNTDLFGDFMSKRQQRLSNSMNIYDLYQCVHNLSPSNNNHQFIARRFSDSHIPSLHHRQQQQKVTTKNFVQPTKDIQ RIASYAADSDQRVKYLPNYHQSAPSTALSAAESKAAVPRKLPDRDSTQNYVLKLQLSSPNSQPMSPRTRPGYRPSCSSSN CSSSSSSSACSSVSISDPNNITAYETNNVNPQFPSNQPLDISSPCARHHHRRNSIAVKFDKALYK |
| 6942 |
PF05955 (Herpes_gp2) |
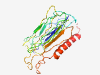 Estimated TM-score=0.41; hard target. |
>Equine herpesvirus glycoprotein gp2 (Length=230) KNFMEASCTVETNSDLAIFWKIGKPSVDAFNRGTTHTRLMRNGVPVYALVSTLRVPWLNVIPLTKITCAACPTNLVAGDG VDLNSCTTKSTTIPCPGQQRTHIFFSAKGDRAVCITSELVSQPTITWSVGSDRLRNDGFSQTWYGIQPGVCGILRSEVRI HRTTWRFGSTSKDYLCEVSASDSKTSDYKVLPNAHSTSNFALVAATTLTVTILCLLCCLYCMLTRPRASV |
| 6943 |
PF13916 (Phostensin_N) |
 Estimated TM-score=0.41; hard target. |
>PP1-regulatory protein, Phostensin N-terminal (Length=89) AERDRLSQMPAWKRGILERRRAKLGLPPGEGSPVPGNAEAGPPDPDESSVLLEAIGPVHQNRFIQQERQRQQQQQQQRNE VLGDRKAGP |
| 6944 |
PF16612 (RGS12_usC) |
 Estimated TM-score=0.41; very hard target. |
>C-terminal unstructured region of RGS12 (Length=138) KGFSKRSATGNGRESASQPGERWEPVQDSSDSPSTSPGSASSPPGPPGTTPPGQKSPSGPFCTPQSPVSLAQEGTAQIWK RQSQEVEAGGIQTVEDEHVAELTLMGEGDISSPNSTLLPPPSTPQDVPGPSRPGTSRF |
| 6945 |
PF12877 (DUF3827) |
 Estimated TM-score=0.41; very hard target. |
>Domain of unknown function (DUF3827) (Length=673) GVPDHQFQVHSVLQFVPSYIDVRFCNFSERIEKGLIMAFSEVRRRYQESPDFIVHIINITMNLPKAQKQLQRVPVDIIFA IRDSTGYLNGSEVSSLLRNLNMVEFSFYLGFPVLQIAEPFHYPELNVTQLLRSSWVKTVLLGVMEQRVNERTFQAKMERR LAQLLGEVLGSARRWKRATNVGNNSVQIVRTTRLDGPDNPLEMIYFVEGPNQERLAATTTSSILNQINVQRAAIVLGYRV QGVLAQPVEKITAPPSETQNNNLWIVVGVVVPVVVVVIIIIILYWKLCRTDKLEFQPDTMSNVQQRQKLQAPSVKGFDFA KLHLGQHNKDDILVIQESAPLPAPVKEVTPSESGDVPSPKSKASSKVSRGAKRRGRISPSDAESTVSEPSSGRESAEEST RPSITPIDGKQHRKTKAGPPQMNGTEEPLSSASIFEHVDRMSRTSEATKRVSNKIQLIAMQPMPASPLHSTAVLEKVSET AKINKEIQTALRHKSEIEHHRNKIRLRAKRKGHYDFPAMDDIGIVDSREHRRIYQKAQMQIDRILDPEVHVPSVFIESKK SARGKRSPKQRRKHQVNGSLTDADKDRLITTDSDGTYKKPPGVNNVAYVSDPDQAPEPRSPSPQDNEVFVGSPPPGHAPP PPAYVPPQPSIEEARQQMHSLLDDAFALVAPTS |
| 6946 |
PF10583 (Involucrin_N) |
 Estimated TM-score=0.41; very hard target. |
>Involucrin of squamous epithelia N-terminus (Length=69) MSQQHTLPVTLPPALNQEPLKPVSPPIDTQQEQVKQPTPLPALCQKMPSKLPGEVPLEHAEKHTTLVKG |
| 6947 |
PF01672 (Plasmid_parti) |
 Estimated TM-score=0.41; very hard target. |
>Putative plasmid partition protein (Length=85) AYDYLKIANAIEEGIIEEEFLVQNGFRQTLFVLRNQESATIKKSKQNPIKPLRFQLKKQESYDFYKSNAKFTSFLMDELF ENQKD |
| 6948 |
PF16935 (Hol_Tox) |
 Estimated TM-score=0.41; hard target. |
