

Go to page (83 pages in total): [First page] << < 64 65 66 67 68 69 70 71 72 73 74 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 6801 |
PF03262 (Corona_6B_7B) |
 Estimated TM-score=0.42; hard target. |
>Coronavirus 6B/7B protein (Length=206) MIVVILVCIFLANGIKATAVQNDLHEHPVLTWDLLQHFIGHTLYITTHQVLALPLGSRVECEGIEGFNCTWPGFQDPAHD HIDFYFDLSNPFYSFVDNFYIVSEGNQRINLRLVGAVPKQKRLNVGCHTSFAVDLPFGIQIYHDRDFQHPVDGRHLDCTH RVYFVKYCPHNLHGYCFNERLKVYDLKQFRSKKVFDKINQHHKTEL |
| 6802 |
PF11214 (Med2) |
 Estimated TM-score=0.42; hard target. |
>Mediator complex subunit 2 (Length=99) HENKLTQCFNDIMRVAVEMMLHQQLKTVQLDSSVVNGFSHNQQRILSEKIHIFHAILDDMETTLGKSKNYVEAIYELGRE KELQKEREREELRKRQEEE |
| 6803 |
PF07343 (DUF1475) |
 Estimated TM-score=0.42; hard target. |
>Protein of unknown function (DUF1475) (Length=236) AASGTTLVVYKAVFAALGVLMVGTLVYTCITDGSPFRTELLTPWMVATLVDFYVNVVAISTWVVYKEVNWISSAFWLVML FCFGSAATCAYIVNKLFEITPSGPSQDPLDLLLLRQGNPPERKCYFVKIGRIVFSILGILMAALVTYTVITDGLPFRKDL LTPWMAATLVDFYINVFAISVWVVHKESTWISSVIWICLLICFGSITTCGYIVVQLLRVSYHDPIDHVLLNSHSKS |
| 6804 |
PF13914 (Phostensin) |
 Estimated TM-score=0.42; very hard target. |
>Phostensin PP1-binding and SH3-binding region (Length=141) PQEAKLPCSGVGATPQYHLHPARPGHTSELQHRGSNTFTVVPKRKPGAFQEPHFSQTNGEPQPQKPEEEEAGSLSGPHAA LENTLKKRYPTVHEIEVIGGYLALQKSCLTKAGSSRKKMKISFNDKSLQTTFEYPSESSLV |
| 6805 |
PF17704 (DUF5550) |
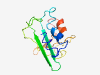 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5550) (Length=123) MMPLAEAGALAQGGGPSATEWACILRRKTPRHKQPILLMVRASRRSGETSAVLKAGRQSVSGRKNSTSKDLVTLGASSLR EERGHPLHPRHGKAVHLRTRGRTRGWVQTRAWMSRRTRGPAER |
| 6806 |
PF08594 (UPF0300) |
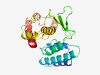 Estimated TM-score=0.42; very hard target. |
>Uncharacterised protein family (UPF0300) (Length=210) FNSFENYFRELTEFLLNGRAPLNVLVHHIMLYHYYPTSMKDVLWKAVEVYLDIHCTEDRYCRVQVEAAKRRLGDIRLYLV YPRDIYKFTNSKDWIAVATRGFMSYIQTRNPLCLGSIPVNNKSISELLSYTTSTDCMAWLSLLICAGSNGSRFPLHKYIN TKTRIMKSKTSITDFLFSDEDLLFFQDKESITEFKPLQQQLLRELADCDA |
| 6807 |
PF09316 (Cmyb_C) |
 Estimated TM-score=0.42; very hard target. |
>C-myb, C-terminal (Length=160) FSPSQFFNTCSGNEQLNMENPSFTSTPICGQKVLITTPLHKETTPKDQKENVGFRTPTIRRSILGTTPRTPTPFKNALAA QEKKYGPLKIVSQPLAFLEEDIREVLKEETGTDIFLKEEDEPAYKSCKQEHTASVKKVRKSLVLDNWEKEEPDTQLLTED |
| 6808 |
PF01606 (Arteri_env) |
 Estimated TM-score=0.42; very hard target. |
>Arterivirus envelope protein (Length=211) MAHQCACFHFFLCGFICYLVHSALAANSSSTLCFWFPLAHGNTSFELTINYTICMPCLTSQAARQRLEPGRNMWCRIGHD RCEERDHDELLMSIPSGYDNLKLEGYYAWLAFLSFSYAAQFHPELFGIGNVSRVFVDKRHQFICAEHGGLNSTLSTEHNI SALYAVYYHHQIDGGNWFHLEWLRPLFSSWLVLNISWFLRRSPVSPVSRRI |
| 6809 |
PF10060 (DUF2298) |
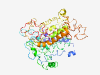 Estimated TM-score=0.42; easy target. |
>Uncharacterized membrane protein (DUF2298) (Length=477) LLGYVIWLPASLGWWRYDSAGLAGGLLVLIGIGLFLWWVGVRPERAAWRSLLPGEAIFLAGFAAMTIIRALNPDLWHPVW GGEKPMEQGFLNAILRSPQMPPYDPFYSNGYINYYYYGLYLMSVPIKLLGIDSAVGFNLALATLYGMLLAGAYELGTRLG GRRRYGIVALSLIGLMGNLTSLIPAGWSLGAQGLRLLLSGQAELLGDWFVGPSRVIPYTINEFPLFSFLYADLHPHLIAL PLALLAIVCGWHLILRPQRPLWLLSALTLGTLAVTNSWDFPTYGLLCGLAIIAGAIRRTGWRWQPLTLAVVQAVGLGLGS LLLFAPFFDRYWPMVRGIGLVTTGVTLPLDYLLIYGVPLIIVIPVIAGGLLQRWRSGFAGRRWALIASGIGLLMIIGAAV PELALRLSLFVVLMVTTILLMRRAAGAPAWYALLLCWVAWAVSLGVEIVYIRDHLDGSDWYRMNTVFKFGLHVWVLL |
| 6810 |
PF06515 (BDV_P10) |
 Estimated TM-score=0.42; very hard target. |
>Borna disease virus P10 protein (Length=87) MSSDLRLTLLELVRRLNGNATIESGRLPGGRRRSPDTTTGTIGVTKTTEDPKECIDPTSRPAPEGPQEEPLHDLRPRPAN RKGAAVE |
| 6811 |
PF05290 (Baculo_IE-1) |
 Estimated TM-score=0.42; hard target. |
>Baculovirus immediate-early protein (IE-0) (Length=139) GHLNNQPKFQYNMYIFLPYVRQLRQINTLFINDYCCSKIVKSNSAALDLLIAHCDKYLHVIASMNERMQLINVFTEPKLY QCNICQDTSVEEHFLKPNECCGYNICNMCYANLWKFCNLYPQCPVCKTSFKGSKQVVER |
| 6812 |
PF05418 (Apo-VLDL-II) |
 Estimated TM-score=0.42; hard target. |
>Apovitellenin I (Apo-VLDL-II) (Length=79) KSIFERDRRDWLVIPDAVAAYIYEAVNKMSPRAGQFLVDISQTTVVSGTRNFLIRETARLTILAEQLMEKIKNLWYTKV |
| 6813 |
PF17670 (DUF5530) |
