

Go to page (83 pages in total): [First page] << < 69 70 71 72 73 74 75 76 77 78 79 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 7301 |
PF15847 (Loricrin) |
 Estimated TM-score=0.37; hard target. |
>Major keratinocyte cell envelope protein (Length=312) MSYQKKQPTPQPPVDCVKTSGGGGGGGGSGGGGCGFFGGGGSGGGSSGSGCGYSGGGGYSGGGCGGGSSGGGGGGGIGGC GGGSGGSVKYSGGGGSSGGGSGCFSSGGGGSGCFSSGGGGSSGGGSGCFSSGGGGSSGGGSGCFSSGGGGFSGQAVQCQS YGGVSSGGSSGGGSGCFSSGGGGGSVCGYSGGGSGCGGGSSGGSGSGYVSSQQVTQTSCAPQPSYGGGSSGGGGSGGSGC FSSGGGGGSSGCGGGSSGIGSGCIISGGGSVCGGGSSGGGGGGSSVGGSGSGKGVPICHQTQQKQAPTWPSK |
| 7302 |
PF15522 (Ntox14) |
 Estimated TM-score=0.37; very hard target. |
>Novel toxin 14 (Length=205) PGVNARFTDLEWAKIRVEIIGGGTDVPIWDSEEYRLSDAFIRRMREMLDAVAGGSVDTWENYTTTTEVSNATFGGLRVGT HGQLTGRPTGDVDLEPVSGEGAGMMSRPARAGRQSHHIPQYLLIQYLQNLRSASTKIAHEGGGRPLFPPGFSVTGNTATG FSGAGRTLDFARLDPGSTTDRGRGLPAISLAAKTHQKGQLHINAA |
| 7303 |
PF06056 (Terminase_5) |
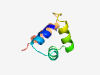 Estimated TM-score=0.37; easy target. |
>Putative ATPase subunit of terminase (gpP-like) (Length=57) VRQRARQLYWQGYPPAEISRLMGINPNTIYAWKKRDQWDGTPPVQRVTQSIDARLIQ |
| 7304 |
PF10268 (Tmemb_161AB) |
 Estimated TM-score=0.37; very hard target. |
>Predicted transmembrane protein 161AB (Length=517) ALLGAQLVITLIMVSVIQKLSPHFSLAKWILCSTGLTRYLHPTDDELRKLSGVPREKGKGKKDKRNGHQANGERSSTFHI PRSLDIRLDTLPISPYDIVHLRFYTEYQWLVDFSLYSAIVYTVSEVYHFFYPLKEEVNLSMLWCLLVVFFAFKLLASLTV QYFKSEESIGERSTCIVTGLAYLLIAMIILIVPEHTLEVGLDKAYHSFNTSASSFLEGQGLNSSGPASKIVVKFFIAVSC GILGALFTFPGLRMARMHWDSLRFCQDRLWLKLLLNVSFAMPFLLVVLWIKPLARDYLTGRVFSGMSSPILSVEAFETMR LGAVVFAVVLRFLLMPVYLQAYLNLAYDRVEEQKKEAGRITNVELQKKISSIFYYLCVVTLQYVAPILMCLFFALMYKTL GEYTWSGVLKESLPLDECSADLEYERAMLATVASEPGAAVDALDFQDITPEGEGEQQVPVEDNILTTAQSFQLSLQSLKS VFTKDVYRGLFGFATWWSCFIWFAASSLGMVYQSYFT |
| 7305 |
PF07376 (Prosystemin) |
 Estimated TM-score=0.37; very hard target. |
>Prosystemin (Length=202) METPSYDIKNKGDDMQEELKVKLHHEKGGDEKEKIIEKETPSQDIKNKDTITSYVLRDDAPEIPKVEHEEGGDGKEKIVE KETISKCIIKIEGDDAQEKLKVEYEEEEYEKEKIVEKGSPSQDINNKGDDPQEKPMVEHEEGDEKETPSQDIIKIEGEGA QEITKVVCEEREKIVVQVDLAVHSTPPSKRDPPKMQTDNNKL |
| 7306 |
PF09574 (DUF2374) |
 Estimated TM-score=0.37; hard target. |
>Protein of unknown function (Duf2374) (Length=42) MSTLESVIWHILGYSAMPVIILSGFLAVAVFCIWLLSLTKDK |
| 7307 |
PF15814 (FAM199X) |
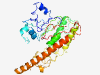 Estimated TM-score=0.37; very hard target. |
>Protein family FAM199X (Length=321) RWNLTSCGTSVASSECSEELFSSVSVGDQDDCYSLLDDQEFTSFDLFPEGSVCSDVSSSISTYWDWSDSEFEWQLPGSDI TSGSDVLSDVIPSIPSSPCLLPKKKNKHRNLDELPWSAMTNDEQVEYIEYLSRKVSTEMGLREQLDIIKIIDPTAQISPT DSEFIIELNCLTDEKLKQVRNYIKEHGPRQRPSRESWKRSSYSCASTSGVSGTSASSSSASMVSSASSSGSSVANSASNS SANMSRAHSDSNLSTSAAERIRDSKKRSKQRKLQQKALRKRQLKEQRQARKERLSGLFLNEEVLSLKVTEEDHEGDVDVL M |
| 7308 |
PF11330 (30K_MP_C_Ter) |
 Estimated TM-score=0.37; very hard target. |
>C-Terminal of 30K viral movement proteins (Length=242) DITKSSVFKSIGDRVSEEINQKRDDYKKKLIENEKLRRREGKGVKIETETRSSSSSDGETLLEEARKSVSLNISKFLADQ RRAPPPPQLEKRTFQWPCGVKMLTMMDTGSSSHYFFSKNITPTSVEMNFGGVAQLEVERAKLSFETFGNKFLLKDVFMFS DQSLGDNILSYTLLKEEGHIDGMRTAGDDVLLEKDGEVVMILDSRDEGRMWIKDDVWAEVTEHGSKSAREYCMKVEKNEI KV |
| 7309 |
PF13071 (DUF3935) |
 Estimated TM-score=0.37; hard target. |
>Protein of unknown function (DUF3935) (Length=70) TRLKQVFGIIISFFVFWFSMLGVQMFAEFLDIESLKFVAGKTEAARAFYSPYPFLIVFLITLLSLYFFVI |
| 7310 |
PF17715 (DUF5560) |
 Estimated TM-score=0.37; very hard target. |
>Family of unknown function (DUF5560) (Length=165) MQAETILEGLEAGLPQAVSSGLSLVPAPGLVLTCLSAPSGPGGMALEPPPTTLRKAFLAQSTLLESTLEGAPEWAAPHPE EQRRSPPACSQHTPPLPSTPTGPPPCSPGGNHPLCALSGRGGGRCSIPSLSSSSTFSLFSSGCWNPRVKLRVRKSQSQGR AGQLI |
| 7311 |
PF15115 (HDNR) |
 Estimated TM-score=0.37; very hard target. |
>Domain of unknown function with conserved HDNR motif (Length=176) MAKGRRFNPPLDKDGKWFPHIGLTSKTPESITCAMLKESHSPCLSQQIERKLPPIYKVREKQAVNNNFPFSMHDNRHSFE NSGYYLDSGLGRKKISPDKRQHISRNFNLWACDYVPSCLDGFSNNQISYVYKEAMVAPNFRRFPRCYNEIQNTFKIVPEQ CCTEFLKKKPKVRFAI |