>Putative Holin-like Toxin (Hol-Tox) (Length=60) MVAILEGGGAYGFKTVNPDPRKGEASCPVSDALQLMLAFGGFILGILTFVVLLIEKISKK |
| 6949 |
PF10838 (DUF2677) |
 Estimated TM-score=0.41; very hard target. |
>Protein of unknown function (DUF2677) (Length=166) LMCMALMARGTYGAYICSPNPGRLRISCALSVLDQRLWWEIQYSSGRLTRVLVFHDEGEEGDDVHLTDTHHCTSCTHPYV ISLVTPLTINATLRLLIRDGMYGRGEKELCIAHLPTLRDIRTCRVDADLGLLYAVCLILSFSIVTAALWKVDYDRSVAVV SKSYKS |
| 6950 |
PF17254 (DUF5321) |
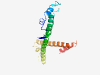 Estimated TM-score=0.41; very hard target. |
>Family of unknown function (DUF5321) (Length=154) STSGHMPKIAQPSLWQSMIPKFIRNRRAKAETSPAKSKEWNPASFYIVIFILIGSQAIRMISLKNEYNAYTRSTDAKIRL LREVIEKIQRGEKVDVEKLLGTGDEAKEREWEEVLREIEEEDSLWHRKNSGSESNRPATPEQGQKQQAEKSRSK |
| 6951 |
PF17522 (DUF5446) |
 Estimated TM-score=0.41; hard target. |
>Family of unknown function (DUF5446) (Length=72) MSGYVCKTDILRMLSDIEDELTRAEQSLNHSVNALEAEMRETENDRLSVLEEEIIDLHICINELYQSPEHRY |
| 6952 |
PF03766 (Remorin_N) |
 Estimated TM-score=0.41; very hard target. |
>Remorin, N-terminal region (Length=52) KDVAEEKASVPPPAEEKPDDSKALAIVEKVEDPPAEKSSGGSTERDAVLARL |
| 6953 |
PF14197 (Cep57_CLD_2) |
 Estimated TM-score=0.41; easy target. |
>Centrosome localisation domain of PPC89 (Length=64) LAANAEELQQRLDTATQQRAELEAQLARLAADRDEARQQLAANAEELQQRLDTATQQRAELEAQ |
| 6954 |
PF15848 (ODAPH) |
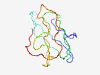 Estimated TM-score=0.41; very hard target. |
>Odontogenesis associated phosphoprotein (Length=108) EEVVNPPGDSQNNANPTDCEIFTLTPPPATRNPVTRIQPITRTPRCPLHFFPPRWPRVYIRFPYRLYPPPKWNHHFQFHP YFCPHSRLPPYYNYFPRRRLWRGSSSEE |
| 6955 |
PF12278 (SDP_N) |
 Estimated TM-score=0.41; very hard target. |
>Sex determination protein N terminal (Length=138) QRRRREWQRQQERERQHEKLKQQKILEYERNRAQALKYAEEITSHHSQSKSGSESPSHVRYRDRSTSTASKSGSLHEELD GSTSGTVPLFRGPQNAQIDTSELRRIKVDIHRNIPVKGPVTELERDILNPEDVIVKRR |
| 6956 |
PF08339 (RTX_C) |
 Estimated TM-score=0.41; very hard target. |
>RTX C-terminal domain (Length=188) SLANINLKEVSFERNGNDLLLKTNNRTAVTFKGWFNKPNSSAGLNEYQRKLLEYAPEKDRARLKRQFELQRGKVDKSLNN KVEEIIGKDGERITSQDIDNLLDKSGNKKTISPQELAGLIKNKGKSSSLMSSSRSSSMLTQKSGLSNDISRMISATSGFG SSGKALSASPLQTNNNFNSYANSLATTA |
| 6957 |
PF17539 (DUF5456) |
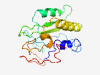 Estimated TM-score=0.41; very hard target. |
>Family of unknown function (DUF5456) (Length=152) MAIYSLHEMAGLLKTGLWQGGEVEKLGYIGTAVPHASPAPACGLTGVYISYDTMLERCEVSMQGGGCELFISQYHPYTGT YMTYVLESNLSDRRLLLAKLEDILADRRNPYLWSHNLYKRNSFGIERRYDTGHSSMWGGRIAAPRQSKFFGR |
| 6958 |
PF12329 (TMF_DNA_bd) |
