

Go to page (83 pages in total): [First page] << < 43 44 45 46 47 48 49 50 51 52 53 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4701 |
PF10769 (DUF2594) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2594) (Length=74) MSETDFSTSEEVQALANEVTCMKALVTLMLKGMGQADAGKVIINMERYIAALEDQQQAEVFSNTVKQIKHAYRQ |
| 4702 |
PF10926 (DUF2800) |
 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF2800) (Length=358) HAFLSPSSSHRWLNCTPSASLESEFENKTSQAAEEGTAAHAWCEHKLKKALRMRSKRPVSSYDSDEMQEHTDAYVDFVLE QLDIAKQNCKDPLVLIEQHVDFSEYVPDGYGTADCVIVSDDRLHIIDFKYGMGVLVDATDNPQMKCYALGALAIYDSLYD IKEVSMSIFQPRRENVSTWTITVDELKTWAEEVLKPKAEMAMNGEGEFCPGEWCTFCRAAVRCRARAEEKLKLAQEEFKL PPLLTDSEIEEVLTIIPDLTKWANEIMAYATESAVNHGKQWNGFKVVEGRSVRKYKDESAVAEAAKEAGYKDIYRQSLIT LTEMQKLMGKATFEKVLGDLIIKPPGKPTLVPDSDKRP |
| 4703 |
PF07663 (EIIBC-GUT_C) |
 Estimated TM-score=0.58; very hard target. |
>Sorbitol phosphotransferase enzyme II C-terminus (Length=93) LSPLLGPGAVIAQVVGVLIGVEIGKGNIPPQYALPALFAIDPQVGCDFIPVGLSLGEAEPETVEVGVPAILFSRLITGPI SVLIAYLFSFGLY |
| 4704 |
PF07506 (RepB) |
 Estimated TM-score=0.58; hard target. |
>RepB plasmid partitioning protein (Length=182) ISRLSAIQEQRMIAKAIERHVPKEKIARALSINVRSLVRKATLVEGICEEAVALLKDKMCPMAVFDVLRKMIPIRQIEMA ELLINANNYSVGYASAILAGTPQAQLVNSTTPKRIKGMTAEALSRMEGELSRLQQAITSIQDTYGQDHLHLTVLRGYLAK LVDNTKVAKYLERRHPEFFSEF |
| 4705 |
PF11250 (FAF) |
 Estimated TM-score=0.58; hard target. |
>Fantastic Four meristem regulator (Length=53) SFPPPLTSISGSNGVQVTSHREGGRLVLQAVNVPSCQTYFHVERSEGRLRLCL |
| 4706 |
PF11889 (DUF3409) |
 Estimated TM-score=0.58; very hard target. |
>Domain of unknown function (DUF3409) (Length=56) EEGATNKKQQKPDRLEKGRMKIVPKESEKDSKTKPPDATIVVDGVKYQVKKKGKVK |
| 4707 |
PF09502 (HrpB4) |
 Estimated TM-score=0.58; easy target. |
>Bacterial type III secretion protein (HrpB4) (Length=225) SPFDRAAAALQAYDRNIRDAICWAHPSWLAGALGVDVRVAETLRAELSSGPRTLLDKASFILCRASGVRMPSMASFLAPG AVMFDALPPEQGLRMLRVRALLFRSAQIRHLIDKGSRMRLADWTGVPLDSIVQASFGAPDIAAFELGGTMLSLDEMDAQT LALEGYFLVIRDEEGAAAMTTSRMLLRLALPRDAGPSRWLNGRVGGLDKQGTRNLITHLSAWLPE |
| 4708 |
PF04540 (Herpes_UL51) |
 Estimated TM-score=0.58; easy target. |
>Herpesvirus UL51 protein (Length=158) ICGWGARPEEQYEMIRAAVPPSEAEPRLQEALAVVNALLPAPITLDDALGSLDDTRRLVKARALARTYHACMVNLERLAR HHPGFEAPTIDGAVAAHQDKMRRLADTCMATILQMYMSVGAADKSADVLVSQAIRSMAESDVVMEDVAIAERALGLSA |
| 4709 |
PF07619 (DUF1581) |
 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF1581) (Length=85) RVVVEDGVSTSYVNGRVLHAETLPENPDPWIDIHCRAASTGGVRRLVITGEPRVPEEIDLLTHPDLSNWITDFYRESGQG DNAEW |
| 4710 |
PF07588 (DUF1554) |
