

Go to page (83 pages in total): [First page] << < 60 61 62 63 64 65 66 67 68 69 70 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 6401 |
PF02989 (DUF228) |
 Estimated TM-score=0.45; very hard target. |
>Lyme disease proteins of unknown function (Length=184) MSDITKIKQEFDKKVAEIKALMKNPQQDSGLLSNSIDFRDQNLIFSNSGGVCTSSKDKIENYPAKGYPYKRGVKLSFGDG TTEVEVEAGGGDDLYGVCSDIDEFSGMATVIPITNNFTGYLTLKKEGQNGVNPGDKLNFNQHGELEKVTGAQKSVNAIAL SKAHKLTEDLFIVLASVFGNRALK |
| 6402 |
PF05716 (AKAP_110) |
 Estimated TM-score=0.45; hard target. |
>A-kinase anchor protein 110 kDa (AKAP 110) (Length=692) TPNKSLSTVASELVNETVSACSKNTTSDKAPGSGDRASGLLRSPPNLKYKTTLKIKESTKESKGPDDKPPSKKSFFYKEV FESRNAGDAKEGGRSLPGERKMFRGQERPDDFTASVSQGIMTYANSVVSDMMVSIMKTLRIQVKDTTIATILLKKVLLKH AKEVVSDLIDSFMKNLHNVTGTLMTDSDFVSAVKRSLFSHGSQKATDIMDAMLGKLYTVMFAKKPPENLRKTKDKSESYS LVSMKGMCDPKHQNTNFASMKSEGKLREKIYSPASKPEKEKTCAETLGEHIIKEGLTFWHKNQQKEGKSPGFQRATFAAP NKQYKPVPDIPLGYPLDTCNLSPPMHHREKPENFVCDSDSWAKDLIVSALLLIQYHLAQGGSMDPQSFLEAAGSTNLPAN KSPVVPDESSLKSPQMGGDQEEAEKKDLMSVFFNFIRNLLGETIFKSDRSSEPKVPEQPIKEEYSHQCERPITPSPIKLS EGDETGGTFAGLTKMVANQLDGHMNGQMVEHLMDSVMKLCLIIAKSCDSPLAELGDDKSGDASRPTSAFPDSLYECLPVK GTGTAETLLQNAYQAIHNELRGISGQPPEGCTAPKVIVSNHSLSDTVQNKQLQAVLQWVAASELNVPILYFAGDDEGIQE KLLQLSAAAVDKGRSVGEVLQSVLRYEKERQLDEAVGNVTRLQLLDWLMVNL |
| 6403 |
PF07655 (Secretin_N_2) |
 Estimated TM-score=0.45; very hard target. |
>Secretin N-terminal domain (Length=100) TETFPLNYLYMERHGLSLTSVNSGRISDSNNSNNNNNSDNNSSNNSSNNDSSSNNSNETTNGTFIRSKTKTDFWGELKET LVSIVGDTGGGRQVVVTPQA |
| 6404 |
PF17398 (NolB) |
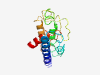 Estimated TM-score=0.45; very hard target. |
>Nodulation protein NolB (Length=150) SPSFGEQAQFEHALAQAAASMKNDTASGPATSAPIPPQLDVQRATAQSSPLGDRVLQTISSMYRDNAVTSAALDHQVGLA KDGLPGPAQKLPVEGGAAGAQMSGVPQAGHDFESMIADLRDLYNGATQVALISKGVSGITSSVNKLLKEG |
| 6405 |
PF05298 (Bombinin) |
 Estimated TM-score=0.45; hard target. |
>Bombinin (Length=141) MNFKYIVAVSFLIASAYARSEENDEQSLSQRDVLEEESLREIRGIGTKIIGGLKTAVKGALKELASTYVNGKRTAEDHEV MKRLEAVMRDLDSLDYPEEASERETRGFNQEEIANLFTKKEKRILGPVLGLVGSALGGLIK |
| 6406 |
PF04767 (Pox_F17) |
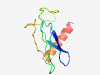 Estimated TM-score=0.45; hard target. |
>DNA-binding 11 kDa phosphoprotein (Length=91) KHTPFYIDTKEGKYLVLKAIKVCDIRKVECNSGSASCVLKVDKPTPSCERPMSPCERGRSPTRRNDVVPFMRTNMLEDIQ NSNRNVVSRIL |
| 6407 |
PF07097 (DUF1359) |
 Estimated TM-score=0.45; easy target. |
>Protein of unknown function (DUF1359) (Length=102) MLSETKIRRNIETIEQKTCRLKNVIHAIKRQTELVELLEDKLQSGEIKKTDKFGAELTDRFSFFESNFNIPVGTLISLLK DNIEGNTTIASELVAKLGIEVE |
| 6408 |
PF04846 (Herpes_pp38) |
 Estimated TM-score=0.45; hard target. |
>Herpesvirus pp38 phosphoprotein (Length=63) ENKNLRKKYNIILDVSTKILVFGTCIMFATGTLIGKGTKIQVPVCNSTNLALAFLAGFLTRHV |
| 6409 |
PF18752 (DAAD) |
 Estimated TM-score=0.45; very hard target. |
>Dictyosteliid AID/APOBEC-like Deaminase (Length=316) FDSNEKVRNDLHKYIMAHTEGVKDMRHGQYAFAFYGSLGAQGGAFPLNRFTSLDLTAQGTSYYVCDINDFRYFKEKEEII VKNPINYNIIREETNQPPIVAAPITVLQTMSKLTTSPYHSESVIIKSLGYFTDPNYPKPKLVYQPIRYLYTQYHPCSVCT TKIMNSSFVNDMTNDNIRIRNLNGLGAMLPSTPPPTTPIAYKINSEESVTNLLLPSDTSKERLYLSFSRQWDIDYCHYIN IRKLAAKPGIQVVSAATNRYGDRHVGYQSTLVTELERSSMIDTRRPDGSIVSELFTFPTTFKSRESNWQYLINQAF |
| 6410 |
PF01021 (TYA) |
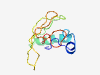 Estimated TM-score=0.45; very hard target. |
>TYA transposon protein (Length=98) ACASVTSKEVHTNQDPLDVSASKIQEYDKASTKANSQQTTTPASSAVPENPHHASPQPASVPPPQNGPYPQQCMMTQNQA NPSGWSFYGHPSMIPYTP |
| 6411 |
PF12277 (DUF3618) |
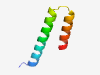 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF3618) (Length=46) ARSPEQIEADIAATRARLASTVDELVDRAHPKNVAKRQVEQAKAQV |
| 6412 |
PF17537 (DUF5455) |
 Estimated TM-score=0.45; hard target. |
>Family of unknown function (DUF5455) (Length=103) PLLWNAIKSVFFAVVSYGIDRIAKKYAPAIVALGVIVSAIVGAVTVYLNFMFEKIAQLNETIPEVVLNVWGWVMPGNAIP CLTILLTARAMSWFVKMGLNVIN |
| 6413 |
PF07913 (DUF1678) |