 Estimated TM-score=0.41; easy target. |
>TATA element modulatory factor 1 DNA binding (Length=67) LRETVKKLRETVDELRETVKKLRETVKKLRETVKKLRETVKKLRETVDELRETVKKLRETVDELRET |
| 6959 |
PF12284 (HoxA13_N) |
 Estimated TM-score=0.41; very hard target. |
>Hox protein A13 N terminal (Length=120) NKNMEGLVGSGNFPANQCRNLMAHSALGGHPSSLVHSSGYSAVDVSGSSSGEPGKQCTPCPAVPQGSSTGPLPYGYFGNG YYPCRMGRGSLKSCTQPSALSYSAEKYMDTPVTSEEYPSR |
| 6960 |
PF07213 (DAP10) |
 Estimated TM-score=0.40; easy target. |
>DAP10 membrane protein (Length=79) MAPPGHLLFLFLLPVAASQTNEGSCSGCGPLSLPLLAGLVAADAVMSLLIVGVVFVCMRLHSRPAQEDGRVYINMPGRG |
| 6961 |
PF15822 (MISS) |
 Estimated TM-score=0.40; hard target. |
>MAPK-interacting and spindle-stabilising protein-like (Length=238) FSLADALPEHSPAKTSAVSNTKPGQPPQGWPGSNPWNNPSAPSSVPSGLPPSATPSTVPFGPAPTGMYPSVPPTGPPPGP PAPFPPSGPSCPPPGGPYPAPTVPGPGPTGPYPTPNMPFPELPRPYGAPTDPAAAGPLGPWGSMSSGPWAPGMGGQYPTP NMPYPSPGPYPAPPPPQAPGAAPPVPWGTVPPGAWGPPAPYPAPTGSYPTPGLYPTPSNPFQVPSGPSGAPPMPGGPH |
| 6962 |
PF11386 (VERL) |
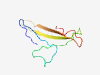 Estimated TM-score=0.40; trivial target. |
>Vitelline envelope receptor for lysin (Length=79) MQMTRGRGINMIRIHYPQTYTSVVPGACVFRGPYSVPTNDSIEMYNVSVALLWSDGTPTYESLECNVTKSQASNAPEPK |
| 6963 |
PF15806 (DUF4707) |
 Estimated TM-score=0.40; very hard target. |
>Domain of unknown function (DUF4707) (Length=447) PLDEPVVLSDEVIFPLTVSLDKLPVSTLKVKVIVTVWKQEVEKAEVQEHGYLTILQHRSPAHIFRDDLNTFKAQVSTTLT VLPPPSVKCQQMTVSGKHLTMLKVLNASSQEEVCIRDVRILPNFNASYLPMMPDGSVLLVDNVCHQSGEVTMASFYRMDS LSCRLPSMLSALEEHNFLFQLQVNEQLQEDTKEGLEVPLVAVLQWSTPKLPFTNSIYTHYRLPSIRLDRPRFVMTASCES PVRVRERFRVRYTLLNNLQDFLAVRLVWTPEGAHSDWAEREGTRRMEERGRRGQAAVSSVVCHTPLSNLGYCRKGSTLSF TVAFQILRAGLFELSQHMKLKLQFTASVSNPPPDARPLSRKNSPSSPAVRDILDRHQASLGRSQSFSHQQPSRSHLMRTG SVMERRAITPPVGSPIGRPLYHPQDKAVLSLDKIAKRECKVLVLESV |
| 6964 |
PF15810 (CCDC117) |
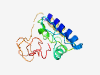 Estimated TM-score=0.40; very hard target. |
>Coiled-coil domain-containing protein 117 (Length=143) EIEAAQRKLQEIEDRITLEDDEDEDLDVEPAPRRPVLVMSDSLKKGLQQGISDILPHTVAQSVSRSCMELVLWRPPEDPF SQRLKDSLQKQRKQQTACRQTPTPCPSPSPHSPLNPSSPPADTHSPLYSFSVAHNSGEEEMEM |
| 6965 |
PF15836 (SSTK-IP) |
 Estimated TM-score=0.40; very hard target. |
>SSTK-interacting protein, TSSK6-activating co-chaperone protein (Length=125) MEQHTSNPTKEEAKEEDNVVCLCRAKPSPSYINLQANSPPATFSNTETTMLPSGAGEKPKECLGLLECMSANLQLQTQLA QRQMAILENLQTSLSQLGSGRESKTCSLPGLCCNLLLNHLPQFHK |
| 6966 |
PF17321 (Vac17) |
 Estimated TM-score=0.40; very hard target. |