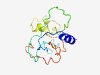 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5530) (Length=140) MTAIRLREFIERRPLIPPSIFIAHQGRDLQGYYPGQLARLHFDHGAKRAPRPLIDLTIPPKRKYHYQPQFDQQTLIRYIC FRRHSKPAEPWYKETTYQRDYSLPFYEIDWNQKLATVSLNPRPLNSLPELYCCEERGGFE |
| 6814 |
PF10039 (DUF2275) |
 Estimated TM-score=0.42; very hard target. |
>Predicted integral membrane protein (DUF2275) (Length=205) RHEAPADFLRNVHARIEREPSWKSLLSRLFLPLKIKLPLEALGVAAAALLIFTFYYSAPETKMDLPRESPVARAPATAEA PMPPGTVAGLPAPAQPPSRAATSVPAPAPTEIHPRPKAAAPAQDRFAAQPSFPEEERGAGRAKPEAEAPAGAVEAGRAGP VERAATGIPAPKPEAGPSKRADRSQAILPAPSAPAVQRRMSPRVV |
| 6815 |
PF04885 (Stig1) |
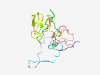 Estimated TM-score=0.42; very hard target. |
>Stigma-specific protein, Stig1 (Length=130) AITLTITFTGSNHEPTPITEPSSSVAKPFPKRVSRFLAESKNPRATDHCYKDDAICYILEGKNSTCCNGKCMDLSQDKGN CGACKNKCKFTSSCCRGECVNLAYDKRHCGSCGNKCMPGGYCIYGLCSYA |
| 6816 |
PF02394 (IL1_propep) |
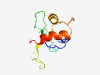 Estimated TM-score=0.42; very hard target. |
>Interleukin-1 propeptide (Length=103) MAEVPELTSEMMAYYSGNEDDLFFEVDGPKQMKCSFQDLDLCPLDGGIQLQISHEHYNKGFRQAVSVVVAMEKLRKMLVP CPQTFQDNDLSTLIPFIFEEEPI |
| 6817 |
PF07125 (DUF1378) |
 Estimated TM-score=0.42; hard target. |
>Protein of unknown function (DUF1378) (Length=59) MRFVQLILLYFCTVVCTLYLVSGGYKVIRNYIRKKIDAAAAEKISASQSAGSKPEEPLI |
| 6818 |
PF17229 (SMG1_N) |
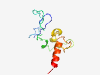 Estimated TM-score=0.42; very hard target. |
>Serine/threonine-protein kinase SMG1 N-terminal (Length=106) SDSLSASQDGVKCSVSRDRGSEVLFEILPSDRPGTPPLHNDAFTATDALEEGGGYDADGGLSENAYGWKSLGQELRINDV TTDITTFQHGNRALATKDMRKSQDRP |
| 6819 |
PF15361 (RIC3) |
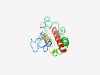 Estimated TM-score=0.42; very hard target. |
>Resistance to inhibitors of cholinesterase homologue 3 (Length=151) LVLCVALLLPKMLLSRGRKDAAERPEGSGHFPPMMHRQAAPEGRSQRAAASGFSRAHNSEAIARAKGAGTGAGTGGKSNL AGQIIPVYGFGILLYILYILFKITSKGNNKPSARRCPPHRSENMKRKITDFELAQLQEKLRETELVMENIV |
| 6820 |
PF15226 (HPIP) |
 Estimated TM-score=0.42; very hard target. |
>HCF-1 beta-propeller-interacting protein family (Length=133) ILQQPLERGPQGRAQRDPRAASGASRGLDASSPLRGAVPMSTKRRLEEEQEPLRKQFLSEENMVTHFSRLSLHNDHPYCS PPMAFPPALPPLRSPCSELLLWRYPGNLIPEALRLLRLGDTPTPHYPATPAGD |
| 6821 |
PF15737 (DUF4685) |
 Estimated TM-score=0.42; very hard target. |
>Domain of unknown function (DUF4685) (Length=123) RTFGPKPEPVLSPRHEEAKHLLQRARMKARTRPLRASHDIVPTIAQGSRDAGRSPALDPRMTLACRDNLQNGNTSDSSSG ESSSGQWPKRGPSPSHVRFEDESARDAESRYLERLQQRQRQVL |
| 6822 |
PF19205 (DUF5877) |
 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5877) (Length=611) MYLNALAKVRRGGGGGRPAAGGRPAAGGRPAAGGRPAAGSRAAAGAAGPPKPAKTVRGPVRPRGFGGGRFPPRRFPPAPV DEAAREAEKILKSIHAHKVVKLLTQRKKHRSLYKAVRHTMPPDYIPVLGAEVLAGISSPDVMQAVLRIAPTRDNTQAADA MLCATILRAMPYKMVHPFLDMAAEGKDRHANVVLRDTFFPQNARTMDEFKAFMRRRAPDRPSPRTPMVKPPFRIPDAKPG LAPSVRNPRLNIPETAMVSIMTSAPWVAGHSIKGTVLPEGSTGALRGGLASRRWYVDNWVSVRMMGNPPVEYLTFSGKRI PETREMYDAYVRHLQSFGATAASSSPSPPPNGDAATALANMYVDTGPEYLAALVRALPSGSGAEYEAALAYTVARAVPVF SNGETLHQIRLREGRYRPDGVHGLSDDIAFAEVYSDPDIVDEWKPRLDRIIKTRVSEVIAAYRRLATGDLTAKRTPELAL SSRIPAPAGLLFPKAFGESAVIRTSRGRRLVIAGARRDAPAVLTDLMSVLITPMYPATTEPQIVASDDPTVLGDEPAPAA VEEPRGQAFRRKWGRGVGGKLLDALENYMDAIQQGRLPPVRQRYGIDVESL |
| 6823 |
PF04704 (Zfx_Zfy_act) |
 Estimated TM-score=0.42; very hard target. |
>Zfx / Zfy transcription activation region (Length=329) DVVIEDVQCPDIMEEADVSETVIIPEQVLDTDVTEEVSLAHCTVPDDVLASDITTATMSIPEHVLTSDSMHVPDVGHVEH VVHDNVVEAEIVTDPLTTDVVSEEVLVADCASEAVIDANGIPVEQQDDDKSNCEDYLMISLDDAGKIEHDGSSEITMDAE SEIDPCKVDGTCPEVIKVYIFKADPGEDDLGGTVDIVESEPENDHGVGLLDQSSSIRVPREKMVYMTVNDSQQEDEDLNV AEIADEVYMEVIVGEEDAAVAHEQQIDDTEIKTFMPIAWAAAYGNNTDGIENRNGTASALLHIDESAGLGRLAKQKPKKR RRPDSRQYQ |
| 6824 |
PF15310 (VAD1-2) |
 Estimated TM-score=0.42; very hard target. |
>Vitamin A-deficiency (VAD) rat model signalling (Length=248) EEDQVSTNHRSICVQTSKHLFWADKLIQASEHSLERMTDTQLGKNKGKTISHLGQQSVPKDTTCSKKQLQTPSAQPAPPA TDSQQLPNPYPSSSSLSPAIGLEELVNFASSLAVASSSKMDLPNLEHMIKAPPQKAEAPSTDPTTQVAVDQPEKKKLTKE LPDRPPLKARESQKAQKPQDKNFSHPYLDFSKPGIKRATIKGEVKLLQPPAMSPLPQGDMKDSVPGTRKGSPLLLKIHFK VSSPTSPE |
| 6825 |
PF15926 (RNF220) |
 Estimated TM-score=0.42; very hard target. |