| 7312 |
PF01516 (Orbi_VP6) |
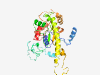 Estimated TM-score=0.37; very hard target. |
>Orbivirus helicase VP6 (Length=324) LLAPGDVIKRSSEELKQRQIQINLIDWIESESGKEDKAEPKEESKAEESKDGQGTQSESSQKKEGSKEAKDADVDRRIHT AVGSGSSAKGPGVRANENVDRGDGKVGGGGGNADAGVGTIGANGGRWVVLTEEIARAIESKYGTKIDVYRDKVPAQIIEV ERSLQKELGISREGVAEQTERLRDLRRKEKSGAHAKAAERGRRKQGKKPHGDVQKEGTEEEKTSEEQASVGIAIEGVMSQ KKLLSMIGGVERKMAPIGARESAVMLVSNSIKDVVRATAYFTAPTGDPHWKEVAREASKKKNILAYTSTGGDVKTEFLHL IDHL |
| 7313 |
PF15626 (mono-CXXC) |
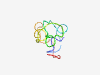 Estimated TM-score=0.37; hard target. |
>single CXXC unit (Length=59) EPTACLECATCREDAPPAVSTPDRSRGGNDGVLSRSLRSSFASDSKTSSSSKCPQATEI |
| 7314 |
PF08151 (FerI) |
 Estimated TM-score=0.37; very hard target. |
>FerI (NUC094) domain (Length=51) SDPEDFSAGAKGYLKVSACVLGPGDEAPLEKKEVSEDKEDIEANLLRPTGV |
| 7315 |
PF15825 (FAM25) |
 Estimated TM-score=0.37; hard target. |
>FAM25 family (Length=65) AVHAVEEVVKEVVDHAKQVGEKAIADALKKAQESGDKVVKEVTETVTNTVTNAVTHAAEGLGKLG |
| 7316 |
PF13677 (MotB_plug) |
 Estimated TM-score=0.37; hard target. |
>Membrane MotB of proton-channel complex MotA/MotB (Length=57) RRGKKHEEEEHENHERWMVSYADMMTLLLVLFIVLYAMSQVDKQKFAQLASGLSDAF |
| 7317 |
PF17320 (DUF5363) |
 Estimated TM-score=0.37; hard target. |
>Family of unknown function (DUF5363) (Length=53) EQTEKKSWFKKALEKYDQFCDELGVNQGACRGCVPVVKFDPEPEKEQKAEKKE |
| 7318 |
PF07818 (HCNGP) |
 Estimated TM-score=0.37; hard target. |
>HCNGP-like protein (Length=94) IPPEPAGRCSSQLQEKIFKLYERRVHGDFDTNSHIQKKKEFRNPSIYEKLIQFCGIDELGTNYPKDMFDPHGWPEDSYYE ALAKAQKVEMDKLE |
| 7319 |
PF14916 (CCDC92) |
 Estimated TM-score=0.37; hard target. |
>Coiled-coil domain of unknown function (Length=53) ENQLQSAQKNLLFLQREHASTLKGLHAEIRRLQQHCTDLTYELTVKSSDQTGD |
| 7320 |
PF15234 (LAT) |
 Estimated TM-score=0.37; very hard target. |
>Linker for activation of T-cells (Length=236) MEAVILLPLVLGFLLLPLLAVLLMALCVHCRELPDSYDSAATDSSIPHSIIIKRPPTLAPWPPATTYPPVTSYPPLSQPD LLPIPRSPQPPGGSHRMPSSRQDSDGANSVASYENDEPACEDEDEDEDEDYHNEGYLVVLPDTAPATGTTVSSAPAPSNP GGLRDSAFSMESGEDYVNIPESEESADASLDGSREYVNVSQELQPTARTKPATPSSQKVEDEEEEEEEGAPDYENL |
| 7321 |
PF04160 (Borrelia_orfX) |
 Estimated TM-score=0.37; very hard target. |
>Orf-X protein (Length=154) MQPENPMIYITKIVILYACNLKFPVKLHGKLLNLALLVLNSLFSLNMVSTLEVYALITSGKQMLLTFLNLDVLNMFMFLL TLFPIFSWPPFTLENQHVTVFNICCFAFLLQESHKPLKQIMDLVILAVLFNVFVFLSKFIIKQEFLIIHRDKVL |
| 7322 |
PF17566 (DUF5472) |
 Estimated TM-score=0.37; very hard target. |
>Family of unknown function (DUF5472) (Length=73) VMLTCHLNDGDTWLFLWLFTAFVVAVLGLLLLHYRAVHGTEKTKCAKCKSNRNTTVDYVYMSHGDNGDYVYMN |
| 7323 |
PF10999 (DUF2839) |
 Estimated TM-score=0.37; hard target. |
>Protein of unknown function (DUF2839) (Length=67) MGEAKRRKSSLGENYGQEERILPWVPITKSQSEQFVQLTTRGAWYGISAMVIIWVTVRFIGPAFGWW |
| 7324 |
PF06773 (Bim_N) |
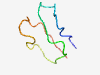 Estimated TM-score=0.37; hard target. |
>Bim protein N-terminus (Length=40) QSSDLNSECDREGGQLQPAERPAQPHHLRPGAPTSLQTEY |
| 7325 |
PF17363 (DUF5388) |
 Estimated TM-score=0.37; easy target. |
>Family of unknown function (DUF5388) (Length=68) DSGVTFPVNVRVDNHIRNKISALLNLGMAKSQKELVKQLVENEIDRLPESDKSRFDRMFEILEEKDNM |
| 7326 |
PF16625 (ISET-FN3_linker) |
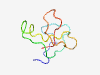 Estimated TM-score=0.37; very hard target. |
>Unstructured linking region I-set and fnIII on Brother of CDO (Length=65) ARPGTTLRPWRDAKLDTATPPMPPSRPGSPEQMLRGRPGLPKPLTSMQPASPQCAGEKGQVAPAE |
| 7327 |
PF17609 (HCMV_UL124) |
 Estimated TM-score=0.37; very hard target. |
>Family of unknown function (Length=126) QTTLPSTVNSTATGVTSDSQQNTTQLPASSAAALSLPSASAVQAHSPSSFSDTYPTATALCGTLVVVGIVLCLSLASTVR SKELPSDHEPLEAWEQGSDVEAPPLPEKSPCPEHVPEIRVEIPRYV |
| 7328 |
PF06370 (DUF1069) |
 Estimated TM-score=0.37; very hard target. |
>Protein of unknown function (DUF1069) (Length=206) MGSKCKAIAGGNDHVQSSSLDGTNSSVRYILPLRSHSQPTRELVEVDVALARLPTCIRWRNVRTNPPVGLHVEQPTPFHH EAMTQLPAHLPCRRPGFEGVRVGKYSLRGRSLSAGSGVIHQPHHTGSGVGVPAGCRVSNEVVVDGNLVRQSLAGVAVAIV RSSGIGVDEVTYASGGDRYHGGPGMLECLDLEGWPIGLAASPLQPS |