 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF1554) (Length=138) AHHGGLGGIYGADEWCMRKVPADLKGSPYYSYKALLADGINRRASVTGNAGDGQINWVLQASTVYSQATTGTPTIGTTNS KKLFTFPLTNKIRPDSITDFGSITWTGLNPNWTTAPYNCNNWTDGADINVGTIGNYNA |
| 4711 |
PF10271 (Tmp39) |
 Estimated TM-score=0.58; hard target. |
>Putative transmembrane protein (Length=430) TGLSSPPLATQTVVPLRHCKIPELPVERSVLFELQLFFCHLVALFVHYINIYKTVWWYPPSHPPSHTSLNFHLIDFNVLT VTTIVLARRLIGAIVKEASQSGKVSLPRSVFLVITRFAVLTGTGWSLCRSIIHLFRTYSFLNLLFLCYPFGMYIPFLQLN CDFRKTGLFSQVANIGPRETGEVNSRSRDYLTVLKETWKQHTRQMYGMEAMPTHACCLSPDLIRNEVEYLKMDFNWRMKE VLVSSMLSAYYVAFVPVWFVKNTQYYDKRWSCELFLLVSISTSVILMQYLLPARYCDLLHKAAAHLGCWQKVDPALCSNV LQHQWTEECMWPQGVLVKHSKNVYKAVGHYNVAVPSDVSHFRFHFFFSKPLRILNILILLEGAVIFYQLYSLISSEKWHQ TISLALILFSNYYAFFKLLRDRLVLGKAYS |
| 4712 |
PF10457 (MENTAL) |
 Estimated TM-score=0.58; hard target. |
>Cholesterol-capturing domain (Length=167) SDVRRTFCLFVTFDLLFISLLWIIELNTNTGIRKNLEQEIVHYRFNTSFFDIFVLALFRFSGLLLGYAVLRLRHWWVIAV TTLVSSAFLIVKVILSELLSKGAFGYLLPIVSFVLAWLETWFLDFKVLPQEAEEERWYLAAQAAVARGPPLFSGARSEGQ FYSPPES |
| 4713 |
PF04797 (Herpes_ORF11) |
 Estimated TM-score=0.58; hard target. |
>Herpesvirus dUTPase protein (Length=365) FNYSFWNCSVHEHFISITNNREIHILPDDLLLPRCPSIRAILFKQIPSFKFSGSRSGPPGTVTSLYVYGHSKNLIKIKPM VVSECEQELIFKITFALECTIPPGSMEIFILPITFLKLDGLYLLCLEDYTSRIMSTSCMQMGTYLASETPQVFLKGGPVL NKHEPMPYLMAQKTKPFDKKMARVHTVQNEVCEVNSIYRGENHVKVAIQKDSEDINFLDTVVVGLTMTNRALVAFEYNPY FSCPWDWKRQSIPIIYDGPCIRIPAGRLAPVKYNNTYSSIYPNATAIITNSDSCADFHISDCEWKPGKTACIVVTNTSTI SVTISSGTHLGEAVFILAPKFFCRKIMSKTHVQKLSSAICLPGNV |
| 4714 |
PF05437 (AzlD) |
 Estimated TM-score=0.58; hard target. |
>Branched-chain amino acid transport protein (AzlD) (Length=99) VFLVIVGTAIVTFIPRVLPLMVLSRFQLPEWATRWLSYVPISVMAALVAQEIFKHDEKISFSTNNVELLAAIPTFFIAIK TRSLLVTVLVGIIVVMVLR |
| 4715 |
PF05977 (MFS_3) |
 Estimated TM-score=0.58; easy target. |
>Transmembrane secretion effector (Length=521) SPLAPFRHDIFRTIWIASLASNFGGLIQAVGAAWLMTSISQSVNMVALVQASTSLPIMLFSLVSGALADNFDRRRIMLVA QSFMLAVSALLTVCAYYGIVTPWLLLIFTFLLGCGTALNNPSWQASVGDMVPRDDLPAAVALNSMGFNLTRSVGPAIGGA IVAAAGAAAAFAANTLSYFAILFALARWKPVAPENRLPRETLGRAVSAGLRYVAMSPNIRKVLVRGFAFGLSASAILALL PLVARDLVGGGPLTYGVMLGAFGVGAIGGALLSARLREFLTSEAIVRYAFAGFAFSALVTAISPQAWLTCLVLAVSGACW VLALSLFNTTVQLSTPRWVVGRALSLYQTMTFGGIAGGSWLWGVTAEQYGAANALIGSCLLMLAGAAIGLRFALPEFKSL NLDPLNRFNEPLLELDLKPRSGPIVVMIDYEIADEDIPEFLKTMAERRRIRIRDGAGHWALMRDLENPTTWTETYHVPTW VEYVRHNQRRTQADAAVGDKLTAFHRGPNPPRVHRMIERQT |
| 4716 |
PF04724 (Glyco_transf_17) |
 Estimated TM-score=0.58; hard target. |
>Glycosyltransferase family 17 (Length=332) DLRALLVLLIGLPILIFVIYLHGQKVTYFLRPIWEKPPKPFKVLPHYYNENVSMGHLCKLHGWKVRETPRRVFDAVLFSN ELDILDIRWHELSPYVSEFVLLESNSTFTGLKKDLHFKENRQRFEFAESRLTYGMIGGRFVKGENPFVEESYQRVALDQL IKIAGITDDDLLIMSDVDEIPSGHTINLLRWCDDIPEVLHLQLRNYLYSFEFFLDDKSWRASIHRYRAGKTRYAHFRQTD DLLADSGWHCSFCFRYISDFVFKMQAYSHVDRIRFKYFLNPKRIQHVICQGADLFDMLPEEYTFQEIIAKLGPIPSTYSA VHLPAYLLETCS |