>E3 ubiquitin-protein ligase RNF220 (Length=250) DSQAPICPICQVLLRPGELQEHMEQELEKLTQLNISKNSILKDAMAAGTPKSILLSASIKREGDSPTASPHSSDDIHHSD RYQTFLRVRANRQTRLNARIGKMKRRKQDEGQVCPLCNRPLSGSEEEMSRHVEQCLAKREGSCMTEDDSVDIENENGNRF EEYEWCGQKRIRATTLLEGGFRGSGFIMCSGKENPDSDADLDVDGDDTLEYGKPQYTEADVIPCTGEEPGEAKEREALRG AVLNGGTPST |
| 6826 |
PF11233 (DUF3035) |
 Estimated TM-score=0.42; very hard target. |
>Protein of unknown function (DUF3035) (Length=139) RRSIGLDRQSPDEFTVVSRAPLTMPPSISNLPKPRPGAPRPQDVTPTAAAAGAVFGGASRAVNKAGGSAGESALVAQAGT KGIDPDVRAKVDQETTALIIADKSWVDTLLFWQKQEQPFSVVDPAKEQQRLREAQAQGK |
| 6827 |
PF16663 (MAGI_u1) |
 Estimated TM-score=0.42; very hard target. |
>Unstructured region on MAGI (Length=59) TGDALQDSLPGSQQSTPRRTKSYNEMQNAGIVPTETEDDDEVPEMNSSFTGDSSEPDDH |
| 6828 |
PF13073 (DUF3937) |
 Estimated TM-score=0.42; hard target. |
>Protein of unknown function (DUF3937) (Length=72) MFTNKKLIRFGLSLFVFLGIINFTISYFQTYLETAADIKWVIPEIWKTILLDVPQGILVLLGAIALYDFTKE |
| 6829 |
PF13259 (DUF4050) |
 Estimated TM-score=0.42; very hard target. |
>Protein of unknown function (DUF4050) (Length=165) SPFRFDSPDAIGAAVKASVADKRARRRRAVREEMAWNEGLACFEARRNAWTGARTRPSNEAATAVSDGSELSKDPSKELT AQKSKESQKSAASPASTTYPIETIVPLSAPLLPPGNPMRASITPAVYMSLYEKVIVHALTPSCPVNLSDMIRACVVGWKR DGEWP |
| 6830 |
PF10839 (DUF2647) |
 Estimated TM-score=0.42; very hard target. |
>Protein of unknown function (DUF2647) (Length=70) AYSSCLNRSLKPNKLLLRRIDGAIQVRSHVDLTFYSLVGSGRSGGGTTAPLFSRIHTSLISVWRAISRAQ |
| 6831 |
PF04618 (HD-ZIP_N) |
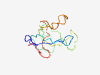 Estimated TM-score=0.42; very hard target. |
>HD-ZIP protein N terminus (Length=101) VEKEDLGLSLSLSFPQNYNSLQLNLMPSLVPSSVISQKPSWNDTFHSSDPNSDSCRADTRSFLRGIDVNRLPSTADCEEE AGVSSPNSTISSVSGKRSERE |
| 6832 |
PF03303 (WTF) |
 Estimated TM-score=0.42; very hard target. |
>WTF protein (Length=238) MKNNYTSLKSPLDEEDELKTDHEIDLEKGPLPEYDSEEEGALPPYSDHALVNNPPNTHRENNPSRSTDNSSPFLIKLLIS FTPIYVLNVLAICYLTYKDAFFKDYGAAEWTLFGFWCLVCTLALIFLTYFYETWVKAVKVTVISLAKCVKVISIGLFNIR REMMIIIWILWLIICCILFVYIKSGDLILNKALICSTCTISAVLLLIVSSVCIPFWTFERTLAKLAKVLLLQSGIVLV |
| 6833 |
PF15470 (DUF4637) |
 Estimated TM-score=0.42; very hard target. |
>Domain of unknown function (DUF4637) (Length=165) KTPLWKKETEELQAEEAEQEEGKEGSEDEDEDNQRPLEDSATEGEEPPRVAEEGEGRERRSVSYCPLRQESSTQQVALLR RADSGFWGWLGPLALLGGLTAPIDRKRSLPEEPCVLEIRRRPPRRGGCACCELLFCKKCRSLHSHPAYVAHCVLDHPDLG KAGAA |
| 6834 |
PF04656 (Pox_E6) |
 Estimated TM-score=0.42; hard target. |
>Pox virus E6 protein (Length=566) MDYIRRKYLIYTIENKIDFIRDEVLSKISNFTLNHVLALKYLILNFSKDVLTKDVLSNPNFYVFLHMVKCPDVYKIVIKQ SFDVPTLYIKALIKNYSSFNHIIETYKKNVQELFFDEKFIEISKHSHNFDNIIGVNYDLRLNPLFFQGEPIKNMELIYSK LFKKSNFLRVKKMEVIRLMIWAYLSKEDTGIDFTDNDNQDIYTLYQKSNGIINSDMTEKFKEYIFEKNKTSYWIWLNEPI SNDKNIYLEGIAESMYDKILSYIYSEIKQGKINKNMLKLTYMFETDEYIQSILLQIIYGVPGDILSIIDSKDETWKKYFI GFYKENFIDGNTFISSKTFSDDLFKVVAKINPEYFDPNKIISIFDHKPEKVKFFDSLDINKTYISNIIYETNDLNLEIME ELQTCQIYNDETKYFIKEYNTYLYLTENDPFVLYNGILVKLSTVPIDKKFSLFSENILKYYLDGKLANIGLIIPNYNGDI ILHILSHLKCVEDVTTFIRFSICINSSILPSIIRTILANFNISIIILFQKFLRDNLFYVENFLEKTNHLTKNDKKYLLDV IKNGRS |
| 6835 |
PF16386 (DUF4995) |
 Estimated TM-score=0.42; hard target. |
>Domain of unknown function (Length=72) MKKKLVLFASVAIALASCQTTPKEDYSWIKKGLDVASAQLQLTAEEIDGTGKLPRSIRTGYDMDFLCRQLER |
| 6836 |
PF16940 (Tic110) |
 Estimated TM-score=0.42; hard target. |
>Chloroplast envelope transporter (Length=594) TPLQSVIDSMSPAVRLGTSVVVVAGALAAGYGLGLKLGGNNKTAGVGGAVVLGVAGAGAAYVANSSVPEIAAVSLHNYVA GSDPGSVKKEDIESIASRKTRFGLADCVENEVGWHAQLHIYGVSKQDEAFNAELCDIYCRYVTSILPSGDEDLKGDEADI IIKFKNSLGIDDPDAAAMHMEIGRRIFRQRLETGDREADAEQRRAFQKLIYVSELVFGKASDFLLPWMRVFKVTKSQVEV AIRDNAQRLYASKLNLISQDIHPEDLISLRDAQLQCRLSDELAEDMFRNHIRKLVEANISTSVNVLKSRTRTADAKLIVE ELDKILAFNNLLISLKNHADADRFARGVGPVSLNGGEYDGDRKMDELKLLYRAYIADALQGGRMEKDKLTALSQLRNIFG LGKKEAESIAIDVTSKVYRKRLGESVTGGVLAAADSKAAFLQNLCEQLHFDTEKAIEIHEEIYRQNLQQYVKDGELSDDQ VQALERLQVMLCIPKQTVEKIHEEICGTLFEKVVKEAIAAGVDGYDADVQQSVRKAAYGLRLTRGVAMSIAGKAVRKVFA SYIQRSRSSGSRTEAARELKKMIMFNNLVVTELV |
| 6837 |
PF09321 (DUF1978) |
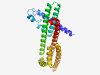 Estimated TM-score=0.42; hard target. |