| 7329 |
PF15760 (DLEU7) |
 Estimated TM-score=0.37; very hard target. |
>Leukemia-associated protein 7 (Length=193) WGWGDGPGAPGNLRDPDHVSTAPARRSGPRRARSGPGREEQGGGVGTRSRRTAARANSPEEEVVRGAEGGAELLPFPRDR GPCTLARMAMRSALARVVDSTSELVSVEQTLLGPLQQEQSFPIHLKDSVEFRNICSHLALQIEGQQFDRDLNAAHQCLKT IVKKLIQSLANFPSDAHVVACASLRQILQNLPD |
| 7330 |
PF12118 (SprA-related) |
 Estimated TM-score=0.37; very hard target. |
>SprA-related family (Length=305) SPVNQGSDARFSQVQNTGLTGAEFTSSDEHSVADIFISPNQSSVDGVTTSSAETGRVVSQGDGGGGSNAVFDTGTPVGVQ FAREDASNSAVAAQEAEKAAKEAKQQKLAEALGGEREPFSEQKVARQQVEQDSNAQAMARENDIERREQVKIRQQEMEVR QLEARQAEVLAHERAHAAVGGQFARAPNFEYELGPDGKRYATGGEVSIDIASVQGNPQATINKMQQVYAAAMAPVNPSQA DLRVAAEALRNIELAKDELASERLAAMPTQDEISPLLDAQNAIDEVPVFEPVTPSLGTNLDKSGA |
| 7331 |
PF09350 (DUF1992) |
 Estimated TM-score=0.37; easy target. |
>Domain of unknown function (DUF1992) (Length=69) LVEDMILQSMKRGDFNNLKGMGKPLKNDESNPYIDVIEYKLNKILINNGFAPPWIMKEMELRTEITSLR |
| 7332 |
PF12551 (PHBC_N) |
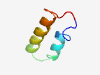 Estimated TM-score=0.37; hard target. |
>Poly-beta-hydroxybutyrate polymerase N terminal (Length=40) DTADRALHAAQARLTGGISPISIASAAFNWTLQLANAPGR |
| 7333 |
PF05597 (Phasin) |
 Estimated TM-score=0.37; hard target. |
>Poly(hydroxyalcanoate) granule associated protein (phasin) (Length=126) KDGSSWMGEVEKYSRQIWLAGLGAYSKISNDGSKLFETLVKDGEKAEKLTKVEAQKQLDDVKNTSKSAKSRVDDVKDLAV GKWNELEEAFDKRLNGAIARLGVPSRSEVQALNSKVDQLTRQIELL |
| 7334 |
PF15820 (ECSCR) |
 Estimated TM-score=0.37; very hard target. |
>Endothelial cell-specific chemotaxis regulator (Length=103) SLRIREDSTILPSPTSETVLTVAAFGVISFIVILVVVVIILVSVVSLRFKCRKNKESEDPQKPGSSGLSESCSTANGEKD SITLISMKNINMNNSKGCPTAEK |
| 7335 |
PF01484 (Col_cuticle_N) |
 Estimated TM-score=0.37; hard target. |
>Nematode cuticle collagen N-terminal domain (Length=49) VGVAVSFTATLVCVIAAPMLYNYMQHMQSVMQSEVDFCRSRSSNIWREV |
| 7336 |
PF15332 (LIME1) |
 Estimated TM-score=0.37; very hard target. |
>Lck-interacting transmembrane adapter 1 (Length=228) LCTACHRPDETLAPRKRVQRQRARPQGGVMPAEASLLRRHHFCTLSKSDTRLHELHRGPAGCRAPRPASMDILRPQWLEV PRGTSRPMAAFSHRELPQAMPAATSPSICPEATYSNVGLAAIPRASLAASPVVAEYACIQKLKETDQGPQSLGQGKAKLT PAVQVDILYSRVRKPKKRDPGPATDQLDPKGRGVILPLGSDQSYETLPLRGLGKDDGLLENVYESIQE |
| 7337 |
PF07416 (Crinivirus_P26) |
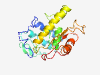 Estimated TM-score=0.37; very hard target. |
>Crinivirus P26 protein (Length=227) MNNFPEIFDDESTCDYDKEIDHQELSDTFWCLMDFISSKHGKSVADINSGMNTLINIRKSLNGSGKVVSITDSYNKTYFH SQRGLTNVDSRINIDILKIDFISIIDDLQIIFRGLIYKDKGFLDSADLLDLDKKTTTRKFQEYFNILKIKIIEKIGMTKT FHFNIDFRNTISPLDKQRKCSISSSHKKTNRLNDLNNYITYLNDNIVLTFRWKGVGFGGLSLNDIKI |
| 7338 |
PF17690 (DUF5537) |
 Estimated TM-score=0.37; very hard target. |
>Family of unknown function (DUF5537) (Length=160) MSVLSPEIKCETSKFTRSSFGSCLIFESSWKKAVLETQKIKKEYTTAFGLEELKECIKMPYLPGLQSCQKSVSSTPLEVP KRLPRADAEVSAVRLKKTKETCSVAPLWEKSKGSGFSDPLTGAPSQYLERLSKIAILEYDTIRQETTTKSKKSKKRDLRD |
| 7339 |
PF07816 (DUF1645) |
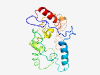 Estimated TM-score=0.37; very hard target. |
>Protein of unknown function (DUF1645) (Length=190) AEELFDGGKIRPFRPPPRSPRKKKEIDTFATEVENTRERTKNDRGRERVSSLSSSNSGRRGARSLSPHRVSKYPWEEEQQ LQQKTEKKSPISSKPALSSTTSKGCKKWSLKDFFLFRSASEGRAADRDPLKKYIALFKRREEVKSSSVRSIEGSNSGSNS KRRGPISAHELHYTVNRAVSEDLKKKTFLP |
| 7340 |
PF09849 (DUF2076) |
 Estimated TM-score=0.37; very hard target. |
>Uncharacterized protein conserved in bacteria (DUF2076) (Length=259) MSPEERQLLTALFDRVRTAAATPRDRDAEALIDQATREQPSATYYLAQAVIVQEKGLEAAANHIKELEERVRQLEAGESE HHQAEQGGGFLSSIFGNTQTQQPAPVPSNPGPSNPGPWGQQSRGYDDSRGYDRDVRQPQQQPTGPWTQQAYAPSAGGSFL RGALGTAAGVAGGMLLANSLSGIFGNHMSSLGWGSPVGANPFGNANAPTEETVINNYYGNDDTRQASDNVDDKDNDNGNL QQADYDDSDDSTDDSSGDD |
| 7341 |
PF17671 (DUF5531) |
 Estimated TM-score=0.37; hard target. |