| 4717 |
PF14194 (Cys_rich_VLP) |
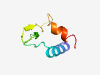 Estimated TM-score=0.58; hard target. |
>Cysteine-rich VLP (Length=56) TPAQRRRANALVRKTCCNYDNGNCLLLDDGEECVCPQTISYSVCCKWFRWAVLPQD |
| 4718 |
PF16501 (SCAPER_N) |
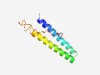 Estimated TM-score=0.58; hard target. |
>S phase cyclin A-associated protein in the endoplasmic reticulum (Length=90) GKKNDLRARYWSYLFDNLKRAVDEIYGTCEADESIVECQEVIMMLDQCRRDFSALIERINVQDAFEKADYKDRPTSLAWE VRKSSPGKTL |
| 4719 |
PF01524 (Gemini_V2) |
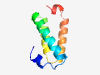 Estimated TM-score=0.58; hard target. |
>Geminivirus V2 protein (Length=78) MWDPLLNEFPETVHGFRCMLAIKYLQLVENTYSPDTLGYDLIRDLISVIRARNYVEATSRYNHFHARLEGTPPSELRQ |
| 4720 |
PF10935 (DUF2637) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2637) (Length=154) AALGFVAMAAVMGWGASFVGLHAYGQDQMVGFSYWSAWLIPATFDGAAFGATLITYRASIHGRSAFRGRLLMWIFTAISS WINWIHQMTPEARIVAAGLPVAAVAVFDVVLAELRADYEERHGKRRLRLRPGLLLLRWLVDRGGTSVAFRRQVT |
| 4721 |
PF09583 (Phageshock_PspG) |
 Estimated TM-score=0.58; hard target. |
>Phage shock protein G (Phageshock_PspG) (Length=63) MLEILFVIGFFLMLLLTGISLLGIIAALLVATVVMFVGGLFAIVIKLLPWLVVAVVAVWLYRA |
| 4722 |
PF14144 (DOG1) |
 Estimated TM-score=0.58; hard target. |
>Seed dormancy control (Length=75) INDIRSAINSQMGDNELHLLVDGVMVHYDELYKLKSIGAKADVFHILSGLWKTPAERCFMWLGGFRSSELLKIIR |
| 4723 |
PF14616 (DUF4451) |
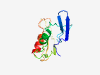 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4451) (Length=118) LDLYAPRFVKGIGRTKMGLCPICIEGVEGEKRWFYMKTSAYNYHMQYYHGISPTTSRPFSPPTAFRTRARENPAPKERRK LVEGRCHKCRKWVACEGVKDVEVKVPEIYWWKHAAACH |
| 4724 |
PF00803 (3A) |
 Estimated TM-score=0.58; very hard target. |
>3A/RNA2 movement protein family (Length=230) QQSSAATSDELQKILFSPEAIKKMATECDLGRHHWMRADNAISVRPLVPEVTHGRIASFFKSGYDAGELSSKGYMSVPQV LCAVTRTVSTDAEGSLRIYLADLGDKELSPIDGQCVTLHNHDLPALVSFQPTYDCPMETVGNRKRCFAVVIERHGYIGYT GTTASVCSNWQARFSSKNNNYTHIAAGKTLVLPFNRLAEQTKPSAVARLLKSQLNNIESSQYVLTNAKIN |
| 4725 |
PF03580 (Herpes_UL14) |
 Estimated TM-score=0.58; hard target. |
>Herpesvirus UL14-like protein (Length=145) HRRNRVKLVEAHNRAGLFKERTLDLIRGGASVQDPAFVYAFTAAKEACADLNNQLRSAARIASVEQKIRDIQSKVEEQTS IQQILNTNRRYIAPDFIRGLDKTEDDNTDNIDRLEDAVGPNIEHENHTWFGEDDEALLTQWMLTT |
| 4726 |
PF17491 (m_DGTX_Dc1a_b_c) |
 Estimated TM-score=0.58; trivial target. |
>Spider Toxins mu-diguetoxin-1 a, b and c (Length=55) AKDGDVEGPAGCKKYDVECDSGECCQKQYLWYKWRPLDCRCLKSGFFSSKCVCRD |
| 4727 |