>Domain of unknown function (DUF1978) (Length=248) EDNKKTIMKEHAEMLESLSSYRKVFLALSDENVVDTPSDPKKWDLSGIPCRDALSEISRDEQWQKKAHLKHQESLYTQAR DRLTDQSSKENQKELEKAEQEYISSWERVKKFEIERVQERIQAIQKLYPNILEREEETTGQETVTPTVQGTTASSDLTDI LGRIEVSSREDNQNQESCVKVLRSHEVEMSWEVKQEYGPKKKEFQDQMGSLERFFTEHIEELEVLQKDYSKHLSYFKKVN NKKEVQYA |
| 6838 |
PF14983 (DUF4513) |
 Estimated TM-score=0.42; very hard target. |
>Domain of unknown function (DUF4513) (Length=132) SGKDTCPILPKLANSCSDESSYKPSNKCNEVHLPRFSLKQGMIPRRYIMPWKENMKFRNMNLKRAEVCGIHAGPLEDSLF LNHSERLCHGEDREVVLKKGPPEIKIADMPLHSPLSRYQSTVISHGFRRRLV |
| 6839 |
PF14724 (mit_SMPDase) |
 Estimated TM-score=0.42; hard target. |
>Mitochondrial-associated sphingomyelin phosphodiesterase (Length=765) QPSFLLATLKADCVNKPFAQRCHDLETVIEEFPAKELHGIFPWLVESIFGSLDGTIVGWNLRCLQGRTNPTEYSVALDFL DPSGPMMKLVYKLQAEEYRYDFPVSYLPGPVKASIQEQVLPECSLYHNKVQFPSSGGLGLNLALNPFEYYMFYFAISLIT QKNSPITHHISPSNSAYFILVDTYLKCFLPTEGSVLPPPSSNPGGAIPSPTPRSPAVPFTSYGMHHTSLLKRHIAHQPSV NADPASQEIWRSETLLQIFVEMWLHHYSLEMYQKMQSPHIKLEVLHYRLSISSKHHGSPAQSSYQALHAYQESFKPTEEH VLVVRLLVKHLHAFSNSLKPEPLSPSAHSHTASPLEEFKRVVIPRFVQQKLYIFLQHCFGHWPLDASFRAVLEMWLSYLQ PWRYAPEKPPQTAELLPRSVSEKWAAFIQENLLMYTKLFVGFLNRTLRTDLVSPKNALMVFRVAKVFAQPNLAEMILKGE QLFLEPELVIPHRQHRLFMTPTLGGSFLSSWPPAITDASFKVKSHVYSLEGQDFQYKQMFGTEVRNLVLKLAQLICQAQQ TAKSISDHSAETTASHSFFSWFKFTPSEMNGSYTGNDLDEIGQDSIKKTDEYLEKALEYLCQIFKLNEAQLTQMMMKCGT AQDENGKKQLPDCIESENGLILTPLGRYQMINGLRRFDVQYQGDTELQPIRSYENATLVRLLYRLSSALNERLADQMDVL CSREDFVGRFCRYHLTNPHLVHKIRYSPVLKDRTSQNWGPRISLR |
| 6840 |
PF06599 (DUF1139) |
 Estimated TM-score=0.42; hard target. |
>Protein of unknown function (DUF1139) (Length=309) MDSERHYEYGSYSNSHGIEFDPNHPYIDLINDDFDENDYLDLETLNLEADYDDVESLALRLKNAPDYTTEIFEKIDRIPN FVYLFETEIIDNWNDYDLFADLRVTDTSDEFYTLSSMLTEHMQSIITLLPSILWPMVSQLTKSNVFQAADDVNITNYWRL MDRRWDFIDEQLRVQFIFRAYDLRAYQNGRVSQILSNSLLFAGLNLIGKRSCIPINSNFSIPDYLDYWFPTDDYHSDNYL IFIKFNETKNSGWKKIVVQYYLRKVFSKIRTGILIAHIDVDFWYNVFMRTLVRKEMIHTKNLIKSVLNF |
| 6841 |
PF15262 (DUF4592) |
 Estimated TM-score=0.42; very hard target. |
>Domain of unknown function (DUF4592) (Length=143) NMKLGPPPPCGIPTKRADDAGMSSEDDGLPRSPPEMSLLHDITATTTTTKFSDPHKHLSSLSLAGTGSEEEEQVPLGSSS RPLSPDQLFSRHASDTMTSPRTSDSSLSPLADFDYPPEFSSCLDNSAAKHKLLVKPRNQRSSK |
| 6842 |
PF05377 (FlaC_arch) |
 Estimated TM-score=0.42; easy target. |
>Flagella accessory protein C (FlaC) (Length=55) RINSLEKKLSKFEMSFNMMERENKDIKDTVQKIDQSVIELLSLYEIVSNQVNPFV |
| 6843 |
PF07327 (Neuroparsin) |
 Estimated TM-score=0.42; easy target. |
>Neuroparsin (Length=102) MKPAAALAAATLLIAVILFHRAEANPISRSCEGANCVVDLTRCEYGEVTDFFGRKVCAKGPGDDCNVYASCGIGMDCRNK KCSGCSVKTLECYFESFPMSEE |
| 6844 |
PF07234 (Babuvirus_MP) |
 Estimated TM-score=0.42; hard target. |
>Movement and RNA silencing protein (Length=117) MALTTERVKLFFEWFLFFGAIFIAITILYILLVLLFEVPRYIKELVRCLVEYLTRRRVWMQRTQLTEATGDVEIGRGIVE DRRDQEPAVIPHVSQVIPSQPNRRDDQGRRGNAGPMF |
| 6845 |
PF16949 (ABC_tran_2) |
 Estimated TM-score=0.42; hard target. |
>Putative ATP-binding cassette (Length=545) LLSGSLLRCLMVLACSAVFWTVLFGIFFEGFMFVGMYVDLKNTIIEHLFSMFFLSLLIMLFVSTGIIVYAGLFQSREAAF LLTTPASTDRIFVYKFFEAIVFSSWGFLLLVSPMMVAFGITVIAKWSFYAIFLVYLLSFVMIPGSLGAVAAILVANFFPR RQKTVLILGGGLILIGAAFVVTQITTTPGSALSTDWLGGLLGRLAFSQHPLLPSRWMSAGLLASARGNWSDGLFYLMVLT AHAGLFYLVATVVARDLYRQGYSRVQGGRSSRRRSGLYWIDALFHRLFFFIPHPVRLLILKDLRTFRRDPAQWSQFLIFF GLLAFYFLNIRRLSYDVQSPYWRNLVSFLNLAVTALILSTFTSRFIFPLLSLEGRNFWVLGLLPLKREAILWGKFAFASG ISLVATEVLVVLSDLMLRMSTAMIALHMGMVAILCLGLSGISVGLGARLPNLKETDPSKIAAGFGGTLNLLVSLVFISAI VTALALPCHLYFAGQDASEFRTFVLSEERFRFWMAIAVVTSLVIGAVGTFLPLRIGIKAFKRMEF |
| 6846 |
PF05093 (CIAPIN1) |
 Estimated TM-score=0.42; very hard target. |
>Cytokine-induced anti-apoptosis inhibitor 1, Fe-S biogenesis (Length=97) IQPPECQPKAGKRRRACKDCTCGLKEKIEAEDAAKRANADKALNTMKLDTDDLAEVDFTVQGKVGSCGNCALGDAFRCDG CPYIGLPAFKPGEEVRL |
| 6847 |
PF08736 (FA) |
 Estimated TM-score=0.42; hard target. |
>FERM adjacent (FA) (Length=43) GFLVMGSKFRYSGRTQAQTQQASALIDRPAPNFKRSASKRYTL |
| 6848 |
PF13054 (DUF3915) |
 Estimated TM-score=0.42; very hard target. |
>Protein of unknown function (DUF3915) (Length=123) MFGSFGCREDFRECHRGHCCDRDREREREREREREREREKEKERHHVCNVLRHIAIGTEISLLTIKGNVTFRNVIFEGFS NGVALFSALNMGDNKDNKDNQNNMNATKFTGLLRVCPEDIIAI |