 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF1678) (Length=196) RIPIPSDPVERIRALRVLREVYRRGKKPSLEVTYRTVNGSTCGPYYVARWRCDSRFKHGRTLYLGKPENESVSFVEWLVS LDRNEVLKLARHLMRNLRSVLKTLLTEISNLPYKRARRVLARGLALTFDARPSNSPRIRDVLEELPDRLESFLIRTLGGW PAHYSSHLNKIIRSRRKSLDGKHEIPDVLLELERWK |
| 6414 |
PF16624 (zf-C2H2_assoc2) |
 Estimated TM-score=0.45; very hard target. |
>Unstructured region upstream of a zinc-finger (Length=95) SKREKGPNNTANSSEIKVKVEPADSVESSSPSITHSPQNELKGTNHSNEKKNTPATQKNKVKQDSESPKSASPSAAGGQQ KARKPKLSVGFDFKQ |
| 6415 |
PF03526 (Microcin) |
 Estimated TM-score=0.45; hard target. |
>Colicin E1 (microcin) immunity protein (Length=55) YYFLASDKMLYAIVISTILCPYSKYAIEHIFFKFIKKDFFRKRKNLNKCPRGKIK |
| 6416 |
PF07547 (RSD-2) |
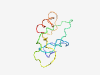 Estimated TM-score=0.45; very hard target. |
>RSD-2 N-terminal domain (Length=82) FGDNPNYLTYNRDLGYADWYGIVDCSDSPPTGPGVYKAFIYVNDLNYQESGNPLFKVTGRLKFLANKDTSDTMMKLEIFE QN |
| 6417 |
PF13498 (DUF4122) |
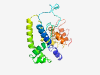 Estimated TM-score=0.45; very hard target. |
>Domain of unknown function (DUF4122) (Length=219) TIVYLSIRLACACYILYKVCGQKKKIREICDLLYAKLPPVKQKEPENVLSEPGGESGVMGSTRFVYLDEDAGKSVAPYMS QPLETAADFIGEEEDVAEEEVECKLPLEEMRMLKEEQEELDGTYPAVDAVSRAVTPDDLDNAGNVLFKLNDADKDEGKSL RAALTLHTIRETDLFEIFSSQVENKNVIDSLMGKYLDGNGNPLPVRKTKSVVPVSDGWK |
| 6418 |
PF13055 (DUF3917) |
 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF3917) (Length=71) MTLKQTGLKRFVPGSILAGIALITYVSSIFIEHVSAEWSTNFVFIGITLFAGSMMVLMVAGIVFFIHMNSE |
| 6419 |
PF02326 (YMF19) |
 Estimated TM-score=0.45; easy target. |
>Plant ATP synthase F0 (Length=80) PQLDQFTYLTQFLWLCVFYMTFYVLLYNDGLPKISRIIKLRKRLVSQEKVGAEQSNDRVEQDVVFKECFQASANYLYSSV |
| 6420 |
PF12375 (DUF3653) |
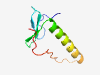 Estimated TM-score=0.45; hard target. |
>Phage protein (Length=68) GYLPNDLRWSGFRICEKRAVLITPSGREFSPKELESFVFWRDEYRQFVEMYGHFEYPKVYPAKENVLP |
| 6421 |
PF15699 (NPR1_interact) |
 Estimated TM-score=0.45; hard target. |
>NPR1 interacting (Length=118) EEELENKKMEMFFNLIKNYQDAKKRRRRDLTQDSGDTASVPTKRSDYGIVPVFRAEDFSHCMDLNLKPSNSVIPTKNQQE EKQEEEEEEDDDDEGEEDDDEEEEVEKVTIKDNGLDLN |
| 6422 |
PF12963 (DUF3852) |
 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF3852) (Length=109) KIRRILSILCVVLLLSCIFGTTAYAAGSGDVAGAIEGTWNDASSQIKTVVNKVVFPAIDLILAVFFFAKLGTAYFDYRKH GQFEWAAPAILFACLVFTLTAPLYIWQIL |
| 6423 |
PF17024 (DMAP1_like) |
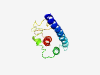 Estimated TM-score=0.45; hard target. |
>Putative DMAP1-like (Length=113) KNFLNLLEKVEELSSKRKQILERHFYELTKYNIDDLEDRDSIITRIVGKAPSHPNAFLASSLGFTKSSWNKKYENMFLKF GLDRKLKHSTYRNMLHFEKLKLLVIKYHEVNKK |
| 6424 |
PF15828 (DUF4710) |
 Estimated TM-score=0.45; hard target. |
>Domain of unknown function (DUF4710) (Length=78) KRSSRQVHFAFLPDKYEPLVEDDETTGRAKEEKKQQKKQKYKKYRKNVGKALRFGWRCLVIGLQEFAAGYSSPMTVAT |
| 6425 |
PF04626 (DEC-1_C) |
 Estimated TM-score=0.45; very hard target. |
>Dec-1 protein, C terminal region (Length=131) PSMMQREVEDEDNKAEDDLVGEAGPQMPENEGTARHKVDALGVGGNKRKKSKSKSAPPTVINYYYAAPQRPVVQSYGTSY GGGGYGSNAYGVPRPVNSYQSQGYRAAVGNDEVDEMLRQHQTMARTINPKQ |
| 6426 |
PF16907 (Caskin-Pro-rich) |
 Estimated TM-score=0.45; very hard target. |
>Proline rich region of Caskin proteins (Length=85) KKRSHSLHRYALSDGEHEEEDGTTSTLGSYATLTRRPGRGQMPRACLQADAKVTRSQSFAIRAKRKGPPPPPPKRLSSVS STPTA |
| 6427 |
PF19185 (DUF5867) |
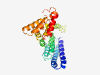 Estimated TM-score=0.45; very hard target. |
>Family of unknown function (DUF5867) (Length=278) DYMAIAMLALKAFARGLSDPVAALCSSSAACDWMALCRGPARVIVRTLRAAIETARAAEWSVPAPKPIHSARKTLAVAID VAGGSRRDALVYMLGLLTSVAVVCTGRGKMEDDMPLQKMAMELQDPACPSRDIAIEGLDIIGDDDEDTIAGAVEPDTPCD PIDGQPPHPQDDLLSNLWGHHPDLFVAVLAHADPWDIGSLALTSRTHYEHVVKAVREDDAAESGRRFRAMGLSGTLWLQQ RLDRRTYDAVAQLITPPPPDRGTLPDLRLLLFLCRPMA |
| 6428 |
PF04876 (Tenui_NCP) |
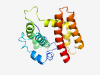 Estimated TM-score=0.45; hard target. |