>Family of unknown function (DUF5531) (Length=152) TLHEILDSSGTIGALVAWMISYKPALFGFLFLLLLLSNWLVKYEVKPTPTEPQQEKTEKRKIPEAEPNCNAMSTREVERL HACFALQDKILERLMFSEMKLKVLENQMFIVWNKMNHHKRSSGQQMFAERKHRPRRQESPFSIDSEGTTNSP |
| 7342 |
PF16611 (RGS12_us2) |
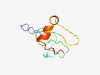 Estimated TM-score=0.37; very hard target. |
>Unstructured region between RBD and GoLoco (Length=72) PSRGKDKQKSAPVKQNTAVNSSSRNHSATGEERTLGKSNSIKIKGENGKNARDPRLSKREESIAKIGKKKYQ |
| 7343 |
PF11747 (RebB) |
 Estimated TM-score=0.37; hard target. |
>Killing trait (Length=68) ITDAVTQANVKVLGEAPAMAMGNLYQAMAHSTGILFQNAVSAQNQQNILAQAATTQGVMQIYSIDTAA |
| 7344 |
PF09592 (DUF2031) |
 Estimated TM-score=0.37; hard target. |
>Protein of unknown function (DUF2031) (Length=208) RVSILKYVLFSIVICSFEYAKNELYFVNDKEIYLGGNVINFRNNRILAYADNQFDLNEFYESTLSLASQLGDCVEGNKEI AHLRNIIDSHIKKHKGSKTSLDLKNVDSKTKKKINELRKELEEVKKELDNKRNDELAIQPIENKKIIKKDGNSSVSEHED FKQLKNNENNENNEITSSNRHMKSKLTKKYRKEAIKYILSWLTLEVKE |
| 7345 |
PF15485 (DUF4643) |
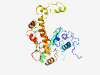 Estimated TM-score=0.37; very hard target. |
>Domain of unknown function (DUF4643) (Length=317) TPTPPIVKVTPKSDSKAVLLSDKDVGKPALPKVSEGKVNAETTKVTEKPTEAKMSLLEKIKKQKIEAASSEKTPEKGEVT DVVKPLAKPKEDQKNLETKDNEPEKVETKASSQQEIRSAVEKEEVEEGSSLTAEPLLKVMQKPKGMKSKLSGWSRLKKHM VVEAEEPKFPEAEPASNKDVPASTDQNVSEMQNKSEAEATPTDSQDSRDSPRAAKMWDAVLFQMFSTKENIMQQIEANKT EEQKKMESEQKMPKEIPTFAHRLPVLLYSPRFDARRLREATSRPATKIATVFEMGLIGRKNKEEEPKDFNRTAKGFS |
| 7346 |
PF17449 (YrzK) |
 Estimated TM-score=0.37; hard target. |
>Uncharacterized YrzK-like (Length=54) MKYHPKMGRAFHDGEANLHYSDHADPDTTGYLTETASEFGLMSKEAVLRKNNRN |
| 7347 |
PF05438 (TRH) |
 Estimated TM-score=0.37; hard target. |
>Thyrotropin-releasing hormone (TRH) (Length=230) LLILASLTVCNLMVSGGQSMSAEDETDRKTIDDIILERAETLLLRSILKKLQDEDGRNDGYYSQPEWVTKRQHPGKRYSD DLEKRQHPGRREEDEDEQYFDVQKRQHPGKREDETHSFLELQKRQHPGKRSTAGHSSDNPILLLSELSKRQHPGKRYLVL HSKRQHPGKRLLEDEDGDSDGDADADGEEDLAEMEKRQHPGKRFWDNSSPDLGTNSPCDVLDPASCSKTS |
| 7348 |
PF12308 (Noelin-1) |
 Estimated TM-score=0.37; hard target. |
>Neurogenesis glycoprotein (Length=99) SPEEGWQIYSSAQDADGKCICTVVAPAQNMCNRDPRSRQLRQLMEKVQNITQSMEVLDLRTYRDLQYVRNTESLMKTVDG KLKTASDNPRSLNPKSFQE |
| 7349 |
PF14937 (DUF4500) |
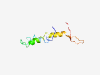 Estimated TM-score=0.37; hard target. |
>Domain of unknown function (DUF4500) (Length=79) DKGFRTPGLRGAQTTTLFRAVNPELFIKPNKPVMAFGLVAITLCVGYLGYMHATRENEQLYEAVDSQGERYMRRKTSKW |
| 7350 |
PF10046 (BLOC1_2) |
 Estimated TM-score=0.37; easy target. |
>Biogenesis of lysosome-related organelles complex-1 subunit 2 (Length=95) DMFTKMSAYLQGELTATCEDYKLLENMNKLTGLKYLEMKEIAVNINRNLKDLNQKYASLQPYLDQINQIEEQVTALEQAA YRLDAYSKKLEAKFK |
| 7351 |
PF15809 (STG) |
 Estimated TM-score=0.37; very hard target. |
>Simian taste bud-specific gene product family (Length=239) LGTNSPLLGQPFLASLSNSEHPQSKPDPGSNDLARAPLKTNASPSDSFQPAGSSEVQRWPPSEELPSMDSWPSEDLWPMM AGAVEDDGGEVLPEELSFLSNAIALPLDSGLVPAGSSSHSADPSLEVSPLHQDSESRPLSRSNVLGDQKQILAQRPPWSL ISRIRKPLLPGHPWGTLNPSVSWGGGGRGTGWGTRPMPCPVGIWGNNKCPSTSWGNINQYPGGSWGSINRYPGGSWGSI |
| 7352 |
PF00951 (Arteri_Gl) |
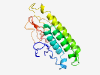 Estimated TM-score=0.37; very hard target. |
>Arterivirus GL envelope glycoprotein (Length=173) SWSFVDGNDNSSTYQYIYNLTICELNGTEWLSSHFDWAVETFVLYPVATHILSLGFLTTSHFLDALGLGAVSTTGFVGGR YVLSSVYGACAFAALVCFVIRAAKNCMACRYARTRFTNFIVDDRGRIHRWKSPIVVEKLGKAEVGGDLVTIKHVVLEGVK AQPLTRTSAEQWE |
| 7353 |
PF03904 (DUF334) |
 Estimated TM-score=0.37; hard target. |
>Domain of unknown function (DUF334) (Length=230) MRKGQLIMSMKDIKNNKENPNTQMNSKSTGTPSSSTQNSLNNEELSELKRQNKLIVKYLAEIQENQKIREKEQKEITSEL KEATKDFRDKSLKIRNDFVDVLQEKLKHVDTEELKDILGRGIYKVREENDRMLQEVKRSHEHYQTRQKYLFTGIGAMLLV FMLFALIMTIGSDFMSFLHVDTLQNAIAGKLKASEGFMTFVWYIAYGLPYVLAIGLFIGLYEWIRAKFHD |
| 7354 |
PF10446 (DUF2457) |
 Estimated TM-score=0.37; very hard target. |