>Vacuole-related protein 17 (Length=423) NLDEISERLFIRSQEAVLQLELWIRRQQRLGEIEHENYGDKLADQYNLYMAQLNSLCVRSEYVRDKMNKGYYRGSSVVNE QTYIEDLVYEFQDLTMKLNELAQKQREHCTPSSKCSSNSDDSFRPRPLKIIERHRADVNATRQSPIKDKKRKNVNFSQMV HEDNSCPSRCTSLPGSPVRLISPTKPLRVAKSYDTGLNPRAKLKKQREDELLSFFKDNQRLSITFCDDIDEDSDSASDQN TVISSSPPCENGFKPLRRYNSYDSILSNKVQPISSAKCPTFLSMPTANRPSMKSVTVSSAPVISKAKNRASSKELLSLFI SDTQKQETKAKANTRSATNSFWSLFKMPDQKSKAKNTLREPHRNLLPESNKERTYSRGVYSSCSRLVVGSHQSTILPAVS QCMVDESMMYEELQEALNTELIL |
| 6967 |
PF12958 (DUF3847) |
 Estimated TM-score=0.40; hard target. |
>Protein of unknown function (DUF3847) (Length=84) TTEKLKQEIADAEKKLAQERSRIQRLQNRKSYYEKGDRKKRAHRLITRGAAVESIAPLSKVLSETEFYTFMEKVFTLPEV RALL |
| 6968 |
PF15029 (TMEM174) |
 Estimated TM-score=0.40; very hard target. |
>Transmembrane protein 174 (Length=234) DDFPHNVFSVTPYTPSTADIQVSDDDKAGATLLFSGIFLGLVGITFTVMGWIKYQGVSHFEWTQLLGPILLSVGVTFILI AVCKFKMLSCQSCKESEERTLDTDQTLGGQSFVFTGINQPITFHGATVVQYIPPYASQDTIGVNATYLPPLVNPFGAANS GGPTVPAPSPPQYCTIYPHDNAAFIDDDLYPAYMDMDHRNDNRSSTDAEQLEEIQLEDNSRDCFSPPSYEEIYS |
| 6969 |
PF04725 (PsbR) |
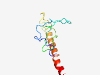 Estimated TM-score=0.40; very hard target. |
>Photosystem II 10 kDa polypeptide PsbR (Length=95) AKKIQTSQPYGPAGGVVFKEGVDASGRVAKGKGLYQFSNKYGANVDGYSPIYTPEEWSSTGDVYVGGKAGLLLWAITLAG ILVGGAILVYNTTSV |
| 6970 |
PF12553 (DUF3742) |
 Estimated TM-score=0.40; very hard target. |
>Protein of unknown function (DUF3742) (Length=119) MNTTTRISTAERLGRALGRGWRAYVRGERRASNWLVSKGVPMAGATVLLWTVKLVVLGLLFYAVFWMALLLVFVMVVAWM ARNADLNDELPEPEWRNGPAGFGLYTYDGFRIDPHVQDD |
| 6971 |
PF17722 (DUF5567) |
 Estimated TM-score=0.40; very hard target. |
>Family of unknown function (DUF5567) (Length=233) MPAQISDLEIMDEDQLIEEVLDKFVNCHEQTYEEFLSTFTHLSKEDNVTKRGAFGTDSSENIFPSVKFAHENEPNDHHLR NKAVFLRTSSQCSEDEQIVIDEGQKVGSSFQGDLNRAGKVKVDNFLDLEDLDVDEETQPQMSKDLLLLPGEVEEDVCPRV PSYIPSAAQPFTPGVKPGPAGRGADKQTEEILGDEVQPFSLDEEFDYDAVMLTPKFTPAEIDAITELSKQERS |
| 6972 |
PF15338 (TPIP1) |
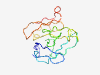 Estimated TM-score=0.40; very hard target. |
>p53-regulated apoptosis-inducing protein 1 (Length=123) GSSSKVSFRSAQASCSGTRRQGLGRGDQNLSVMLPNGRAQTHTLGWVSGLLVLGAQVHGGCRGIEAPSVSSGSWSSATVW CLTGLGLGLSKPFLPGVIVLRDRPLESACELSYDQKKALLSLQ |
| 6973 |
PF15475 (UPF0444) |
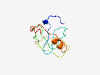 Estimated TM-score=0.40; very hard target. |
>Transmembrane protein C12orf23, UPF0444 (Length=91) KDHPQQQPGMLSRVTGGLFSVTKGAVGATIGGVAWIGGKSLEITKTAVTSVPSMGVGLVKGSVSAVAGGVTAVGSAVASK VPLTGKKKDKS |