>Tenuivirus major non-capsid protein (Length=173) QRTIEVSVGPIVGLDYTLLYDTLPETVSDNITLPDLKDPERVTEDTKKLILKGCVYIAYHHPLETDTLFIKVHKHIPEFC HSFLSHLLGGEDDDNALIDIGLFFNMLQPSLGGWITKNFLRHPNRMSKDQIKMLLDQIIKMAKAESSDTEEYEKVWKKMP TYFESIIQPLLHK |
| 6429 |
PF06618 (DUF1148) |
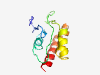 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF1148) (Length=114) MRSHTPPEGTVIMVVPECWSVWIWRDGLSALLPALFSPAEEGFFLETRLFQFESSPRFLLDVDMADCLSRAWNKWPGIAL TWWCWYIRDRVAPIHSLSTKYHLLRGSALASLQN |
| 6430 |
PF13705 (TRC8_N) |
 Estimated TM-score=0.45; hard target. |
>TRC8 N-terminal domain (Length=523) FTDVILRVPPLFIIDELLRIGLGFSSENVVLDNTEHAYSEYLYRYLYCQTYFIIVLKFLCCCIGCIAAICTFMLWTKHLI MVYLYLIPVGAIFVSYWSNVSTMKAIISYVSTQDSTPSILDSILRLDLKILFNEGAGFLVIQNYTLQCMLASLFCYLHLA PRHPILQKLLILSFMAPSILGICPVPTQVLYHAPVFAALLPLAVCKFIIWYNGITMLKTIYTGYQHAHNFISNYGLSALV ETEWLRLNVPCVLRMFWILRVGGQIIEILANHYGEETFNYYMMMKTLLVNGCETLTAVLGMTSIISYICHYIGCFFQWVL LTEDGDEKSIGTVSAILFYMLALQTGLTSLDKDKRLVRLCRNFCLLFTAVLHCVHNIVNPLLMSLSASHNPALHRHLRAL GVCVFLILFPVSLLIFLWSHYTVSTWLLAVSVFSVEVIVKVLVSLAIYSLFLIDAYRSVFWEQLDDCVYIIRSFGNTIEF AFGIILFFNGFWILVFESGGTIRAIMMCIHAYFNIWCEAQAGW |
| 6431 |
PF06770 (Arif-1) |
 Estimated TM-score=0.45; hard target. |
>Actin-rearrangement-inducing factor (Arif-1) (Length=205) LAYLIVFVLSIMGVIDGRYAFLLEIEGKQSVINLSILIMLAFGMWILFYMFYFIWKIIVWTKNRSSSSNTNVNFNLKENF YIAITCITVNVITGLCWMLFAAFQIYVFKNGHLPALDVLYRHYDLESMCWNSIVYLEIDYANTETLSQNCVYVNIYKKCI MCRAIVSDHEPTVFNQNYPVIIMGVLTILAVQCWNLYVQLKEMRR |
| 6432 |
PF16098 (FXMR_C2) |
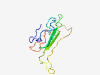 Estimated TM-score=0.45; hard target. |
>Fragile X-related mental retardation protein C-terminal region 2 (Length=90) GYKGNEEIQWNESRARNSRDPKPRQQEDSLQIRIDSNNERSVHTKALQSSFADSGTSNSSSRPRPGKDRGPKKEKQDSLD APRVLVNGVS |
| 6433 |
PF15937 (PrlF_antitoxin) |
 Estimated TM-score=0.45; hard target. |
>prlF antitoxin for toxin YhaV_toxin (Length=97) ESKVTIRGQTTIPAPVREALKLKPGQDSIHYEILPGGQVFMCRQGDEQEDHTMNAFLRFLDADIQNNPQKTRPFNIQQGK KLVAGMDVNIDDEIGDD |
| 6434 |
PF12428 (DUF3675) |
 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF3675) (Length=118) PGYTAPSPVFRLGGIPLNFRGHWQIARRDMNNNPQIIAVVSPERSFVDQEYDEYADSTSRSVFCFRSVAAIFVVLLFLRH ALPVIANRAGSYSFSVFMLWFLRTTAIVLPIYVMMKAM |
| 6435 |
PF10491 (Nrf1_DNA-bind) |
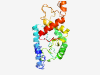 Estimated TM-score=0.45; very hard target. |
>NLS-binding and DNA-binding and dimerisation domains of Nrf1 (Length=211) GPVGVAAAAAVATGKKRKRPHSFETNPSIRKRQQTRLLRKLKATIDEYTTRVGQQAIVLIVTPGKPQNNFKVFGAHPLEN VVRNCRGVIMQELEAALAQQAPPQTQEDPTLHELPPLVIDGIPTPVEKMTQAQLRAFIPLMLKYSTGRGKPGWGKETTRP VWWPKDLPWANVRSDARSEEEKQKVSWTHALREIVINCYKYHGREDLLPAF |
| 6436 |
PF08172 (CASP_C) |
 Estimated TM-score=0.45; hard target. |
>CASP C terminal (Length=264) NLQEDMSRTNAELERSQNLNAQLENDLANLQAEGQNAFPSGASVAGTYTTRIAPSIAPGRRGGRTSPTSSIISGFNPRDS GSEPMGGGSGILPMITAQRDRFKKRNAQLETELSEVHRTVSQLRQEIAALQKDNLQLYEKTRYVSTYNRGGPSSLSSSPA AVSSASAYAPNPNPSTVSIGSGGGNQGGIGLDRYRQAYESNISPFAAFRGRESARAYKRMSIPERAVYSVTRMVLASRTS RNLFAAYCVALHLLVFLSLYWLGS |
| 6437 |
PF15665 (FAM184) |
 Estimated TM-score=0.45; hard target. |
>Family with sequence similarity 184, A and B (Length=211) VIYALNTKNDEHEEEIESLKEAHEDEVQHIVTETRDKIMQYKSKMADEADLRRRLASLEESVELHEHMKRQALAEFEMYR QRMEDSQLCTEAQHTQRVVSMSREVEEMRRDFEEKLRAFSQAQAQFEADKRRALEEMRATHRQEVEELLNNQQNQSATST EDQEKLAELHRQEVEALMERVEELTKDKVRLVEEYEAKLGKAQEYYERELE |
| 6438 |
PF11759 (KRTAP) |
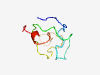 Estimated TM-score=0.45; hard target. |
>Keratin-associated matrix (Length=66) MSYYSGYYGGLGYGHGGSRGLGCGYGCGCNSFRRLGYGCGYGYGSGYGNGSGYGYGCCRPSCCGGY |
| 6439 |
PF14777 (BBIP10) |
 Estimated TM-score=0.45; easy target. |
>Cilia BBSome complex subunit 10 (Length=63) MPEVKSMFREVLPKQGQLCVEDVPTMVLCKPKILPLKSVTLEKLEKMQRDAQEAIRQQEAGQS |
| 6440 |
PF16445 (DUF5042) |
 Estimated TM-score=0.45; hard target. |