| 6849 |
PF12440 (MAGE_N) |
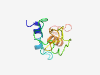 Estimated TM-score=0.42; very hard target. |
>Melanoma associated antigen family N terminal (Length=93) QKSKLRAREKRRQAREELKDLVGAQATVPEEEESPSSSSPSFQDVPQSSPGTETPRNPKVPGRVRSTTTTTAAAVSNTRF DEGANNQVEERPN |
| 6850 |
PF10863 (NOP19) |
 Estimated TM-score=0.42; very hard target. |
>Nucleolar protein 19 (Length=151) MSRSKEYQERLNLQAKLQSAFSNNTAAVLGWLKELEETDRSEDRIDPGHSKQSDDHTELEDSKKAFFELPVVQVGSGLHF STQDDVSTKEDIHTIGEFIESDKKLSSLAKKKKRNDQGPQRNNMYMITKDDTKAMVALKRKMRKGEREGVR |
| 6851 |
PF03187 (Corona_I) |
 Estimated TM-score=0.42; very hard target. |
>Corona nucleocapsid I protein (Length=207) MESSRRPLGLTKPSVDQIIKIEAEGISQSRLQLLNPTPGVWFPITPGFLALPSSKRERSFSLQKDKECLLPMESPLQSKR DIGIDTTAVLLKHLMGSRSNYCPDGIFTILAQGPMLEPVMETALKVSSGLQTAKRTPIPALILSKGTQAVMRLFLLGLRP ARYCLRAFMLKALEGLHLLADLVRGHNPVGQIIALEAVPTSASLPLL |
| 6852 |
PF17542 (RP853) |
 Estimated TM-score=0.42; hard target. |
>Uncharacterized RP853 (Length=317) MNNAIVLNNVNTNLFKKDIINNFVNNLTKSTCSTIANMLAIDYTLRLNSKPEAEKYIKKCVTRLKSYLGMRVINDDNFSV ALELFAIMITYEYENEQNKDNILEQSQNVIAANLVEVYFSDAKITSSEKEKIKNIFKTLLKEKNFDKIINYAESHKKLFK MSVMYAMQKYNNIDQATIYIKKEFNKILDMSLAFTQKCNIFKQTTGKIVGAVCALLVGAISVATAGAAFSIIIVPTLIFA IRYAPALGEKIGEFILNNDNVIKLEQSNIDEFMKTLQNNKENLLSQEKVKKIKNNITVVPPTINSKLIKKVVNNKKN |
| 6853 |
PF10854 (DUF2649) |
 Estimated TM-score=0.42; hard target. |
>Protein of unknown function (DUF2649) (Length=67) MQNDWQEFKEFFIYIFLFIDKANVESITMWNLTQNEYLTLMVGVWIVILFLTWFFLWMVFKIVGYFK |
| 6854 |
PF06517 (Orthopox_A43R) |
 Estimated TM-score=0.42; very hard target. |
>Orthopoxvirus A43R protein (Length=195) MMMKWIISILTMSIMPVLAYSSSIFRFHSEDVELCYGNLYFDRIYNNVVNIKYIPEHIPYRYNFINRTFSVDELDDNVFF THGYFLKHKYGSLNPSLIVSLSGNLKYNDIQCSVNVSCLIKNLATSTSTILTSKHKTYSLHRSKCITIIGYDSIIWYKDI NDKYNDIYDFTAICMLIASTLIVTIYVFKKIKMNS |
| 6855 |
PF16858 (CNDH2_C) |
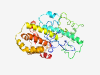 Estimated TM-score=0.42; very hard target. |
>Condensin II complex subunit CAP-H2 or CNDH2, C-term (Length=289) PWQSLDPFDSLDSKPFKKGKPYSVPTCVEETPGQKRKRKGSTKLQDFHQWYLAAYADHADSRRTRRKGPTFADMEVLYWK HVSEQLETLRKLQRRETAEQWLPRTEEEMWPAEDDHLGDSLEGLGAADDFLEPEEDVEIEGAKPGEAADMDAEAMPESLR YEELVRRNVELFIATSQKFVQETELSQRIRDWEDTIQPLLQEQEQHEPFDIHTYGDQVVSRFTQLNEWCPFAELVAGQSA FEVCRSMLASLQLANDYTVEIRQQPGLETAVDTMSLRLLTHQRAHKRFQ |
| 6856 |
PF11960 (DUF3474) |
 Estimated TM-score=0.42; very hard target. |
>Domain of unknown function (DUF3474) (Length=141) MASWVLSECGIRPLPQIYTRPRTGHISNSKNNHSRIRFFHSNQSKTDLKFREPIGCSKERNWALKVSAPLRVEYGEEEER GEIINRVNGVEEQPEFDPGAPPPFKLADIRAAIPKHCWVKDPWRSMSYVVRDVVVVCGLAA |
| 6857 |
PF01673 (Herpes_env) |
 Estimated TM-score=0.42; very hard target. |
>Herpesvirus putative major envelope glycoprotein (Length=535) LFSELLFASHLINVPRAIENNVTYEASSAVGVDNEMTSSTTEFIEEIGDVLALDRACLVCRTLDLYKRKFGLTPEWVADY AMLCMKSLASPPCAVVTFSAAFEFVYLMDRYYLCRYNVTLVGSFARRTLSLLDIQRHFFLHVCFRTDGGLPGIRPPPGKE MANKVRYSNYSFFVQAVVRAALLSISTSRLDETETRKSFYFNQDGLTGGPQPLAAALANWKDCARMVDCSSSEHRTSGMI TCAERALKEDIEFEDILIDKLKKSSYVEAAWGYADLALLLLSGVATWNVDERTNCAIETRVGCVKSYWQANRIENSRDVP KQFSKFTSEDACPEVAFGPILLTTLKNAKCRGRTNTECMLCCLLTIGHYWIALRQFKRDILAYSANNTSLFDCIEPVINA WSLDNPIKLKFPFNDEGRFITIVKAAGSEAVYKHLFCDLLCALSELQTNPKILFAHPTTADKEVLELYKAQLAAQNRFEG RVCAGLWTLAYAFKAYQIFPRKPTANAAFIRDGGLMLRRHAISLVSLEHTLSKYV |
| 6858 |
PF05271 (Tobravirus_2B) |
 Estimated TM-score=0.42; very hard target. |
>Tobravirus 2B protein (Length=116) MTNWATLWPNDRLFLSDTYQLIWFDIEADRIEHKHFKAQNSEDISMIPKGFVSFVDNRLPMCINHKGEVYIRVGSFDTAY YQKFGDLDVSDFDDQVLPPDRDFTFNKVVFGDVPQE |
| 6859 |
PF04259 (SASP_gamma) |
 Estimated TM-score=0.42; hard target. |
>Small, acid-soluble spore protein, gamma-type (Length=78) GTDVQHVQKQNSGAASGQYGTEFASETNVQEVKQQNQQAEAGKSYGAQGSGQYGTEFASETNVQEVKQQNQQAEAGKS |
| 6860 |
PF12527 (DUF3727) |
 Estimated TM-score=0.42; very hard target. |
>Protein of unknown function (DUF3727) (Length=98) NLTLKRTAFTLTVAGELPPVNEDEMLTIEMEEDDVELEPEQFQLLASFYHEEQEYGIYTPLDPLLFFAKRNKAGQPELLS PEEFKKVQPLLEELLFDE |
| 6861 |
PF08591 (RNR_inhib) |
 Estimated TM-score=0.42; very hard target. |