| 6974 |
PF12516 (DUF3719) |
 Estimated TM-score=0.40; very hard target. |
>Protein of unknown function (DUF3719) (Length=64) MLFEGKVSPQTQNLWAECSEWARRSLHLRVLGRQLVLPTDEGVQHFQGSKPASAVHRPFSDNCE |
| 6975 |
PF12885 (TORC_M) |
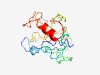 Estimated TM-score=0.40; very hard target. |
>Transducer of regulated CREB activity middle domain (Length=165) RTNSDSALHTSAMNPNPQDPFGMNQQMGRGPPQRNASMNEAEVDSSGDVFSFPAVPNEENLLGVNKPMPKQLWEAKKVQS LSSRPKSCEVPGINIFPSPEQNAGLSHYQGTLNTGGSLPDLSNLHFPSPLPTPLDPDEAGYPNLSGGSSTGNLPAAMMHL GIGNS |
| 6976 |
PF15319 (RHINO) |
 Estimated TM-score=0.40; very hard target. |
>RAD9, RAD1, HUS1-interacting nuclear orphan protein (Length=239) MPPRKKRRQRSQKAQLLFQEPPLEGPKHHYGSPQLTITHTRQVPSKPIDHNTATSWVSPQFDTTAESWFPVNQKRHHRNQ AKHSSRKSTTSKFPHLTFESPLSSSGSAMLGIPLTRECPRQSEKRLSRRPLVPMLSPQSCGEVSAHTLQNFPYVFIPPDI QTPESSLVKEGPNPSDQRESSLPSCSFHTSTPKGPEPGPILVEDTPEEKYGIKVTWRRRHHLFAYLRERGKLNKDQFLV |
| 6977 |
PF04838 (Baculo_LEF5) |
 Estimated TM-score=0.40; hard target. |
>Baculoviridae late expression factor 5 (Length=158) YALFKLFKEFRINKNYSKLIDFLTENFPNNVKNKTFNFSSTGHLFHSLHAYVPSVSDLVKERKQIRLQTEYLAKLFNNTI NDFKLYTELYEFIERTEGVDFCCPCQLLHKSLLNTKNYVENLNCKLFDIKPPKFKKEPFDNILYKYSLNYKSLLLKKK |
| 6978 |
PF15118 (DUF4560) |
 Estimated TM-score=0.40; hard target. |
>Domain of unknown function (DUF4560) (Length=64) ATALSGLAVRLSRSAAARSYGVFCKGLTRTLLIFFDLAWRLRINFPYLYIVASMMLNVRLQVHI |
| 6979 |
PF13097 (CENP-U) |
 Estimated TM-score=0.40; very hard target. |
>CENP-A nucleosome associated complex (NAC) subunit (Length=176) GDITRKVKSAQKINTQQHEAVPATASSEPSEKPAGPVTPRKTGPLRAQSSVEKETCATQSQPKIQKKRKLSPGKRKKSRS KAKDSDISDSVHIWCLEGKRTSDIMELDIILSAFEKTIIEYKQKIESKVCMEAINKFYVNIKEELIRMLKEVQMLKNLKR KNAKMISDVEKKRQHL |
| 6980 |
PF06608 (DUF1143) |
 Estimated TM-score=0.40; very hard target. |
>Protein of unknown function (DUF1143) (Length=147) ASQCLCCSKFLLQRQNLACFLTNPHCGSIINADGHGEVWTDWNNMSKFFQYGWRCTTNENAYSNRTLMGNWNQERYDLRN IVQPKPLPSQFGHYFETTYDTSYNNKMPLSTHRFKREPHCFPGHQPELDPPRYKCTEKSTYMNSYSK |
| 6981 |
PF17461 (DUF5423) |
 Estimated TM-score=0.40; hard target. |
>Family of unknown function (DUF5423) (Length=348) KGLLSRFLTAPDRHPKLRYVYDIALIAISILCIVSIILWTQGSGLALFAIAPALAIGALGVTLLVSDLAESQKSKEIADT VAAVSLPFILTGTAAGLMFSAIAVGGGAVILANPLFLMGSMTLGFALMSLHRVTYQYLSNREQWKQQKKLEQVELAAWES HLPKESKSSALEEVRYSPRLMKRGKTWRKRAIRRKNYTPIPLVDKTLQTMQPDALFSSTTTHSTDSEQILTSVSPQSSDT ESSSSSSFHTPPNSDKELSDSNSSDSSSSSEYMDALETVAAGDVSGITPPSKPSSSPKTTRRVVKLSRSERNAQHHRNKD QEQRQDSSESSEEDSSSDSSQKKKPSRK |