>Protein of unknown function (DUF2457) (Length=445) FHEFASDIPREDDWIRQAFSSQGRRITIDDTLQKENAIRKLATEVEEEAEQEEEEMATALDDDEDLDNLDDGDDVEDEDE DEDEDQEDEDDNSDEDDNSDGYKTDEETGFADSDEEEDEDEDLRLWTPGRSTVQNIDSTPIARRPSLPEQLSDSSIASRT SLRARRGRAHRIKIRPGTPELPDSTDFVCGTLDEDRPMEDAYMSCLAARRREKLQLIPQDIDPSFPASDPEDEGEEEIYN PVHHSSDDEMFFHGKMDDLHHANDRSRRRKKTEHTSPKRFHSPPPLKRRASPAPKARGRSPRRLFDGPSPRRLRSPAPQM LRSPPASPRNGHGAIAFKTLASRPGLTVTKSLPRPPSMFPHMAKGRRVKAPANDIHIRGAIDIVKGLERKRQRRKEKFYQ KYCNRARKGQVPERKPQRGLGCERMRELGLLMAGKIDQGNYVLSV |
| 7355 |
PF15344 (FAM217) |
 Estimated TM-score=0.37; very hard target. |
>FAM217 family (Length=233) SDLSDSERIPIPPSPLTPPDLNLRAEEIDPVYFDLHPGQGHTKPEYYYPDFLPPPFSSWDLRDMALLLNAENKMEAVPRV GGLLGKYIDRLIQLEWLQVQTVQCEKAKGAKARPLTAPGTSGALKSPGRSKLIASALSKPLPHQEGASKSGPSRKKAFHH EEIHPSHYAFETAPRPTDVLGGTRFCSQRQTLEMRTEEKKKKSSKSTKLQRWDLSCSESSSKVETNGNIRIPK |
| 7356 |
PF11044 (TMEMspv1-c74-12) |
 Estimated TM-score=0.37; hard target. |
>Plectrovirus spv1-c74 ORF 12 transmembrane protein (Length=49) MPTWLTTIFSVVIILGIFAWIGLSIYQKIKQIRGKKKDKKEIERKESNK |
| 7357 |
PF04936 (DUF658) |
 Estimated TM-score=0.37; hard target. |
>Protein of unknown function (DUF658) (Length=186) KVFDAYIEGEKKATGTIDEIADYFDISRNSISLWIKNGKDPKKANPKYKHAILNKEKTKELMEQKKKEERKLPASVYDYY DKGEFIMTGTAREISQFLNIGKHNVYSYIQVGKHAFDYRKTRKHAILNEAETRKRFPLLSVSSEEELIETKEKERRKHET KEERRLRRNIRAQMAIENSRKDDSVF |
| 7358 |
PF16628 (Mac_assoc) |
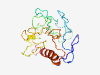 Estimated TM-score=0.37; very hard target. |
>Unstructured region on maltose acetyltransferase (Length=186) KPSSGKTPVPLQSKDGYTDVGSQYVHEISQQHDRQQSLTEQLEAGKMRSIIVDDSDRTTTQFNNSSPTGVGSSRGSWSKR SWPNTGHTAYITDHITKSDYRDVPSIHELPREGHPKTDYQIVPPIRELSHGTAHGKPGVSLFQGGIDQRPALATNVDTSS PQAQARMALSIEHQLSARTVPGEETE |
| 7359 |
PF15840 (ARL17) |
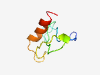 Estimated TM-score=0.37; very hard target. |
>ADP-ribosylation factor-like protein 17 (Length=62) MWKGGRSHPFLPHSSRCAGSGGQLDSILPHQSPAWGPWGCKDLSSGFPSFLTSSILWKSAVV |
| 7360 |
PF17502 (DUF5434) |
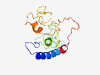 Estimated TM-score=0.37; very hard target. |
>Family of unknown function (DUF5434) (Length=186) MDVFGRGRAATADDYRRFLERNSRAAAKLAAATTPVRTDSVISQPEAEERPRHRSLSRRNSVSSKDTHVVAGGDKHRDRP DRPSGTLVALAMAAQRRKSISRQEPLVNASASNTLRSSQRSRGEGRNSRSEGHSSSSSSYTQRSNSHGHRSESQYHRRKS ITQSTSARLPQQQGHPRPRRNTLRHM |
| 7361 |
PF01683 (EB) |
 Estimated TM-score=0.37; easy target. |
>EB module (Length=56) CKSGYFSNSGSCDPVRSAGEACTGSGQCVENAECSSSSGAGVCRCKTGFYDVTGSC |
| 7362 |
PF05334 (DUF719) |
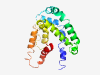 Estimated TM-score=0.37; very hard target. |
>Protein of unknown function (DUF719) (Length=181) GWGYWGSWGKSILSTATATVATVGQGLTQVIEKAETSLGIPSPEELSAQAEKDKTDDSDSSKENKDEEEKTYGMGGAMGM ISSGSMGMLSSLTTVVQSTGKTVISGGLDALEFIGKKTMDVIAEGDPGFKKTKGLMNRNSTLSQVLREAKEREEQQKAEN VTSETDKKAHYGLLFDEFQGL |
| 7363 |
PF05760 (IER) |
 Estimated TM-score=0.37; very hard target. |
>Immediate early response protein (IER) (Length=325) ECAIDAQNIISISLQKIHNSRTQRGGIKLHKNLLVTYVLANARDFYMSVEFAGMYSIQQNGEMRTVSDEKQDSFELTGNC DDIAGEFCCNYSGDSLDTYHCAAPQSAGYPAMVPEAHTQTVPACPMAPVRHCDLNPAMVPEAHTQTVISCPMAPVRHCDS NLLDSEIRWDYYSEPYTFKRDILISNTPNNQKTVLDLDTHVVTTVDYGYLHADYCLSFKQHGQCLQGSPKKRKIDSSYVS DSDYLSDFLPMSCKQIRSEDVYCCNSEQLDTNNISNLMSVLDSGFNELLNWQTDLEQVGLVNGQTNCCKQALACSGAWTR AIEAF |
| 7364 |
PF15144 (DUF4576) |
 Estimated TM-score=0.37; hard target. |
>Domain of unknown function (DUF4576) (Length=91) MAVSVLRLTVVLGLLVLILTCYADDKPDKPDDKPDDKPDDSGKDPKPDFPKFLNLLGTEIIENAVEFILRSMSRSTGFME FDDNQGKHSSK |
| 7365 |
PF05281 (Secretogranin_V) |
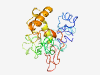 Estimated TM-score=0.37; very hard target. |
>Neuroendocrine protein 7B2 precursor (Secretogranin V) (Length=234) DLLNRMDNDMQVGYYDVDNEGDLAAVAAAAAKDNVDLVSRSEYARLCNGGSDCLLQSNNAHPSLRDQEFLQHSSLWGHQY VSGGMGEVPNRYNTIVKTDASLPAYCNPPNPCPEGYDTEQQAGNCISDFDNTAIYSREFQAAQECTCDSEHMFDCADQEN AAGVGPGNSDLNSAVEKFLLHQFGNENVLDNANVVAKKAGHMPALRMDSIPNPFLQVAPGDRLPIAAKKGNMLF |
| 7366 |
PF04360 (Serglycin) |
 Estimated TM-score=0.37; very hard target. |
>Serglycin (Length=149) QVLLQGSRLILALALILVLDSSIQGIPVQRARYQWVRCSPDSNSANCIDEKGPVFDLLPGESNRILPPRTGPFSMTRSQN LNDVFPLSEDYSGSGSGSGAGSGSGSGNGFLTDIEQEYQPVDENDAFYYKFRSRERNLPSDNQDLGQDG |