>Ribonucleotide reductase inhibitor (Length=101) NLLSVGMRVRKSVPEGYKTGTYSSFKLWSDQSTVSRPTASTKKRSTAASSQRELMPFCGINKIGGLSAQPVGSSDDEDYE RDIPGLDDIPGLSSSQESVES |
| 6862 |
PF05841 (Apc15p) |
 Estimated TM-score=0.41; very hard target. |
>Apc15p protein (Length=134) IALFNDIRRNQDPAGAKHEKYSMRDLRSKKNQHGNCMNDDDDNSLERFIRRKKSRVIKYIPSLSAYNVFSEFPYFPTSSG QLQDGKLDEFMMLSEQYKSKLPKIRKLGWNRFKPIGINKTMYDSEMLKSRVQAQ |
| 6863 |
PF14866 (Toxin_38) |
 Estimated TM-score=0.41; easy target. |
>Potassium channel toxin (Length=55) EILDKVKKVFSKGVADLNNMSELGCPFIEKWCEDHCESKKQVGKCENFDCSCVKL |
| 6864 |
PF12938 (M_domain) |
 Estimated TM-score=0.41; very hard target. |
>M domain of GW182 (Length=239) SGGFAGRYPVNSGQPSMSFPHNNLMNNMAGTAVTGGGNNNANMTALQVQKYLNQGQHGVAVGPQAVGNSSAVSVGFGQNT SNATVAAAASVNIAANTNNQPSGQQIRMLGQQIQLAIHSGFISSQILTQPLTQTTLNLLNQLLSNIKHLQAAQQSLARGG NANPMAVNVAISKYKQQIQNLQNQINAQQAVYVKQQNMQPTSQQQQQQQPQQQQHPSVHLSNSGNDYLRGHDAINNLQN |
| 6865 |
PF13131 (DUF3951) |
 Estimated TM-score=0.41; easy target. |
>Protein of unknown function (DUF3951) (Length=52) MILLTIGAILLTLFIFFIIGFITFMMFVDRATPQIYYTPCESVTVKSKGKHK |
| 6866 |
PF15934 (Yuri_gagarin) |
 Estimated TM-score=0.41; easy target. |
>Yuri gagarin (Length=231) ILKELADLICRLRSMEFSYSDIYTESSTRNPFCAAVMDIFENNNRQEQQLKTFNQRLACEIDNLHTTLKDRDHQIKQLHD MITGFNDIHENTRLKNEIYDLKQKNCVQARVVRKMGLELKGEEEQRIELCDKYEKLLGSFEEQCKELKRANSRVNTLQER LSQVEKLQEELRTERNILREEVIALKEKDAKRNGHERALCDQLKCCRTEIEQSRTLIRNMQCHLKLEEAHH |
| 6867 |
PF13169 (Poxvirus_B22R_N) |
 Estimated TM-score=0.41; hard target. |
>Poxvirus B22R protein N-terminal (Length=95) NESCLRKESRYHRIVNNIKPKELTDHKAIAAMRYLSIIRTRERERLHNCFDWKSIREEIKRSFISMCDTRSNQDRLDAVY RYNYTYSVSLYIHNK |
| 6868 |
PF10779 (XhlA) |
 Estimated TM-score=0.41; easy target. |
>Haemolysin XhlA (Length=66) VKDKLDTHERRLNNHSERIDKLEQDGRELKTELKNLCENLKSLTSMMKWFIATLVGALISFFFYAV |
| 6869 |
PF01621 (Fusion_gly_K) |
 Estimated TM-score=0.41; very hard target. |
>Cell fusion glycoprotein K (Length=349) MSIRTSIALIGILFAISIYTVLLVVYVSTLSQNGSGCIYATLVDSSLYDAKNFTWEQYNSTLIYTALGNKLPLDGGFDDF SDVCRTYLVNLTSISGLASHVSTKPKIRSVVGTRNCVTYLWRIHIQSLSSSLGLYTIFYVIREWRRMFGVVRFEDDAIST ARYTKNYAARVISSVLLKTTYTKMSRFMCEIMIYKNALSRTFKDDPISFLFHHPIAAVLIITEGLVRLGAQCLCLATLSM YFVPCEKVLSKWFLSITGIFIGIIICIELSLLLAPGPVDGAAMLGETKQVKKDECALETSPSGVHVFCSNCCASLISNIL IKVLYILFMIILIVTIVRYERTLQIALFG |
| 6870 |
PF15682 (Mustang) |
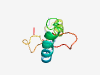 Estimated TM-score=0.41; hard target. |
>Musculoskeletal, temporally activated-embryonic nuclear protein 1 (Length=74) APVKKKRPPVKEEDLKGARGKLSSNQEIKSKTYQVMKQCEQSGAAAPSIFSRDRTGGETVFDKPKEEPPKSVFG |
| 6871 |
PF12314 (IMCp) |
 Estimated TM-score=0.41; very hard target. |
>Inner membrane complex protein (Length=93) KPVIHERIKKCPKTIFQEKIVEVPQIKVVDKIVEVPQYVYQEKIIEVPKVMVQERIIPVPKKVIKEKIVEIPQIELKNIN IEKIQEVPEYIPE |
| 6872 |
PF06129 (Chordopox_G3) |
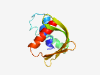 Estimated TM-score=0.41; very hard target. |
>Chordopoxvirus G3 protein (Length=107) LIYLLFFIIFLVLTYYFNKHPTNKLELSVDKLNRENKIIKQRDDAFPVVLNTTVFTRPETPVPTKVHTYYDSATGVVTML SNNKKRIFRLDFDDDVRTLLPILLLSK |
| 6873 |
PF15687 (NRIP1_repr_1) |
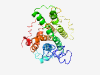 Estimated TM-score=0.41; very hard target. |
>Nuclear receptor-interacting protein 1 repression 1 (Length=304) HQAAGGSGTAVDKKSAGHNEEDQNFNISGSAFPTCQSNGPVVNTHTYQGSGMLHLKKARLLQSSEDWNAAKRKRLSDSIV NLNVKKEALLAGMVDNVPKGKQDSTLLASLLQSFSSRLQTVALSQQIRQSLKEQGYALSHDSLKVEKDLRCYGVASSHLK TLLKKSKAKDQKPDTNLPDVTKNLIRDRFVESPHHIGQSGTKVMGEPLSCAARLQAVASMVEKRASPATSPKPSVACSQL ALLLSSEAHLQQYSREHALKTQNANQAASERLAAMARLQENGQKDVGSFQISKGMSSHLNGQAR |
| 6874 |
PF15013 (CCSMST1) |
 Estimated TM-score=0.41; hard target. |
>CCSMST1 family (Length=75) DKPLQYFNSPASRWRAEHTRSGQDNEDMPWYQPYVVIASMAVFMLYFCVLREENDIDRSLEKSLYEHIPGLEEKQ |
| 6875 |
PF09879 (DUF2106) |
 Estimated TM-score=0.41; hard target. |
>Predicted membrane protein (DUF2106) (Length=148) LGRIWNSLANPKSIPRVYAAILGIILIAGFIIPLALNDQQLYPKPAPQSEVAAQNPLAPYDRGGTPFVEKGTVKAQYPQF SPAIGQVTAYLSPIAIWVKSTTRYFGTSIYSSPGGLIDEILYYTRGFDTVLESTILMMAFTIASWVAI |
| 6876 |
PF16636 (APC_u15) |