| 6982 |
PF15260 (FAM219A) |
 Estimated TM-score=0.40; very hard target. |
>Protein family FAM219A (Length=125) QKHREMAKAMLRRKGVIAGAIRHQPKSTAKRSVKFNKGYAALSQTADETLVSLDSDSDGELESRCSSGYSSAEQVNQDLS RQLLQDGYHLDEIPDDEDLDLIPPKPISSSPCACCFGESSSCSIQ |
| 6983 |
PF05586 (Ant_C) |
 Estimated TM-score=0.40; very hard target. |
>Anthrax receptor C-terminus region (Length=86) VRWGEKGSTEEGARLEKAKNAVVSVPEDADEPMVKRPVPRAAPAPHGTENKWYTPIKGRLDALWALLGRQYDRVSLMRPT SADKVR |
| 6984 |
PF05541 (Spheroidin) |
 Estimated TM-score=0.40; very hard target. |
>Entomopoxvirus spheroidin protein (Length=945) SNVPLATKTIRKLSNRKYEIKIYLKDENTCFERVVDMVVPLYDVCNETSGVTLESCSPNIEVIELDNTHVRIKVHGDTLK EMCFELLFPCNVNEAQVWKYVSRLLLDNVSHNDVKYKLANFRLTLNGKHLKLKEIDQPLFIYFVDDLGNYGLITKENIQN NNLQVNKDASFITIFPQYAYICLGRKVYLNEKVTFDVTTDATNITLDFNKSVNIAVSFLDIYYEVNNNEQKDLLKDLLKR YGEFEVYNADTGLIYAKNLSIKNYDTVIQVERLPVNLKVRAYTKDENGRNLCLMKITSSTEVDPEYVTSNNALLGTLRVY KKFDKSHLKIVMHNRGSGNVFPLRSLYLELSNVKGYPVKASDTSRLDVGIYKLNKIYVDNDENKIILEEIEAEYRCGRQV FHERVKLNKHQCKYTPKCPFQFVVNSPDTTIHLYGISNVCLKPKVPKNLRLWGWILDCDTSRFIKHMADGSDDLDLDVRL NRNDICLKQAIKQHYTNVIILEYANTYPNCTLSLGNNRFNNVFDMNDNKTISEYTNFTKSRQDLNNMSCILGINIGNSVN ISSLPGWVTPHEAKILRSGCARVREFCKSFCDLSNKRFYAMARDLVSLLFMCNYVNIEINEAVCEYPGYVILFARAIKVI NDLLLINGVDNLAGYSISLPIHYGSTEKTLPNEKYGGVDKKFKYLFLKNKLKDLMRDADFVQPPLYISTYFRTLLDAPPT DNYEKYLVDSSVQSQDVLQGLLNTCNTIDTNARVASSVIGYVYEPCGTSEHKIGSEALCKMAKEASRLGNLGLVNRINES NYNKCNKYGYRGVYENNKLKTKYYREIFDCNPNNNNELISRYGYRIMDLHKIGEIFANYDESESPCERRCHYLEDRGLLY GPEYVHHRYQESCTPNTFGNNTNCVTRNGEQHVYENSCGDNATCGRRTGYGRRSRDEWNDYRKPH |
| 6985 |
PF13390 (DUF4108) |
 Estimated TM-score=0.40; very hard target. |
>Protein of unknown function (DUF4108) (Length=143) SRFVTVNQLMDALSPILENVKQVDVYFDDYVESLYYKGKFNIKPIAFAFDNKLIENAKIWELIPNIEFITNINDKWFKRI PTTKVLCKLMIKTEEKEFNGFKYHPNKVSELENEKLQKKLNDRLSNDRIEKINKLAEVAFNNE |
| 6986 |
PF19017 (DUF5746) |
 Estimated TM-score=0.40; very hard target. |
>Domain of unknown function (DUF5746) (Length=181) ILSLLAALIFIGIVVWIIWTRSSYSYPWSDEGPLGPENPPAICYKCSNYFHMMDGREVNSNIDVEKGESGGIEISGYLGV AIDVEDPRQELCQEVVNRTALAEIQVPLDDDCGDSLYDGCFKMVTKSFRMVPNIGREALSVTVVTRNCAELPKNLPLGCY KTFGGAGMEREICYCKGNYCN |
| 6987 |
PF17037 (CBP_BcsO) |
 Estimated TM-score=0.40; very hard target. |
>Cellulose biosynthesis protein BcsO (Length=201) MKSYDDMQRFKDKAQIKDINFMDMSGQVLQSETVVWPIMKQLLRNQTGDDTLSGAQPVSIAQPVPVSQHVLSAPASPRRV TPSSSLNVNADPSGGNQHSLFNAIAASLPSAESVSAPAPQPIAPPTPVSQPQPAALTIETVPPATVRPHPLASAPAVSEA SAASEAGRFRQLFSRHHHDEIHSVSKTMLLKPLLEKIALCR |