>Domain of unknown function (DUF5042) (Length=433) EENIMEQFSDEDNVTVRLNLSTRSVEAGESDMLDEYSSDPANWYKTGTLFPKPPALPRIALMIFNKDLNRYVYNSLISFN PDGTSGRYQIKVRIPKGETEFYAFYAPNQTGQDLPYYTPEGDFRNKIPWDFLKMDIEEINREAIVGAAFPAILKEQENGS IGLPSNSPYDGVDPSPANEKIIDWTETVAKPWEDVPGMSNRLHMGMLSGKAEATVQPSTGEQVQEVTVPLFRDFSRIRIY IASAAAHDRINYIYKRIAFLNFPVMMSPSFRENDSDESQLAASSPSTENIMEHTGVYSYGIKNGGAMSIYEAPFTTDGKI DHTQMETRKYEQFFLPQYLAPYIPESNTWQKGYPHPKIQLTVEHSDGGNNSQLKTKTFLFDVGEETSPGVYSGPIYPNRD YKVFIVLPESSDKEIIYRVEPWVRKNVDFPPFQ |
| 6441 |
PF06547 (DUF1117) |
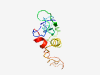 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF1117) (Length=112) VGLTIWRLPGGGFAVGRFSGGRRAGERELPVVYTEMDGGFNNSGAPRRISWASRRSRSRESGGSLGRAFRNIFSFLGRFR SSSSSSSRSTSEPGSITRSHSHSSSVFSRYVR |
| 6442 |
PF06465 (DUF1087) |
 Estimated TM-score=0.45; hard target. |
>Domain of Unknown Function (DUF1087) (Length=61) EDAEEEPQREIIKQEENVDPDYWEKLLRHHYEQQQEDLARNLGKGKRIRKQVNYNDTSQED |
| 6443 |
PF11081 (DUF2890) |
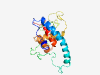 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF2890) (Length=177) MAPKKKLQLPPPPPTDEEEYWDSQAEEVLDEEEEMMEDWDSLDEASEAEEVSDETPSPSVAFPSPAPQKLATVPSIATTS APQAPPALPVRRPNRRWDTTGTRAGKSKQPPPLAQEQQQRQGYRSWRGHKNAIVACLQDCGGNISFARRFLLYHHGVAFP RNILHYYRHLYSPYCTG |
| 6444 |
PF08074 (CHDCT2) |
 Estimated TM-score=0.45; hard target. |
>CHDCT2 (NUC038) domain (Length=171) YDIWHRRHDYWLLAGIVTHGYARWQDIQNDPRYMILNEPFKSEIHKGNYLEMKNKFLARRFKLLEQALVIEEQLRRAAYL NMTQDPNHPAMALNARLAEVECLAESHQHLSKESLAGNKPANAVLHKVLNQLEELLSDMKADVTRLPSMLSRIPPVAARL QMSERSILSRL |
| 6445 |
PF03821 (Mtp) |
 Estimated TM-score=0.45; hard target. |
>Golgi 4-transmembrane spanning transporter (Length=234) HVIMSVLLFIEHSVEVAHGKASCKLSQMGYLRIADLISSFLLIAMLFIISLSLLIGVVKNREKYLLPFLSLQIMDYLLCL LTLLGSYIELPAYLKLASRSRSGPSKVPLMTLQLLDFCLSILTLCSSYMEVPTYLNFKSMNHMNYLPSQEGMSHDQFIKM MIIFSIAFITVLIFKVYMFKCVWRCYRFIKCLNSAEERSNSKMLQKVVLPSYEEALSLPSKAPEGGPAPPPYSE |
| 6446 |
PF11025 (GP40) |
 Estimated TM-score=0.45; hard target. |
>Glycoprotein GP40 of Cryptosporidium (Length=164) ATPKEECGTSFVMWFGEGTPAATLKCGAYTIVYAPIKDQTDPAPRYISGEVTSVTFEKSDNTVKIKVNGQDFSTLSANSS SPTENGGSAGQASSRSRRSLSEETSEAAATVDLFAFTLDGGKRIEVAVPNVEDASKRDKYSLVADDKPFYTGANSGTTNG VYRL |
| 6447 |
PF03010 (GP4) |
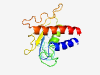 Estimated TM-score=0.45; very hard target. |
>GP4 (Length=152) MKIYGCISGLLLFVGLPCCWCTFYPCHAAEARNFTYISHGLGHVHGHEGCRNFINVTHSAFLYLNPTTPTAPAITHCLLL VLAAKMEHPNATIWLQLQPFGYHVAGDVIVNLEEDKRHPYFKLLRAPALPLGFVAIVYVLLRLVRWAQRCYL |
| 6448 |
PF15692 (NKAP) |
 Estimated TM-score=0.45; hard target. |
>NF-kappa-B-activating protein (Length=81) LRARSWNKKLYECEANMPDRWGHSGYKELYPEEFDTDSDQQEGDEQKAINGKRKSHQGKQTARESHKRKKTKKSHKKKQK K |
| 6449 |
PF16638 (Tristanin_u2) |
 Estimated TM-score=0.45; very hard target. |
>Unstructured region on methyltransferase between zinc-fingers (Length=131) KLDVFSRTRGRGRGRGKRRFGPGRRPGRPPKFIRLEITSENGDKCDDGTQDLLHFSTKEQFDEAEPATLNGLDQPEQTTL PIPQLPQETQPSLEHEPETHSLHLQPQHEESVLPTQSTLTADDMRRAKRIR |
| 6450 |
PF17624 (US30) |
 Estimated TM-score=0.45; very hard target. |
>Family of unknown function (Length=282) CTVDVGRNVSIREQCRLRNGATFSKGDIEGNFSGPVVVELDYEDIDITGERQRLRFHLSGLGCPTKENIRKDNESDVNGG IRWALYIQTGDAKYGIRNQHLSIRLMYPGEKNTQQLLGSDFSCERHRRPSTPLGKNAEVPPATRTSSTYGVLSAFVVWIG SGLNIIWWTGIVLLAADALGLGERWLRLALSHRDKHHASRTAALQCQRDMLLRQRRRARRLHAVSEGKLQEEKKRQSALV WNVEARPFPSTHQLIVLPPPVASAPPAVPSQPPEYSSVFPPV |
| 6451 |
PF10305 (Fmp27_SW) |
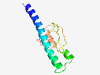 Estimated TM-score=0.45; hard target. |
>RNA pol II promoter Fmp27 protein domain (Length=102) ARQKLDAYNATSWKKRIDRHYAQGKNSMRDLRGLFWGPDHLPDDLEEAERILEVPQRPALMAAIITDLHLLIDKPTFPMK DLPEFIHSVGKGMPKDMKYGLL |
| 6452 |
PF15055 (DUF4536) |
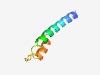 Estimated TM-score=0.45; hard target. |
>Domain of unknown function (DUF4536) (Length=47) NCWSCRVLSGFGLIGAGGYVYWVARKPMKLGYPPGPGTIAQMVFGIS |
| 6453 |
PF17496 (DUF5431) |
 Estimated TM-score=0.45; very hard target. |
>Family of unknown function (DUF5431) (Length=70) MSSQHQYGRLPAERQGEVSYETAAKYCYLVRINRVLNTLNIHIPYPKIALRNPAEGRRTGGRCRSGLRIR |