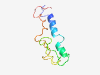 Estimated TM-score=0.41; hard target. |
>Unstructured region on APC between APC_crr regions 5 and 6 (Length=81) FDDDDVDLSREKAELRKGKENKESEAKVTNHTELTSNQQSANKTQAVPKHPINRGQPKPVLQKQSTFPQSSKDIPDRGAA T |
| 6877 |
PF15843 (DUF4719) |
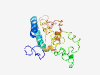 Estimated TM-score=0.41; very hard target. |
>Domain of unknown function (DUF4719) (Length=202) CGERERCCDATNATAVRCCKLPLHAFLDNVGWFVRKLSGLLILLVLFAIGYFLQRIICPSPRRYPRGQARPGQRPGPPGG AGPLGGAGPPDDDDDSPALLRDEAAAGSQDSLLDSGGGGRGRGGGGRSDPSCASEHEMRVVSPVFLQLPSYEEVKYLPTY EESMRLQQLSPGEVVLPVSVLGRPRGGVAAEPDGGEGRYPLI |
| 6878 |
PF04690 (YABBY) |
 Estimated TM-score=0.41; hard target. |
>YABBY protein (Length=159) PSEQLCYVQCNFCDTILAVSVPCSSLFATVTVRCGHCTNLLSVNMRAHLLPTAANHLQLGHNFFSHQSIMEEMRNNPSSM LMNQPLPDPFGPVRVDELLKPPVVNRPPEKRQRVPSAYNRFIKEEIQRIKAGNPDISHRQAFSAAAKNWAHFPHIHFGL |
| 6879 |
PF17549 (Phage_Gp17) |
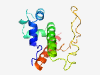 Estimated TM-score=0.41; very hard target. |
>Gene Product 17 (Length=140) MNNYQLTINEVIDIINTNTEINKLVAKKENLFPTDLYDLDKQELIAIILNSDFALSSIKRVLLEVTVEELGTQDNDEDDE LEDLDGEIDRVDYIDKDGIRFDVPRETSPHVDKSIVTFNDELLDEANKIAKSIQEHDFND |
| 6880 |
PF10339 (Vel1p) |
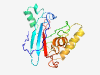 Estimated TM-score=0.41; very hard target. |
>Yeast-specific zinc responsive (Length=201) IFTFFSVLISVATAVRFDLTNVTCKGLHGPHCGTYVMEVVGQNGTFLGQSTFVGADVLTESAGDAWARYLGQETRFLPKL TTIASNETKNFSPLVFTTNINTCNPQSIGDAMVPFANTVTGEIEYNSWADTADNASFITGLANQLFNSTDYGVQVASCYP NFASVILSTPTVNIFGKDDTLPDYCTAIQLKAVCPPEAGFD |
| 6881 |
PF12097 (DUF3573) |
 Estimated TM-score=0.41; hard target. |
>Protein of unknown function (DUF3573) (Length=417) KKTCLIVSSLLACSGLAYSEDSPQVVSQGGPLGATSIGDQNLGQPDPNASGASSTTQTTGSNLNDRELLLKLQQQVQQLQ GQLQQLKAQGNGGGLQNTYNGSSQFTTYSSKVDGNKNPRTLGGNGESKDLSQALIGGQTSSDIMGNVNASNSIINLASEP LGGVFNQKGGIDVGGAPAITTQGQVTYLGSYSGNNSIPIGQISSNLFASTLLGQREKFDDYSVFFGGFIEADAQAWFGSA VTKVQNAGQLSSNGQNIYLTSANLYFLSNLGHYVTAQFDFDTNESGSFSLGNAFVIFGNLDISPFFVTAGRNKLSVGSYG GGGTWTSGITKFLSPNQVTNVSIDYKDQVWNANIAVFGSDDRRANFSTGLFYADSWTPNLAAGFNVGYVFNIAGAGNSSI ANSLANLNRSSDNVGAL |
| 6882 |
PF16869 (CNDH2_M) |
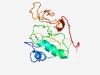 Estimated TM-score=0.41; very hard target. |
>Condensin II complex subunit CAP-H2 or CNDH2, mid domain (Length=155) RPPANLLVFEGDCVDSEASELDSYLLATCGFYGDFVLLDPCDAPAVFAFLQGKKSCKEDILPHRGGSAPSKARHKAFTSP NARSGGTARRKTPGEVLGNAVQPEIEKPQEMNPGKSPAKDWSDHPGDLTPDPSMSQPDYVDPGSPDPGDDSDDED |
| 6883 |
PF17040 (CBP_CCPA) |
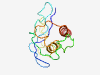 Estimated TM-score=0.41; very hard target. |
>Cellulose-complementing protein A (Length=119) TDDWAPVPKAQQRRGQRPTGPGFFFARAGDRTQMGRLFQPTPVPTAQPVLKPASKVTTMTKLDKNSWNKSAGRQPAPTDN SPTLTEVFMTLGGRATDRLIPKPSLREALLRKREDENGQ |
| 6884 |
PF15450 (CCDC154) |
 Estimated TM-score=0.41; hard target. |
>Coiled-coil domain-containing protein 154 (Length=526) ASTASVLEQDTPKHWKQLEQWLVNMQTQVVSLQEHKDRCECTTLRLLQELCQVQASVQLQDSELKQLRQEMQQAAWAPEK EILEFPSPQNQNQMQALDKRLVEVRESLTQIRRKQALQDSERKGTEQEANLRLTKLAGKLEQEEQDRQVACSALQKSQEE ASQRVDYEVARMQAQVTKLGEEMSLRFLKREAKLCGFLQKSFLALEKRMKSSESTRLKAESSLREELENRWQKLQELAEE RVRALRGQCKQEESHLLEQCWGLDRAVVQLTKFVRQNQVSLNRVLLAEQKARDAKGHLEESQAGELTAYLQENLEAVQLA GEMARQEMHNALELLREKSQALEVSVAELVRQVKDLSDHLLALSWRLDLQEQTLIQRLHEAKSEWEGAERRSLEGLAQCR KEAQAHLREVQETVDSLPRQIEAVSDRCILHKSDSDLKISAEGKAREFEVGAMRQELAALLSSVHLLREGNPGRKIAEIQ GKLATFQNQIMKLENNVQDNKTIQNLKFNSETKLRMEEMATLRESL |
| 6885 |
PF14875 (PIP49_N) |
 Estimated TM-score=0.41; very hard target. |
>N-term cysteine-rich ER, FAM69 (Length=157) ARFSYLRMKYLFFSWLAVFISSWVLYVQYSSYTELCRGHECKNTICDKYRKGVIDGSACSSLCDKGTLYMGKCLSAKPNN QVYTGSWGDLEGVIKCQIDDILHYNLGTEMEPRKEAVLFDKPTRGTSVEKFKEMVFSHLKEKVGEQANLQELVSHVL |
| 6886 |
PF09731 (Mitofilin) |
 Estimated TM-score=0.41; hard target. |