| 7367 |
PF03533 (SPO11_like) |
 Estimated TM-score=0.37; hard target. |
>SPO11 homologue (Length=43) AFAPMGPEASFFEVLDRHRASLLAALRRGGGEPAGGGTRLASS |
| 7368 |
PF17628 (IncD) |
 Estimated TM-score=0.37; very hard target. |
>Inclusion membrane protein D (Length=141) MTKVYANSIQQERVVDRIALLERCLDPSNSLPTAKRLVAVAVATILAVALLVVAGLLFSGVLCSPVSVLAASLFFGVGAF LLGGALVGGVLTTEAVTRERLHRSQTLMWNNLCCKTAEVEQKISTASANAKSNDKTRKLGE |
| 7369 |
PF15553 (TEX19) |
 Estimated TM-score=0.37; very hard target. |
>Testis-expressed protein 19 (Length=159) MCPPVSMRYEAEGMSYLHMSWMYQLQHGSQLRICFACFKAAFLDLRQLLELEDWEDEDWDPELMDHTEAGSEQEASPGMG PSWGQGQGQPAQGGSVDWGSGTLASGPVESEEVDLDDHFVPTELEPQDATPLGLGAEDADWTQSLPWRFGGLPTCSHWP |
| 7370 |
PF10472 (CReP_N) |
 Estimated TM-score=0.37; very hard target. |
>eIF2-alpha phosphatase phosphorylation constitutive repressor (Length=410) MEPGTGGSRKRLGPRAGFRFWPPFFPRRSQAGSSKFPTPLGPENSGNPTLLSSAQPETRVSYWTKLLSQLLAPLPGLLQK VLIWSQLFGGMFPTRWLDFAGVYSALRALKGREKPAAPTAQKSLSSLQLDSSDPSVTSPLDWLEEGIHWQYSPPDLKLEL KAKGSALDPAAQAFLLEQQLWGVELLPSSLQSRLYSNRELGSSPSGPLNIQRIDNFSVVSYLLNPSYLDCFPRLEVSYQN SDGNSEVVGFQTLTPESSCLREDHCHPQPLSAELIPASWQGCPPLSTEGLPEIHHLRMKRLEFLQQASKGQDLPTPDQDN GYHSLEEEHSLLRMDPKHCRDNPTQFVPAAGDIPGNTQESTEEKIELLTTEVPLALEEESPSEGCPSSEIPMEKEPGEGR ISVVDYSYLE |
| 7371 |
PF15003 (HAUS2) |
 Estimated TM-score=0.37; hard target. |
>HAUS augmin-like complex subunit 2 (Length=190) GGLSDALSIASDLGFAVAPPPSHEELQSLATSNGEKGDDLIRVLRELSAVQRKIADLQVELQGRKDDKNVAHLTHVSEMQ KKIETLSRITQILKDVIQNKDRIIARLQQPYSLDCIPVEAEYQKQFSELLMKAASDYGALTASVSDFQWSQNFKEPPSVW GEMLRPIPVALASCTRFFEAMSAMRESFAT |
| 7372 |
PF10533 (Plant_zn_clust) |
 Estimated TM-score=0.37; very hard target. |
>Plant zinc cluster domain (Length=49) KKRCHSHAHSEDFAGKFAANGGRCHCSKRRKNRVKRTIRVPAISSKMAD |
| 7373 |
PF08604 (Nup153) |
 Estimated TM-score=0.37; very hard target. |
>Nucleoporin Nup153-like (Length=509) PSTTSTASNYPDVLTRPSLHRSHLNFSMLESPALHCQPSTSSAFPIGSSGFSLVKEIKDSTSQHDDDNISTTSGFSSRAS DKDITVSKNTSLPPLWSPEAERSHSLSQHTATSSKKPAFNLSAFGTLSPSLGNSSILKTSQLGDSPFYPGKTTYGGAAAA VRQSKLRNTPYQAPVRRQMKAKQLSAQSYGVTSSTARRILQSLEKMSSPLADAKRIPSIVSSPLNSPLDRSGIDITDFQA KREKVDSQYPPVQRLMTPKPVSIATNRSVYFKPSLTPSGEFRKTNQRIDNKCSTGYEKNMTPGQNREQRESGFSYPNFSL PAANGLSSGVGGGGGKMRRERTRFVASKPLEEEEMEVPVLPKISLPITSSSLPTFNFSSPEITTSSPSPINSSQALTNKV QMTSPSSTGSPMFKFSSPIVKSTEANVLPPSSIGFTFSVPVAKTAELSGSSSTLEPIISSSAHHVTTVNSTNCKKTPPED CEGPFRPAEILKEGSVLDILKSPGFASPK |
| 7374 |
PF04697 (Pinin_SDK_N) |
 Estimated TM-score=0.37; very hard target. |
>pinin/SDK conserved region (Length=133) MAVAVRTLQDQLEKAKESLKNVDENIRKLTGRDPNELRPGQARRLTIPGPGGGRGRGIGLLRRGLSDSGGGPPAKQRDIE GALLRLAGDQRARRDSRHDSDAEDDDDVKKPALQSSVVATSKERTRRDLIQDQ |
| 7375 |
PF18615 (SMYLE_N) |
 Estimated TM-score=0.37; very hard target. |
>Short myomegalin-like EB1 binding proteins, N-terminal domain (Length=382) MRDACRICARELCGNQRRWIFHPASKLNLQVLLSHVLGKELCRDGKAEFACSKCAFMLDRVYRFDTVIARIEALSIERLQ KLLLEKDRLKFCIASLYRRNNGEELSPDDKVGDGTVDLSSLPDVRYAALLQEDFVYSGFECWTEHEEQASESHSCHASES GASRPRRCRGCAALRVADSDYEAICKIPRKVARSISCGPSSRWSASVCNEEPPIGELGATESPNAKVPADGDSMEEGTPG SSIESLDTTVEVGSLQQKEEEMDKGGKGSGKCDCCSDDRTTLNSSLNSNRLELALSLIKTCDYKPVQSPKGSRLPVLVKS KPSHDTMDGSPHAGLVTTGSGFLNADVKSFSRPPLGFPLEMSDLQELWEDLCEEYMPLQVQN |
| 7376 |
PF15838 (DUF4717) |
 Estimated TM-score=0.37; hard target. |
>Domain of unknown function (DUF4717) (Length=71) PNERSQKEKTSSTDQVQEQFEEHFVASSVGEMWQVVDMAQQEEDRTSETATVRDHLFDLAFCFNLASIMVF |
| 7377 |
PF08196 (UL2) |
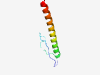 Estimated TM-score=0.37; hard target. |
>UL2 protein (Length=59) MAEDSVTILIVEDDDAYPSFGSLPASHAQYGFRLLRSIFLIMLVIWTAVWLKLLRDALL |
| 7378 |
PF05950 (Orthopox_A36R) |
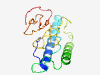 Estimated TM-score=0.37; very hard target. |
>Orthopoxvirus A36R protein (Length=158) MMLVPLITVTVVAGTILVCYILYICRKKIRTVYNDNKIIMTKLKKIKSSNSSKSSKSTDSESDWEDHCSAMEQNNDVDNI SRNEILDDDSFAGSLIWDNESNVMAPSTEHIYDSVAGSTLLINNDRNEQTIYQNTTVVINETETVEVLNEDTKQNPNY |