| 6454 |
PF06756 (S19) |
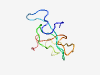 Estimated TM-score=0.45; very hard target. |
>Chorion protein S19 C-terminal (Length=71) GNHGGNYGGHQRGYGRPRWSVQPAGATLLYPGQNSYRRYASPPEYTKVVLPVRAAPPVAKLYLPENNYGSH |
| 6455 |
PF12687 (DUF3801) |
 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF3801) (Length=180) AKQLAILIYAILKEQKKTRGKARMNTMLRSGKELRVFAVKPEDLKTFCQEAKRYGVLYCVLKSKNSKDSVVDVMVRAEDA SKINRIFEKFEMATVDMGTIEQDLERGLEKAGEGEIQTHQQEERGQSTNPTKGREAKSSPSEPTYEARRAAGGTSKPNAK PSVKQELRNIREQQGKNAAR |
| 6456 |
PF15539 (CAF1-p150_C2) |
 Estimated TM-score=0.45; very hard target. |
>CAF1 complex subunit p150, region binding to CAF1-p60 at C-term (Length=291) AKEWDEFLAKGKRFRVLQPVKIGCVWAADRDCAGDDLKVLQQFAACFLETVPVQEEQTPKASKASKRENRDQQILAQLLP LLHGNVNGSKVIIREFQEHCRRGLLSKHTGSPPSPSTTCLHTTSEDAAIPSKSRLKRLISENSVYEKRPDFRMCWYVHPQ VLQSFQQEHLPVPCQWSYVTSVPSAPREDSGSVPSTGPSQGTPISLKRKSAGSMCITQFMKKRRHDGQVGAEDMDGFQAD TEEEEEEEGECMIVDVPDAAEVQAPCGATSGAGGGVGVDTSKAALPASPLG |
| 6457 |
PF15074 (DUF4541) |
 Estimated TM-score=0.45; very hard target. |
>Domain of unknown function (DUF4541) (Length=92) SYFHREAKTGDVSTYDSIFKRPEGYDKKLHRCDREHAKSRGLNINEEEMARPVAVLSSSEYGRRINKPIEQPIKDHARIN HVQAEFYRKNGI |
| 6458 |
PF07956 (DUF1690) |
 Estimated TM-score=0.45; hard target. |
>Protein of Unknown function (DUF1690) (Length=134) WKGSAPTSVSQDMLDSLQSSTETDASRAQTLELQIQARVAEELKRLQAKEDAKLKEAEEKLSELTDKDEKDSPSRQTVSK EVEALRAKLEGRKKVRELPDTVESARSEVIRCLRENDRRPLDCWKEVEKFKDEV |
| 6459 |
PF05771 (Pox_A31) |
 Estimated TM-score=0.45; very hard target. |
>Poxvirus A31 protein (Length=113) METFHMLRSMERKFYADLALALPAHKSAFAHRDMLFIFYAPKAATVARYLAAADVCHSDMTVLGKLHIGGRKLLLVHMDL FYYGLSRAGAMYRLGRSIARLSLDSRKVYAHIS |
| 6460 |
PF03509 (Connexin50) |
 Estimated TM-score=0.45; very hard target. |
>Gap junction alpha-8 protein (Cx50) (Length=66) IQKAKGYKLLEEEKPVSHFYPLTEVGVEARCLPTPFETFEEKVNDVGPLEDISKVYDETLPSYGQT |
| 6461 |
PF19218 (CoV_NSP3_C) |
 Estimated TM-score=0.45; very hard target. |
>Coronavirus replicase NSP3, C-terminal (Length=450) YFCKEHVEGYGNSTFVKDEYCASTMCKVCLFGYQELADLPHTKVVWKYVGFPIFVNWLPFLYLAFLFIFGGIFVKGLVCY FLAQYVNNFGVYFGMQETFWPLQVIPFNVFGDEIVVTFLVYKALMFIKHVCFGCDKPSCVACSKSARLTRVPMQTIVNGA NKSFYVVANGGSSYCHKHKFFCLNCDSYGPGNTFINETVARELSNVVKTNVQPTGDSFIEVDKVSFENGFYYLYSGETFW RYNFDVTDAKYGCKEVLKNCNVLADFIVYNNTGSNVSQIRNACVYFSQMLCKPIKLVDATLLSTLNVDFNGALHSAFIQV LNDSFSKDLSSCASMTECKQALGFDVSDEEFVNAVSNAHRFNVLLSDNSFNNLLTSYAKPEEQLSTHDVATCMRFNAKVV NHNVLIKENVPIVWLARDFQQLSEEGRKYLVKTTKAKGVTFLLTFNSNAM |
| 6462 |
PF05314 (Baculo_ODV-E27) |
 Estimated TM-score=0.45; very hard target. |
>Baculovirus occlusion-derived virus envelope protein EC27 (Length=283) RTDERRVENYRTVTEIVDADNSYKKEFDVSDLVYQNEAYLRKINRRELYLMVAKYYVEVVRELNLPDMRVLFGNRSMLEK VYSFVYYSLAFVNNQMVPHSVKFVDMKFVDVRDRSMSIPTDPIVFYKSLHSDDSTITCFIDSANILRILEKPVDVEAKFE MDDGKSQVFKMLDKIKNVEKKQSLATQRIIMYPFMETMEATPKMNEAYVTQFVTLLTIFSNAYLGLFKLMRSDFQQYFHY LLNHESLVREQSLPNVNNLIIGHFSFNVEAGASGKKGSGGLVF |
| 6463 |
PF09135 (Alb1) |
 Estimated TM-score=0.45; hard target. |
>Alb1 (Length=102) HSRAAKRAASPSLDADKSLASVPRPEPTATVKPHILSAQHNSGINKKSKPKHMTRAQKRRQEKGLERAELVMDRTEKKVA KSVGRGKTVQARRATWEELNQK |
| 6464 |
PF17701 (DUF5547) |
 Estimated TM-score=0.45; very hard target. |
>Family of unknown function (DUF5547) (Length=123) QVLPRICNKVEETFPQLKKVPANHCGICSAVPMGFSLSKSVTQVTAMHTDSKMDDQLIQGTEKTKLEPVTQLFQNTKKIR FEDTSQVNFSRNEGTGTGYLSEKALGSVVYVKESDGIEMTDVE |
| 6465 |
PF12280 (BSMAP) |
 Estimated TM-score=0.45; very hard target. |
>Brain specific membrane anchored protein (Length=193) RGCRLFSICQFVRDSEDLNQTKSECESTCHEAYTHSDEQYACNLGCQNQLPFAEQRHEQLEAMMPRIHLLYPLTLVRGFW EDVMSQAHSFITSTWTFYLQADDGRVVIFQTEPQIKFFSQFQVETDVKEEQKTFPVYKEYHRTLIQERDRDMTGDRSYDD EYNLFSCLSRNPWLPGWILTTTLILSVLVLIWI |
| 6466 |
PF05452 (Clavanin) |
 Estimated TM-score=0.45; very hard target. |
>Clavanin (Length=80) MKTTILILLILGLGINAKSLEERKSEEEKVFHLLGKIIHHVGNFVYGFSHVFGDDQQDNGKFYGHYAEDNGKHWYDTGDQ |
| 6467 |