| 6988 |
PF06830 (Root_cap) |
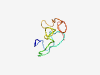 Estimated TM-score=0.40; very hard target. |
>Root cap (Length=56) GFQFHNLTKDVDGVLGQTYRHDYVSKVDISAKMPIMGGAPKYLSSGLFSADCAVSR |
| 6989 |
PF05650 (DUF802) |
 Estimated TM-score=0.40; hard target. |
>Domain of unknown function (DUF802) (Length=53) LSARLEQTSAAVAGNWSHALAEQQQTNQILNQDLRATLEGFSAGFDTRAGALL |
| 6990 |
PF15180 (NPBW) |
 Estimated TM-score=0.40; very hard target. |
>Neuropeptides B and W (Length=118) LVCVAISLLISCQPSEAWYKQATGPSYYSVGRASGLLSGIRRSPYIRRSESETGTETGDTTGNNLVSEPRQTAILKSMAI CVKDISPNLQSCELLQDGMNTFQCKADVYLSLDSQDCM |
| 6991 |
PF17705 (DUF5551) |
 Estimated TM-score=0.40; very hard target. |
>Family of unknown function (DUF5551) (Length=155) MATSERRIYSMESMAPEISEDIGCLLAYMKESVCVLLISQSFFSCVAEMYIPDMNRPGKAERRAEKGLRPPRELAGETRR LQSWPTLARWLRRVSFSLYKPSIQAAPEPDPGNWAKSPRLSLGPEGITKGKHGFKFQGIKEKFNVSKKVLKMTFL |
| 6992 |
PF15852 (DUF4724) |
 Estimated TM-score=0.40; very hard target. |
>Domain of unknown function (DUF4724) (Length=91) MFTRHTLFRQFAVLADTSFNYVKVKPLIVQSKLNLTGVISSSTGEHGSSLDNKYVDIVNDQLSYAPSGREKRKGKEKKNK ENQFSHTSKPE |
| 6993 |
PF05440 (MtrB) |
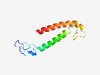 Estimated TM-score=0.40; hard target. |
>Tetrahydromethanopterin S-methyltransferase subunit B (Length=93) VHVAPEVNLVMDPMSGLVATEREDIISYSMDPILEQLDELDKVADDMVNSLSPNMPLLNSNPGRERTSYYAGIASNAFYG IIVGLIFATIIII |
| 6994 |
PF10113 (Fibrillarin_2) |
 Estimated TM-score=0.40; hard target. |
>Fibrillarin-like archaeal protein (Length=503) MHDIVKEAVNNLDAAWELAKMKKTPEEIVDAVSDLSREETMQLGKNFKRFPIGCDLTEILVGACASDIKKIDLLGNCIVA DMLGASIHVCAYAFADIAENYGMRGVDLVKEVRELVDVPLDIDHFGKYGPMRFPSDITRCWGQCYYEGPPFKECPRGRIH KRLLEKEKKGIKERDEWIKLASSVAINLISVQGGEAHAAPLDEAIEVAKLTKKYGRGLEAIMCVGDGYEDLIKAFEAALE IGVDVFVIEGGPFNQAKDRLDAFARAITCARILAPGKVVGTNGAYEDECRIGLRSGLNAIITGFPKNHHGYMCGYSPGTA RRGNFGMPRVIKIIKEEVKNGLTRAPIQKDELIALTKSVKVIGKENIYPNKIGYTTVGDAHWVCVSLTPLYEKLHVNKTV DDIVKMAENGLIGEKVALLGGRFIAWVLAKKLDKYVDQILISDVDPWVEKITVENLRSEIKADVHRAHSNDKEAYNESDA TIISSTIPGVVRKLTKKLPESVA |
| 6995 |
PF06126 (Herpes_LAMP2) |
 Estimated TM-score=0.40; very hard target. |
>Herpesvirus Latent membrane protein 2 (Length=497) NYKKYLWGTWFAALITCCGCLSIMFCLLTINLQNTIFLLSNISVYYQLFCTITNIYVQSKKQRFQASPPIGPSIIVGCIA FASWSFSTQSTLSTVCVCIISLLSIITAILSLGGTLRVVKCTIDSGLLCIVMVLVLIFSMGLQIYNNWTHCQFFLPLWTL LLVFFIHIFATDNGPCLKLAACVFAICGGILKATPAFFCVSHSCLSVIIAGCISCIHIGMTGLFITMKRHWIGSTKGLMS FLLLQGGVLVTLTTTIGILFIKREQDTNNEGSITLLAGCGFLLYCFFCWQSFHKASLSGGFLFLFLAWTCAGCCVKLVLL YTDGWTTGVTSGLICVIVILSTGQAVLVGYLYRESRLVSFNNVTTRLPIYTPHDTPHAHAGRICPDVNHLARRLPPLPSR NVIHSRILSSTTDMALSPVRVCNTEVTTQLEMQQLHSERTVTYASILGDNTPPPTRASACINQSGISNVSNCGVRSLDPP PFQPADEVYEEVLFPTD |