| 7379 |
PF15908 (HMMR_C) |
 Estimated TM-score=0.37; easy target. |
>Hyaluronan mediated motility receptor C-terminal (Length=150) EEERQDYHKQLAEAQLQSTSEAETEHWRNLYEELYAKVKPFQEQLDRFAAERNALLNEKGATQAELNKLADAYANLMGHQ NQKQKIKHMVKLKEDNLELKQEVSKLKAQVGKQKQELDRLKSSQVQRRFDPSKAFKHDLKENQQPTSALT |
| 7380 |
PF10940 (DUF2618) |
 Estimated TM-score=0.37; hard target. |
>Protein of unknown function (DUF2618) (Length=40) KGRSLMAHIRRTRHIMMPSYRSCFSYSLFASQNKPSNRAL |
| 7381 |
PF13863 (DUF4200) |
 Estimated TM-score=0.37; easy target. |
>Domain of unknown function (DUF4200) (Length=118) LLEKRREMSEVEQALGAQKEEFQMKMESLQQRREELERKEYQLRESLLKFDKFLKENDSKRARALKKAQEERDMRRAKDC EIARLKEDTSGLMKGRDKVQHRLEKYIIYQQYLEKVLE |
| 7382 |
PF04501 (Baculo_VP39) |
 Estimated TM-score=0.37; very hard target. |
>Baculovirus major capsid protein VP39 (Length=252) MALVPVGMAPRQMRVNRCIFASIVSFDACITYKSPCSPDAYHDDGWFICNNHLIKRFKMSKMVLPIFDEDDNQFKMTIAR HLVGNKERGIKRILIPSANNYQEVFNLNSMMQAEQLIFHLIYNNENAVNAICDNLKYTEGFTSNTQRVIHSVYSTTKSIL DTTNPNTFCSRVSRDELRFFDVTNARARRGGAGDQLFNNYSGFLQNLIRRAVAPEYLQIDTEELRFRNCATCIIDETGLV ASVPDGVELYNP |
| 7383 |
PF06503 (DUF1101) |
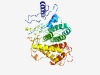 Estimated TM-score=0.37; very hard target. |
>Protein of unknown function (DUF1101) (Length=360) MERSSREHSKYSKANTLNETYQMRMYKDDSTPDYCYSEISVGLSSSSPKMSLSDYFSAVSVTYGDEARLDEYKPLLYSDL LFAESYELDVDINLLVWQLLSSNQDSKSLCVNVLRMLHTYSLGNAYMGGGIYHFSQGTNTETLSDIVDILRLIGRLAKII IKTKFSQMELKCVQTHLIYYFTGKAYKSLSLSWDSKSILSTSNGYSTSEGLLDYYIRNKLDLFKALYSKNLVYGGNYYLI YQVLVYYYIITNGRYSTGFNLRKDSIKHYNIPNDNPKMCNSILPRKPNLSMMYIRAILIMVMIKDYSPIKLVPLYLNALE IEDPAYMSSRITDGGIRMETDNMASTPDISRVLPAYFNGV |
| 7384 |
PF14757 (NSP2-B_epitope) |
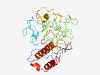 Estimated TM-score=0.37; very hard target. |
>Immunogenic region of nsp2 protein of arterivirus polyprotein (Length=272) VKSYPRWTPPPPPPRVQPRKTKPVKSLPENKPVPAPRRKVGSDCGSPILMGDNVPNGWEDFAVGGPLDFPTPSEPMTPLS EPVLMPASQHIPRPVTPLSGPAPVPAPRRTVSRPMTPLSEPIFVSAPRHKFQQVEEANPAATTLTYQDEPLDLSAFSQTE CEASPLAPLQNMGILEAGGQEAEEVLSGISDILNDINPAPVSSSSSLSSVRITRPKYSAQAIIDSGGPCSGHLQREKEAC LSIMREACDAAKLSDPATQEWLSRMWDRVDML |
| 7385 |
PF07188 (KSHV_K8) |
 Estimated TM-score=0.37; very hard target. |
>Kaposi's sarcoma-associated herpesvirus (KSHV) K8 protein (Length=238) MPRMKDIPTKSSPGTDNSEKDEAVIEEDLSLNGQPFFTDNTDGGENEVSWTSSLLSTYVGCQPPAIPVCETVIDLTAPSQ SGAPGDEHLPCSLNAETKFHIPDPSWTLSHTPPRGPHISQQLPTRRSKRRLHRKFEEERLCTKAKQGAGRPVPASVVKVG NITPHYGEELTRGDAVPAAPITPPYPRVQRPAQPTHVLFSPVFVSLKAEVCDQSHSPTRKQGRYGRVSSKAYTRQLQQ |
| 7386 |
PF16037 (DUF4790) |
 Estimated TM-score=0.36; hard target. |
>Domain of unknown function (DUF4790) (Length=91) DPSYALHKPYSLTCAIRRLRNFRKYNQQLEYEMNQLALYQSTAIDEALFFRLMSYTSLWPPLHTYKEIDYTSRHFFPRSR KYMRRLNDLLK |
| 7387 |
PF19186 (DUF5868) |
 Estimated TM-score=0.36; very hard target. |
>Family of unknown function (DUF5868) (Length=202) MKYITGNYLDLGKKESEIRYNLKSIIKYIYCQIPLDHDKMKKLYKYVEVDCDYDYKKDDDYDYLGPDFMPNKMQDDEYVY KFKHELNESYFPEPHSGEDINNELIREMHEFLIEKSPKKLILKDIRTIKLIKNKGVNKDRRGVRDLDWKLHFWQEYIISS KRSLKLHDLIIAAYKIKSHKFEKWYEMFCRVSYDDFIIFHNS |
| 7388 |
PF17621 (DUF5508) |
 Estimated TM-score=0.36; very hard target. |
>Family of unknown function (DUF5508) (Length=263) MESRNSYENKIDEISSLSESKEHPIDIQEKKDAFVNEFKGVLFDKNTRSSELLFNFYECCYKFLPRAQPQDKIDSYNSAL QAFSIFCSSTLTHNNIGFDFKLFPEVKLSGEHLETVFKYKNGDDVREIAKINITLQKEEGGLYNLRGLDFKGCFFSGQNF SNYDIQYVNWGTSLFDVDTPCIFNAPAYNKSNEKSLKPVSENGLSGVLTDRNNKIKLITGVAPFDDILFMDDDFDDSSSE DDPVENSPVVTSPVVSSSKSSFQ |
| 7389 |
PF08374 (Protocadherin) |
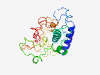 Estimated TM-score=0.36; very hard target. |
>Protocadherin (Length=212) SLSNATVVESQVARSLHTPLAQDIAGDPSYELSKQRLSIVIGVVAGIMTVILLILVVVMARYCRSKGKHGYEAGKKDHED FFTPQQHDKAKKPKKDKKGKKGKQPLYSSIVTVEASKPNGQRYDSVNEKLSDSPGMGRYRSVNGGPGSPDLARHYKSSSP LPTVQLHPQSPTAGKKHQAVQDLPPANTFVGAGDNISIGSDHCSEYSCQASS |
| 7390 |
PF15333 (TAF1D) |
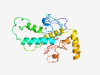 Estimated TM-score=0.36; very hard target. |