PF02010 (REJ) |
 Estimated TM-score=0.45; hard target. |
>REJ domain (Length=463) ADMRVNCPGPKSTKVSWNIYKVSNLSDKPDWSKPFNPTGIEGRHFIRLKIPCSSLDTGLYLFNFTVQLISPTFPQMIERS DAVLIEVGSDLWGAVIMGGHFRSVGFSDRWILKGSVLSNGEMVSLSQGLSYTWYCTKQERDYTSMALSTVGKCYPDQVDL KWTTSSNSIQMVLPETLQANNTYHFLLLVQNGSRITQAQQTVHVRAHGALVLNIACTENCGTSVIPTERFSLSGKCGNCR EIKSWYYWSLRSAQSHEIKFDWSSRTTTGRSSSYLHFKAFALVSMAEEFYILSLKVVTKNGQSSSCEYFFYINAPPQIGN CVLHPKEGIAFLTKFIIQCHGFEDRNQPLAYKVIAASDQMTISSISSIQNNTLGIIIYVGHESKTPWSFLPPGRPSQTFA LPIYVQVFDALGASSQVTLQATVLDQRQPPTNAVLHQQLYDLMNGSSALMTKLLITKDYLSIG |
| 6468 |
PF07069 (PRRSV_2b) |
 Estimated TM-score=0.45; hard target. |
>Porcine reproductive and respiratory syndrome virus 2b (Length=73) MGSMQSLFDKIGQLFVDAFTEFLVSIVDIIIFLAILLGFTIAGWLVVFCIRLVCSAVLRARPTVHPEQLQKIS |
| 6469 |
PF08493 (AflR) |
 Estimated TM-score=0.45; very hard target. |
>Aflatoxin regulatory protein (Length=301) NSTSNSTSNSNSNSNSNSNTPPATPATKNVTQAVPPLNGWFTPNSTISNADPPPSPGIIHPSPKPTTSGGSANLFPNLLP PVDQSLASAFTDLTTDLDDFFASPISLSLPDTSDTDILGPPHFFPTGVDSSSNGSTNLFDTFPLFEDAISEFFAPTSNPR SPPNSRASPTSDAQSYQDTHVNDSPCFCLLRALGLMKQLFPNPSTACMTSATQGLNKSSSLPTIQDVIAQNAQTIEAVSA MLHCSCSQDAYLLTIISLIVFKVLGWYAAAARKAPSSGDGNRSMQSPCISLSRSSSHLEEV |
| 6470 |
PF08065 (KI67R) |
 Estimated TM-score=0.45; very hard target. |
>KI67R (NUC007) repeat (Length=112) KITKMPYKSLQPEPIDTPIRMKQRPKTSLGKVDVKGELLAVSNLTRTPGEITYTHTEPAGDGKSIRTFKESPKQILNPVA YVTGMKRWPRTPKEEAQSLEDLAGFKELFQTP |
| 6471 |
PF15764 (DUF4693) |
 Estimated TM-score=0.45; very hard target. |
>Domain of unknown function (DUF4693) (Length=285) PARGLPECRLPSVGDRTHMGRGARATKPGPGLRDQQIPPSAAPQAPEAFTLKEKGILLKLPMAYRKAASQNSRLWAQFSS TQTSDSRDAATTAKTQFLQKMKMTSGWPSSGLSAAEAEAEVGLLQKACSLLRRRMEEELSADPADWMQEYHCLLTLEGLQ AITGRCLHRVQGLRAAVAGQQLGPWPEGRCPRASVPCEGRTDPSWSPQLLLYSSTKELQTLAALRLRVAMLEQQIHLEKV LMAELLPLVSTRDPQGPSWLVLCRAVHSLLCEGGERFLTILRDEP |
| 6472 |
PF14931 (IFT20) |
 Estimated TM-score=0.45; hard target. |
>Intraflagellar transport complex B, subunit 20 (Length=118) GLFIDDIYRLRVQDPRIANETNELREECLEYANKLQEFKSLAEEFHKLLHNYGKDVEREKLRAIATQNHLKTMAKQRLAE QQMYQSKIHEKNLELERLKSEYQYLQRIESEQQEVINC |
| 6473 |
PF17605 (DUF5501) |
 Estimated TM-score=0.45; very hard target. |
>Family of unknown function (DUF5501) (Length=107) MNIIINTLGLNIFIKTVYNFSDHTMERYHPYQRAPWSASIFKKNYNNVKVASYVEIPYTTNNADVEITYTTNAPSRPESK ARRRLDFTNMTPLPCSIDDGSFAKPLM |
| 6474 |
PF12879 (SICA_C) |
 Estimated TM-score=0.45; very hard target. |
>SICA C-terminal inner membrane domain (Length=141) KYFGPLGKGGPRLRRSPAEIHGPSVQEQVLDHVQQDASHEYQLVKERKPRSVPARTKRSGRVNRRTIIEIHFEVLDECQK GDTQVNQKDFLELLVQEFMGSELMEEEEQVPKEEVLMEGVPLELVPIEEVPMERVPNLGSG |
| 6475 |
PF17325 (SPG4) |
 Estimated TM-score=0.45; very hard target. |
>Stationary phase protein 4 (Length=112) SFWDAFAVYDKKKHADPSVYGGNHNNTGDSKTQVMFSKEYRQPRTHQQENLQSMRRSSIGSQDSSDVEDVKEGRLPAEVE IPKNVDISNMSQGEFLRLYESLRKGEPDNKVN |
| 6476 |
PF15105 (TMEM61) |
 Estimated TM-score=0.45; very hard target. |
>TMEM61 protein family (Length=188) ASTLRYCMTVSGTVLLVAGTLCFAWWSEGDAGTQPGQPAAPTGHPMPEAPNPLLRSVSFFCCGAGSLLLLLGLLWSIKVS TRGPPRRDPYHLSRDLYYLTVEPSEKESYRVPKVAAVPTYEEAVCCPLAAGPPTPPAQPVEEGLEDRASGDALLGAQCPP PPSYESIVGAGQGICGQTVPGPGCSCPG |
| 6477 |
PF06762 (LMF1) |
 Estimated TM-score=0.45; very hard target. |
>Lipase maturation factor (Length=463) LYFQWDNLLLETGFLCILVAPLTLIRGSRAVREHDSVTFWLIRWLLFRLMFASGVVKLTSRCPTWWGLTALTYHYETQCI PTPLAWFAHQLPVWWQKLSVVATFVIEIPVPFLFFSPLRRLRLAAFYMQLLLQVLIILSGNYNFFNLLTVTLCLSLLDDR HVNFWLRKADKSGDESNDSKLLLWLSRLVELLVWTLMIFGTIICFDLQVDSDKNTVSSRTAFTYHQFNQFLKTVTIPCIW IGVLSLTWELVASMFRCACISSSLKRFWGMIQWTMFGAAAVSMFAVSLVPFSFIEYESHSRLWPGVRQAYGLVDRYQLVN SYGLFRRMTGVGGRPEVVIEGSIDGNTWTEIEFMYKPGNVSASPPVVTPHQPRLDWQMWFAALGPQTQSPWFTSLMYRLL QGKREVIELIQTDVSKYPFHEEPPTYLRAHRYRYWFTEPKADGSFPERWWRRVYDEEFYPRVH |
| 6478 |
PF13997 (YqjK) |
 Estimated TM-score=0.45; hard target. |
>YqjK-like protein (Length=73) LLQEIQQQRLDLANSAAHWLEVTAPYDRGWAKIVGMRKYLMVGSGIVALYGIRHPSKLVRWSRRAVSIWGTIR |
| 6479 |