| 6996 |
PF15340 (COPR5) |
 Estimated TM-score=0.40; very hard target. |
>Cooperator of PRMT5 family (Length=151) AGFATADHSSQERETEKAMDRLARGAQSVPNDSPARGEGTHSEEEGFAMDEEDSDGELNTWELSEGTNCPPKEQPGDIFN EDWDLELKADQGNPYDADDIQESISQELKPWVCCAPQGDMIYDPSWHHPPPLIPHYSKMVFETGQFDDAED |
| 6997 |
PF15345 (TMEM51) |
 Estimated TM-score=0.40; very hard target. |
>Transmembrane protein 51 (Length=239) GSGAHYALCALGLGLIALGVVMIVWTVVPMDGEASSGSTTTPASNSSNPGDGGDEVQKDTTKSSTIAMVLVAVGAILLVL SISLGFRNKRRARARGNQTTAAVLLASGLPERQEEPAVDPAAFSVPSYDEVVGSGEFPVRQSNLRQSTSQLPSYEDIIAA VENEGADPAENSRLTGSTKAPAPSAPARSSSRASRLLRPLRVRRIKSDKLHLKDFRLQIRSPTQNPVTIEPITPPPQYD |
| 6998 |
PF07218 (RAP1) |
 Estimated TM-score=0.40; very hard target. |
>Rhoptry-associated protein 1 (RAP-1) (Length=782) MSFYLGSLVIIFHVLFRNVADGINVNGDNNYGKTIINNDFNFDDYNYWTPINKKEFLNSYEDKFSSESFLENKSSVDDGN INLTDTSTSNKSSKKGHGRSRVRSASAAAILEEDDSKDDMEFKASPSVVKTSTPSGTQTSGLKSSSPSSTKSSSPSNVKS ASPHGESNSSEESTTKSSKRSASVAGIVGADEEAPPAPKNTLTPLEELYPTNVNLFNYKYSLNNMEENINILKNEGDLVA QKEEFEYDENMEKAKQDKKKALEKIGKESDEEPFMFSENKFLENQVKERNVAGSFSRFFSKLNPFKKDEVIEKTEVSKKT FSGIGFNLTEKEAKVLGVGVTYQEYPETMLYNCPNNSNLFDTIESLQGRVIDIKKRESMISTTFEQQKECLKNMGVLDLE LNDTQCKFGTCIGSFGEHHLRLYEFENDLLKFHPNIDYLTLADGYKLQKNDIYELSHVNFCLLNPKTLEEFLKKKEIKDL MGGDDLIKYKENFDNFMSISITCHIESLIYDDIEASQDIAAVLKIAKSKLHVITSGLSYKARKLVYKIYSEIQKNPDELY EKLTWIYDNIYMIKRYYTAYALEGVCSYLEHDKSQMYTELHIYNKIVDSVRYYSSCFKNVIVYNAIISGIHEKIKHFLKL VPRHNFLLDYHFNSIFEKEIKPAKKYSTSHIYFDPTVASYAYYNLDRRTMVTIINDYFEAKKKELTVIVSRMKTDMLSLQ NEESKIPNDKSANSKLATRLMKKFKAEIRDFFKEMRIQYAKLINIRYRSHLKKNYFAFKRLD |
| 6999 |
PF05555 (DUF762) |
 Estimated TM-score=0.40; very hard target. |
>Coxiella burnetii protein of unknown function (DUF762) (Length=239) YFFTLPKIDIKEKSASQTPEKVLREWLRGASNQEDASLFIGNSPIFGEHQDTSGSSLPKVEIPEFINSIKAVEAVFKKEG LKDEEILNYILHFGGINGYMTLGQVVFEEISMNKGKSSRMENRRYSYRVKNKGEVEYIEQFDFYKKNTCDRNEIGDFIGQ VKIVSIISKDGDIQHACKKVEIAVPGEKLNSFRKHFGDQLNFVNRMKLRLVELYNSLVERFRAAINKADRTNNNCFPFC |
| 7000 |
PF03021 (CM2) |
 Estimated TM-score=0.40; very hard target. |
>Influenza C virus M2 protein (Length=139) MGRMAMKWLVVIICFSITSQPASACNLKTCLKLFNNTDAVTVHCFNETQGYMLTLASLGLGIITMLYLLVKIIIELVNGF VLGRWERWCGDIKTTIMPEIDSMEKDIALSRERLDLGEDAPDETDNSPIPFSNDGIFEI |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417