>TATA box-binding protein-associated factor 1D (Length=215) SGSSLFKTQCVPYSPKRRQRNSIRKFVHSPESVQARDSSSDSSFEPRPLTLKAIFERFKKKKRKKRKYKPTGRPRGRPKG RRSTRCSQITKKHIRDRGPGFPFLESKNGRRPLPWRKILTFEQAVARGFFNYLEKLKYEYHLKESLKQMNVGEDLEKDDL DSRRYKYLDDDGSLSPIEELAAEDEVAANLEHDDECDIKLVEDSCFIISSELPKK |
| 7391 |
PF17685 (DUF5533) |
 Estimated TM-score=0.36; very hard target. |
>Family of unknown function (DUF5533) (Length=139) MPVIPIPRRVRSFHGPHTTCLHSACGPVRTTHLVRTKYNNFDIYLKSRWMYGFIRFLLYFSCSLFTSILWVALSVLFCLQ YLGIRIFLRFQYKLSIILLLLGRRRVDFSLMNELLIYGIHVTMLLVGGLGWCFMVFVDM |
| 7392 |
PF17730 (Centro_C10orf90) |
 Estimated TM-score=0.36; very hard target. |
>Centrosomal C10orf90 (Length=512) IVISQMIDENKSRENRASLPLPCAIAQSRAHHAKQSLANRSGVNIHRAFALLPGRLGIPAPSDERGPEAELPPKEERPCG GPRRGFASITITARRVGPPARALVWGTAGDSLCPKCRAEDTLFQAPPALANGAHPGRHQRSFACTEFSRNSSVVRLKVPE AHTGLCERRKYWVTHADDKETSFSPDTPLSGKSPLVFSSCVHLRVSQQCPDSIYYVDKSLSVPIEPPQIASPKMHRSVLS LNLNCSSHRLTADGVDGLVNREPISEALKQELLEGDQDLVGQRWNPGLQESHLKETPSLRRVHLGTGACPWSGSFPLENT ELANVGANQVTVRKGEKDHTTHCHASDHANQLSIHIPGWSYRAVHTKVFSGSSKRQQGEVCMTVSAPPVEQKPTRHFLPI GDSSPSDDCLSRDLSEPTERRHQSFLKPRILFPGFLCPLQDVCASLQEDNGVQIESKFPKGDYTCCDLVVKIKECKKSED PTTPEPSPAAPSPAPRDGAGSPGLSEDCSESQ |
| 7393 |
PF08039 (Mit_proteolip) |
 Estimated TM-score=0.36; easy target. |
>Mitochondrial proteolipid (Length=58) MLQSIIKNVWIPMKPYYTQVYQEIWIGMGLMGFIVYKIRSADKRSKALKAPAPARGHH |
| 7394 |
PF15551 (DUF4656) |
 Estimated TM-score=0.36; very hard target. |
>Domain of unknown function (DUF4656) (Length=358) QQMRDRKGRKAELKDPNGREAETITFISGTAEAPPNQSFCCSSLSQAWNTYKAVFCCIVTCGGCFQDCSVCIPYPGPAET STEDGKNGDYNGRLPNSPTNTSPAEKNGNQIKKSSMGSSFSYPDVKLKGIPVYPNRNTNHHPESDSCCKELLEKPFRNSI EKPPLPSSHRSSEEYYSFHESDVDISELNGSMSSRQIDVLIFKKLTELFSVHQIDELAKCTSDTVFLEKTNKISDLINSI TQDYNLDEQDAECRLVRGIIRISTRKSRVRPHIPLPSQSHEEKSGRGNAPDSGNETMLESMVISQDELAVQISEETPADV LARNMRRHSSAGSPTSRDSSFQDTETDSSGAPLLQVYC |
| 7395 |
PF17069 (RSRP) |
 Estimated TM-score=0.36; very hard target. |
>Arginine/Serine-Rich protein 1 (Length=294) SKYVNDMWPGSPQEKASPSTSGSGRSSRLSSRSRSRSSSRSSRPHSRSSSRSSSRSHSRPRRSRRSRSRSRSRRRHQRKY RRYSRSYSRSRSRSRGHRYYRDSRYEQPRRYYQSPSPYRSRSRSRSRGRSHHRRSYYAITRGRRYYGFGRTVYPEDRPRW RERSRTRSRSRSRTPFRLSEKDRMELLEIAKANAAKALGTANFDLPASLRAKEASQGAVSCSGPKTEHSEKQTEVGTKNA SEKSAAAQRNIAFSSNNSVAKPLEKTTKAAVEETSSGSPKIDKKKSPYGLWIPV |
| 7396 |
PF06295 (DUF1043) |
 Estimated TM-score=0.36; hard target. |
>Protein of unknown function (DUF1043) (Length=122) ALIGLVVGFVIGALVMRFGNAKLRQQKALQTELENNKSELEEYRKELVGHFARSAELLDNMARDYRQLYQHMAKSSHELM PNMPVQDNPFNYRLTESEADNDQAPVKMPPKDYSDGASGLFR |
| 7397 |
PF05501 (DUF755) |
 Estimated TM-score=0.36; very hard target. |
>Domain of unknown function (DUF755) (Length=123) TKETFPCPVTCKKQYKLQTQKNKVQKPCSMTGITEGALLHRQLLKECKKTSKLIHLSNLMIQKHPPKRKDAQRNSSSTKH TKKRSRDVSAHSAKKIPAKKKKRASDSSSTSSSSNSSTSKESS |
| 7398 |
PF15047 (DUF4533) |
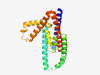 Estimated TM-score=0.36; very hard target. |
>Protein of unknown function (DUF4533) (Length=225) DKERLIQAAKMFFFHVQDLASVTNTLTELFNRSMNTQILLMAVKNNSNIKDFFEQMLKIFKEMQSVVDARHDKIQKESLC SKVAMAMCSVVQKSTNVEELHQSAKEVFKSAYTPVIISVLNSSNILGSLESSLSHLMKFPIMNLQLSDFYTEDTKEQSDV TTSERSRSPGSSKTTMIDTLKKLQDVLKTEDSRNPTKSAADLLEQIVKAMGPILEILQKAIKTME |
| 7399 |
PF15559 (DUF4660) |
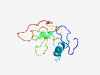 Estimated TM-score=0.36; very hard target. |
>Domain of unknown function (DUF4660) (Length=110) SDDEGPKSSKTKDGASASRSDVASQQGTKRAAGGMPLPKPDELFRSVSKPTFLYNPLNKQIDWESRIVKAPEEPPKEFKV WKTNAVPPPESYATEEKKKGPPPGMDMAIK |
| 7400 |
PF15282 (BMP2K_C) |
 Estimated TM-score=0.36; very hard target. |
>BMP-2-inducible protein kinase C-terminus (Length=267) DDFDVFTKAPFSKKVSQQDGQVVGTEPLLSPASPRSVDIFGCTPFQPSLLPKTGGSKEDLFGLVPFEEIAGSQQQKVKQQ RNLQKLSSRQRRMKQEVSKNNGKRHHGTPTTTKKALKPSYRTPERARRHKKAGRRDSQSSNEFLTISDSKENISVALTDS KDRTNPLQTDESLLDPFGAKPFHPPDLMRHPQHQGFSDNRGDHGAVVVAGRPRQSSLHGAIHGGDGLKTDDFGAVPFTEL VVQSAQSQQQQALELDPFGAAPFPSKQ |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417