PF17088 (YCF90) |
 Estimated TM-score=0.45; very hard target. |
>Uncharacterised protein family (Length=388) LSSVLSLLINIDQETILDFLGINGPFNSEDYSYLFGTSQVSTPVSLETFKLIIQSIWEHYQHGLGFVDIENIALFILVIR FIFLSKKYNIKTGFFITCIGLGAGYLWYMHLRDLAYYYMRTLWMCPLTHNLAYDFSEIFYQQRSEFSKAGTRFDSNSFYG PAVRAFTDITSNDNYRYDPISMLWSYLPTDIKFLSDKVYYFITLKAIPQTYRFIDTQITNLSGMLWYTFIVRINKRYCPY LLRWHWTFLIGLEFLERPFIFVQDRLIYYLNNILIPNSYFREAELVTNLLITLVAAQYIFIILGMLHALCGQYFYFPFLT ENTELHIGLRPKDSIYSGGHTIWQNKRAYMISRQASSKLKKYRFGTIRSAYSKLPRLWYGWLGRGTLD |
| 6480 |
PF12259 (Baculo_F) |
 Estimated TM-score=0.45; hard target. |
>Baculovirus F protein (Length=632) ITMMALTNGLPESIIDSNTIIEVNPLPHTSGFYYQPINKMQFVENIWTFVIEMDHGDIFMELDNLYEETHKFIKFISSET ISNCSSNKIVQTELHTFVIKQILSLVKKHNDIDSKIPKSGVFDDHHELTLSRESAERGHGLRRKRGLLNFVGSVDKYLFG VMDSTDADLLHKVAAGENALNNQVKQLTEELISLSNYTEHMACIEKHRSDVCVYIEAKMSLFVAQINDIASLYTNLDRAV DDALSNRINSLFMTPKRLYTEMLNVTAHLPPKLSWPVPLEQKNMHHLINNNIVKSHVFITKDRTLLFIVEVPLISQQVFN VYQVVPIPLCSNDGKCAIIVPDSKYLGLSTNLQNYIRLDDDATKICKITLNNLLCYKPNIVYDSNGALLCDIKILLDNKN NGVVDVAKDCDVRIGKFDTEIFYPISQYNHWLYVLQADTKIVFDCDTEALIAPRIIKAGTGIIIGKLLDFNCKISTSKVE MSLVQMKSSLFSVVNLPIKTSFNLSTILNDLDKFEIDNFKSNTDLDHKNLNGMTQRLIDLRKRMENNTIFSAKETAFGTD NDEDNWFCWFVSIFNVSCHLAKSIVATLILMIVFVAAFRLYKCMCPGLCTDIFSCFRRNRATVVRVNSKLQF |
| 6481 |
PF15194 (TMEM191C) |
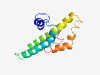 Estimated TM-score=0.45; very hard target. |
>TMEM191C family (Length=121) MEAAAALDASRGGREPWDSQPRRVQDCAGSLMEEVAKADCEKRLFGGVGAGGIRLWALGALQTLLLLPLGFLALPLLYVA LLNPDAIRRGLPRLGSDAAFRRLRYTLSPLLELRARGLLPA |
| 6482 |
PF15466 (DUF4635) |
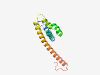 Estimated TM-score=0.45; very hard target. |
>Domain of unknown function (DUF4635) (Length=134) MVTQQVGSRGRVAAELFERRRGSRCEDKKQTLLVLLILVLYVGTGISGRSWEVSERIRECNYPQNPVASQGFEYQTNEPA EEPIKVIRTWLKENLHVFLEKLEKEVQELEQLVQDLEEWLDALLGDGHAEDPCS |
| 6483 |
PF12718 (Tropomyosin_1) |
 Estimated TM-score=0.45; trivial target. |
>Tropomyosin like (Length=146) KMQAMKLEKDNAQDKADTCEGQYKDANAKAAKVEEEVADLIKKLAQIESDLEAHKNALEQANKDLEEKEKLLTSTESEVA ALNRKVQQVEEDLEKSEERSGNALAKLLEATQAADENNRMCKVLENRSQQDEERMDQLTNQLKEAR |
| 6484 |
PF15996 (PNISR) |
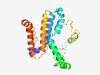 Estimated TM-score=0.45; very hard target. |
>Arginine/serine-rich protein PNISR (Length=206) EIDANKRKMLPAWIREGLEKMEREKQRQLERQQPPTSETDSPAFNVKQVSSKTLTTSVNILNMSNVASDSEDSIAAPEEA AHAPNLAQLIGNDVLSKGSNSDEEQDDDEEEERQRQQNNRQNVGIVARAANAEAALSGKNYEERLADLMLVVRRTLTEIL LETTNEEIAAIATETLKAHRAKASSAQVIRKSALSSITGNLGLAVY |
| 6485 |
PF15457 (HopW1-1) |
 Estimated TM-score=0.45; very hard target. |
>Type III T3SS secreted effector HopW1-1/HopPmaA (Length=321) MMPSQITRSSHSSLPEVAPASGDATGVSEQTPQQARTVAFFASGELAVAFGRTSAAPAQDSVRLLSALQRELDKQHPSWH TVAQLCQSLAEVGTTEQGWHQLASEDQVPALRELLDRCTHRLADMPASHASHDSLSLACEGLRTARLHQSVARLTARPSA LARAVPDLLTLTHLDPETLGAEPPVVSSYTLFSHFVRTAKQRTADLNDSLQRQPDAVVSLLRSHADTLNDLEMLPGALQA LTENCQDAPACNELRELAEVAGGLLQLLREHDLLPRLEEISIEPGEAPAPGHEPAEPRLTTRQALLKAGGNLVRKFDAYG A |
| 6486 |
PF15786 (PET117) |
 Estimated TM-score=0.45; hard target. |
>PET assembly of cytochrome c oxidase, mitochondrial (Length=70) ASKITLGLSCLISFSTIIGVHYVQELERQTLHGGPIKDAKRMEEKERKRNLNRNDHEMQQELKRKYESVQ |
| 6487 |
PF17230 (DUF5304) |
 Estimated TM-score=0.45; hard target. |
>Family of unknown function (DUF5304) (Length=152) DDDAWATACAEDLAEEKARRRAEYGPPPGSAAEELRKFFDTVTDKLSGLQSPLLGAVAGPAAQQVVRQVVEQAKAAVEPV MERNPDVFDHLAAAGNELLAAYRSAVQAQEQRWTARAGDPKRPDKPADDPSDPRDDRDNGEGTGPGQRIDLD |
| 6488 |
PF04731 (Caudal_act) |
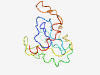 Estimated TM-score=0.45; very hard target. |
>Caudal like protein activation region (Length=133) MYPGSARCTSNNLPVQNFVSAPAYSDYMGYHPVPALDTHGQPAPAWGSHYGPQREDWSAYGPGPSSAVPAAHINGSSPGQ GSYSSADYSALHPAAAAGLPPVDAINAQQISPNSQRHSSYEWMRKTVQSTSTG |
| 6489 |
PF05304 (DUF728) |
 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF728) (Length=139) TCVLKGCVNEVTVLGHETCSIGHANKLRKQVADMVGVTRRCAENNCGWFVCVVINDFTFDVYNCCGRSHLEKCRKRVETR NREIWKQIRRNQAENMSATAKKSHNSKTSKKKFKEDREFGTPKRFLRDDVPFGIDRLFA |