>Mitochondrial inner membrane protein (Length=659) SVGKIIGASVLFVGGGVGGTVLYASYDPRFRENVEKAIPYSDKLFEMALGPAPYSVPIPKKPVQQGPLKISSVAEVMKES KQPVPKPQTSKTEAVAPSEEKEDKEHEGSHAIKERSPDEVAARLAQQDKEEQEKISALAATLEGALSSTARITLQAITAQ ESAVQAVNAHLQILKEAMDDSETAGEKKSAQWRTLEEALKDRRRAVDEAADVLLKAKEELEKTKGVIENAKKSQIPGAKS HIIAAEENLHHMIVDLDNVVKKVQAAQSEAKIVAQYHELVATAREEFQRELDSITPDVQPGWKGLTGQLSTDDLNSLIAH AHRRIDQLNKELAEQRVREQQHIELALEKQKLEDKKALEAAVAKALEHHKSEIEIEQEKKVEEVREVMESEMRTQLRRQA AAHTDHLRDVLKIQEQELKVEFEQNLSEKLSEQEMQFRRLTQEQLDNFTLDINTAYARLKGIEQAVESHAVAEEEARKAH QLWLSVEALKYCMKTASGDSPTEPLESAVKAIKASCSDNAFTEALTAALPQESLTRGVYSEEALRARFYTVQKLAKRVAM IDETRNSLYQYFLSYLQSLLVFHPQQLKPPAELSPDDLDTFKLLSYASYCIEHGDLELAAKFVNQLRGESRRVAHDWLTE ARMTLETKQIVDILTAYAS |
| 6887 |
PF14992 (TMCO5) |
 Estimated TM-score=0.41; hard target. |
>TMCO5 family (Length=275) INSLNMDLERDMQRIDEANQELLLKIHEKENEIQRLENEITQTRDLAEDEEWEKENCTAMEREKALQELEEETARLDRKN ETLVHSIAELQRKLTRKSHKATKCEQDNTKGSPEESKAKLQQLEASCADQEKELAKVMEDYAFVAQLCEDQALCIKKYQE TLRKIEEELETRFLEREVSKVLSMNSVRKESNSQNNKDHSLQKKGTWFCKRIFQYVFFTTLFFIRLLGYLFFHISFINPD LLINILPKILSRGILWKLRCFLFPSLTLETEDMLP |
| 6888 |
PF14946 (DUF4501) |
 Estimated TM-score=0.41; hard target. |
>Domain of unknown function (DUF4501) (Length=176) VAQQPECCVDVVDANATCPGTSLCGPGCYRRWNADGMVSCVRCRNGTFPAYNGSECRSPAGRGVQFPMNRSTGTPGRPHL GGPHVAASLFLGTFFISSGLILSVAGFFYLKRSSKLPRVFYRRNKAPALQPGEAAAMIPPPQSSVRKPRYVRRERPPDRA TDPAAFSSVETRVSNV |
| 6889 |
PF15331 (TP53IP5) |
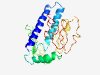 Estimated TM-score=0.41; very hard target. |
>Cellular tumour antigen p53-inducible 5 (Length=220) QPVSKVIERNRLRTVLKNLSLLKLLKSSNRRIQELHKLAKRCWHSLLSVPKILRISSGENSACNKAKQNNEEFQEIGCSE KELKSKKLESTGDPKKKEYKEWKPQVQSGMRNKAKTSLAAMPQKEKHIEPEVPKTSRDDSLNPGAQGRQPLTEGPRVIFI KPYHNRIPMGHMKQLDIADQWIWFEGLPTRIHLPAPRVMCRSSTLRWVKRRCTRFCSASL |
| 6890 |
PF14379 (Myb_CC_LHEQLE) |
 Estimated TM-score=0.41; easy target. |
>MYB-CC type transfactor, LHEQLE motif (Length=46) EVKEALRAQMEVQRRLHEQVEVQKHVQIRMEAYQKYIDSLLEKACK |
| 6891 |
PF15208 (Rab15_effector) |
 Estimated TM-score=0.41; very hard target. |
>Rab15 effector (Length=240) MGQKISHEDNQESKAETLVIREVFSQGVVYASQRLKDYLGFTDPQSKFQTATDTLNEIFLVNFISFCVEKGVEDHITTSK TTKQQSLLFGVDWIWTLLGVDKQIKLQIAVQALQLAELFHSESSPTKALDDCCWEATLADEHFQNRSRFEKLAEFCHLVG QDCLGLFIVFSVPGKPKDIRGVMLDSIAKEKQKCCLLGRNALRQFVMSTESFLSTKEMLEKCLSTRNGLKEVGNVYVNFL |
| 6892 |
PF06286 (Coleoptericin) |
 Estimated TM-score=0.41; very hard target. |
>Coleoptericin (Length=144) MMKLYIIFGLIALSTAYAVPERYFQPPYPDTAAVHAYRDEPFTVTPTEFRSYLGITDEDEIDMPVVYIRERRSLQPGAPN FPLPGSQLPTSITSNVERQGPNTAATINAQHKTDRYDVGATWSKVIRGPGKSKPNWSVGGTYRW |
| 6893 |
PF14982 (UPF0731) |
 Estimated TM-score=0.41; very hard target. |
>UPF0731 family (Length=78) FRFGTQPRRFPVEGGDSSIGLESGLSSSAACNGKEMSPNRQLRRCPGSHCLTITDVPITVYATMRKPPAQSSKEMHPK |
| 6894 |
PF15206 (FAM209) |
 Estimated TM-score=0.41; very hard target. |
>FAM209 family (Length=149) FMFSSLRDKAKEPPGKVPCGGHFRIRQNLPEHAQGWLGSKWLWLFFVFVLYVILKFRGDSEKNKEQNPPALRGCSLRSPL KKNQNASPSKDYAFNTLTQLEMDLVKFVSKVRNLKVAMATGSNLKLPSSEVPGDPHNNITIYEIWGEED |
| 6895 |
PF04394 (DUF536) |
 Estimated TM-score=0.41; easy target. |
>Protein of unknown function, DUF536 (Length=43) DEQLRVKDIQISEKDKQLDQQQQLTLKAMADKDVLKLELEEAK |
| 6896 |
PF13110 (DUF3966) |
 Estimated TM-score=0.41; hard target. |
>Protein of unknown function (DUF3966) (Length=42) MIVYISRKFSQERNLQKSEITAELDMLAGESDKKQKIKEDHQ |
| 6897 |
PF15141 (UQCC3) |
 Estimated TM-score=0.41; hard target. |
>Ubiquinol-cytochrome-c reductase complex assembly factor 3 (Length=83) MDTLRKALIVGALLGAGAGVGSALFVLVTPGEQRKQEMLKEMPEQDPRRRDEAARTKELVLATLQEAAATQDNVAWRKNW TVG |
| 6898 |
PF06403 (Lamprin) |
 Estimated TM-score=0.41; very hard target. |
>Lamprin (Length=138) MAATMQALLVIALLHLATATPVVTKHKVSTFSTGFLGHPVGGLGYGGLGYGGLGYGGLGVAGLGYGGLGYPGAALGGAYT HHAAGLVHPYGGLGYHAAPYHHALGGLGYPLGIGAGVVAPHVVNSKIAAPLAPVVAAI |
| 6899 |
PF11947 (DUF3464) |
 Estimated TM-score=0.41; hard target. |
>Photosynthesis affected mutant 68 (Length=147) KNKRLPFEPSKKRQKPAKASKQTPVVKKTQEVATQSDRAPSVTREQMAVPKVVSDRMARRMAAFCGIPTALGMSTFIVSY LIVSHGWFKLPNVAVLLVSMGFFGLGVLGLSYGVLSASWDEEIVGSMLGWQEFTSNWGRMLSAWRSR |
| 6900 |
PF17697 (DUF5543) |
 Estimated TM-score=0.41; very hard target. |
>Family of unknown function (DUF5543) (Length=118) MYCQDSNICAVFAVQGGKVGRKHGIKRGRRPSIRSPAQRARGPWIHESKHPAFAKQQINLEMPNSRATTELAWVCRSLTL STAPLSPPPSLVHCEDCSCLPGCHSGDLYNLAPAERTC |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218