| 6490 |
PF15045 (Clathrin_bdg) |
 Estimated TM-score=0.45; very hard target. |
>Clathrin-binding box of Aftiphilin, vesicle trafficking (Length=79) SRKLWRALQNTNSTSASRCLWSESRCQENLFLVLGIDAARKNLSGDPGHILECSDLKEPEELGVPTFRLQPCRALIQTK |
| 6491 |
PF02459 (Adeno_terminal) |
 Estimated TM-score=0.45; hard target. |
>Adenoviral DNA terminal protein (Length=575) MLEDLAPGAPATLRWPLYRQPPPHFLVGYQYLVRTCNDYVFDSRAYSRLRYTELSQPGHQTVNWSVMANCTYTINTGAYH RFVDMDDFQSTLTQVQQAILAERVVADLALLQPMRGFGVTRMGGRGRHLRPNSAAAVAIDARDAGQEEGEEEVPVERLMQ DYYKDLRRCQNEAWGMADRLRIQQAGPKDMVLLSTIRRLKTAYFNYIISSTSARNNPDRHPLPPATVLSLPCDCDWLDAF LERFSDPVDADSLRSLGGGVPTQQLLRCIVSAVSLPHGSPPPTHNRDMTGGVFQLRPRENGRAVTETMRRRRGEMIERFV DRLPVRRRRRRVPPPPPPPEEEEEGEALMEEEIEEEEAPVAFEREVRDTVAELIRLLEEELTVSARNSQFFNFAVDFYEA MERLEALGDINESTLRRWVMYFFVAEHTATTLNYLFQRLRNYAVFARHVELNLAQVVMRARDAEGGVVYSRVWNEGGLNA FSQLMARISNDLAATVERAGRGDLQEEEIEQFMAEIAYQDNSGDVQEILRQAAVNDTEIDSVELSFRFKLTGPVVFTQRR QIQEINRRVVAFASN |
| 6492 |
PF16890 (DUF5083) |
 Estimated TM-score=0.45; hard target. |
>Domain of unknown function (DUF5083) (Length=157) MEKIKKINKIFLSFEVIPFAMMILLAFPPIIASLINNRLQYGRDFSDRVSFYCYISIMVAYAITILLEIIFYFILLKNYQ YDQSNIELANRVKVTKVLSILYMIPGLYAIPFYFRFLHVRSFRENKKSGQPAYHFWEFFIKPNRKPIELKDFFIKNR |
| 6493 |
PF15397 (DUF4618) |
 Estimated TM-score=0.45; hard target. |
>Domain of unknown function (DUF4618) (Length=258) LRNRRTSLQELRSHENFLTRLNGELVRNIQDMEDSAAQKVRAMLQQQDIFATVMDILEYSNNKKLQQMKCELQEWQEKEE AKMRHLERQVEQLNARIHKTKEEVSFLSTYMDHEYPVRSVQIASLARQLQQVKDSQQDELDDLNEMRRKVLESLSNKIQK KKRKLLRSLVVKTQSPYEEALLQKTRDSQDILKCTGRFREFIKHFEEEIPILRAEVQQLQVQLQEPRDIIFADVLLRRPK CTPDMDVILNIPVEELLP |
| 6494 |
PF05984 (Cytomega_UL20A) |
 Estimated TM-score=0.44; hard target. |
>Cytomegalovirus UL20A protein (Length=103) MARRLWILSLLAVTLTVALAAPSQKSKRSVTVEQPSTSTNSDGNTTPSKNVTLSQGGSTTDGDEDYSGGDYDVLITDTDG GNHQQPQEKTDEHKGEHTKENEK |
| 6495 |
PF15471 (TMEM171) |
 Estimated TM-score=0.44; very hard target. |
>Transmembrane protein family 171 (Length=320) SPTAAAEPDGEQQDRLVSKLIFFFFVFGAILLCVGVLLSIFGFQACQYESLPHCSMVLKVAGPACAMVGLGAVILARSRA RLRFHQGRRQGHQADPDQAFICGESRQFAQCLVFGFLFLTSGMLISVLGVWVPGCGSGWEQEPLNDTDSADEEPQICGFL SLQIMGPLIVLVGLCFFVVAHVKKRNELNADQDAPDREERQTQSTEPVQVTVGDSVIVFPPPPPPYFSESSAAAVTRSPG ANGLLPNENPPPYYSIFNYGRTPTPESQGTVSERNRESVYTISGTSPSFEISHTPHLSSESPPRYEEKDNTAGTPLSPSS |
| 6496 |
PF09718 (Tape_meas_lam_C) |
 Estimated TM-score=0.44; hard target. |
>Lambda phage tail tape-measure protein (Tape_meas_lam_C) (Length=74) QWLLQMNSVWGAYRAPLQDISGMTDELFRNASEKLEKSLFNFATTGKLSLSNFAKTVIDDVARIAARQLSMLAL |
| 6497 |
PF15994 (DUF4770) |
 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4770) (Length=168) LSRLGLIPAVRDKDLKMLMKLSGGNDLAFLWFLMELYYKKPGDEYSSLNEQLILTSIAHLDMITTLRELDNILPPGHQSK RQLEKRRKQKLRDKSWLSPPHKLVKKHSSPYFNKLERPVQYGKPLEVERPDFKVKFFRYEVYKDRNYVPPNEENRWYAKH SFNRAKRI |
| 6498 |
PF15256 (SPATIAL) |
 Estimated TM-score=0.44; very hard target. |
>SPATIAL (Length=195) FFSRHHPHPQRVTHIQDLTGKPVCVVRDEFSLTSLPQPALLSRCLMGMPTISVPIGDPQSNRNPQLSSEAWKSELKELAS RVAIFTKENELKNKEQKEEPAREQGAKYSAETGRLIPTSTRAAGRRSSHQGQRNHPSSRDGGLQTFLLQDQELLVLELLC QILQTDSLSAIQFWLLSAPPKEKDLALGLLQTAVA |
| 6499 |
PF05549 (Allexi_40kDa) |
 Estimated TM-score=0.44; hard target. |
>Allexivirus 40kDa protein (Length=271) QRTFFSNAGVAIDATRALLGYLPPTRYDVPPATLPLDELYGQLHALHQNSLEWLTHINHNVESIIDFFNPNSLFSQGTPL SRLRDAITTLTRTVDDIHSVLTYTELSPEHASSSKAQTKFEVIERSLEALHQKVDKLALLAESNPAPPQVSPNSAKTPPL GAPEDLPVYQAVHPTTSCRTYGTIIFDGTSSRIPMDILGRPASTALRLDMTVATSSQNTTVSYKMFDDGYLLLSENVETA HKLQHCPNDCLALLHQRCQNFIYKIRSHGLC |
| 6500 |
PF14989 (CCDC32) |
 Estimated TM-score=0.44; very hard target. |
>Coiled-coil domain containing 32 (Length=150) LWTEICSSLPEVRPEGAPENNTEFKDSFHPAAQVGVQVNGQANGTDTSSSSAKWEPMEDSEIYIATLENRLKKIKGQSSD VTSRDMLRSLSQAKKECWDRFLHDAQTSELFQGGEMDESALEHFKRWLIPEKVAISAEELEYLLRPSHTK |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417