PF12554 (MOZART1) |
 Estimated TM-score=0.58; easy target. |
>Mitotic-spindle organizing gamma-tubulin ring associated (Length=46) RESLDLAFGMSNILDTGLDRHTLSVLIALCDLGLNPEALAAVVKEL |
| 4728 |
PF15651 (Tox-SGS) |
 Estimated TM-score=0.58; very hard target. |
>Salivary glad secreted protein domain toxin (Length=95) CYAPDSEGNQIICPQRESIVNVFSKSATFDNPEVFGQDLFSKCLPLTWHDRPSVVCDGQETTFIYTPIQSLRVFDMVDGW LLLARIAPAALRNLK |
| 4729 |
PF06979 (TMEM70) |
 Estimated TM-score=0.58; very hard target. |
>Assembly, mitochondrial proton-transport ATP synth complex (Length=132) VKFFSYSTSMFSLCVMPYVLTKTGLGASSLAMKVAFCGFIGFFTFLTPALLHLITKGYVVRLYHNKDTDMYTAVTYSVLL MEKKTVFHQSDVKIPDVSRMFTSFYAKKRSMLVNPMLFELPHDYNHLMGYDR |
| 4730 |
PF08135 (EPV_E5) |
 Estimated TM-score=0.58; easy target. |
>Major transforming protein E5 family (Length=43) MTYGLLLFLGLTFGLQLMLLVFLLFFFLVWWDQFGCRCENMQL |
| 4731 |
PF11998 (DUF3493) |
 Estimated TM-score=0.58; hard target. |
>Low psii accumulation1 / Rep27 (Length=72) PEQYARLKAEITAPYRGLRQFIYIACGASGFIGALIFFSQLLAGRDVEKTLPSLAVQIGVVALMVFLWRWEQ |
| 4732 |
PF17455 (LtuB) |
 Estimated TM-score=0.58; very hard target. |
>Late transcription unit B (Length=79) NYFPASIEGETKKEHKHHYSTASKEKESLRKRAKEFDVLVHSLLDKHVPQNSDQVLIFTYQNGFVETDFHNFGRYSVKL |
| 4733 |
PF15140 (DUF4573) |
 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4573) (Length=175) NIDPLRTVSNIDPLRAVSNIDPLRAVSNIDPLCAVSNIDPLRALSDIDPLRTVSNIDPLRAVSNIDPLRAVSNIDPLRDV SNIDPLRAVSNIDPLRDVSNIDPLRAVSNIDPLRAVSNIDPLRAVSNIDLLRAVSNIDPLRAVSNIDLLRAVSNIDPLRA VSNIDPLRAVSNIDP |
| 4734 |
PF07151 (DUF1391) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF1391) (Length=48) QKIDLGNNESLVCGVFPNQDGTFTAMTYTKSKTFKTETGARRWLEKHT |
| 4735 |
PF17943 (HOCHOB) |
 Estimated TM-score=0.58; easy target. |
>Homeobox-cysteine loop-homeobox (Length=119) DVTYFGVNKHPSIHDMIDISEEVFVPYDQVFHRFSELRIINHERCEKNDTCERVSKYFISKIGYNGLLTENTVAMMHHEF KNFIHLGPMTDTGRIHLIVDKLDLAPDTVRKFYYKWFIK |
| 4736 |
PF09703 (Cas_Csa4) |
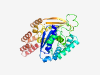 Estimated TM-score=0.58; very hard target. |
>CRISPR-associated protein (Cas_Csa4) (Length=342) KEIYTPGHDVFADTLIMHGVVRHLAASGIVEGYVERLGERYKISFEGEPRALADIESVELLLFEIERYVVDGEEPVGRLR KIFDANINISATRSWLAQLSEALVNAELSVFTLDHKAKFREGRTASRRLSTVYLTLSPIYGKYFIENRVAKENKAFGVCH TCFAYLNIGFLYGATTIVIITRDGRRVVQLTPAPRGRISIADLALLQRLTEGYLVRSQEMPTLAYPLYWLSTGETLYATE ESLDILVWVTEKRGNFQRVTDAMALDLSRIMEFIAEVKNKTRGWPKFVECLALEGPEILAQLTEVVLFGGDEYTVVRALS TSGKCAEDWRQVDILAEILFTW |
| 4737 |
PF05734 (DUF832) |
 Estimated TM-score=0.58; hard target. |
>Herpesvirus protein of unknown function (DUF832) (Length=225) VVPVVGLADDDVEQWRRALDSFLVHGKTADTIRKLSRLFKGDGHYGLLASLVIMQSAFGHSSKFRHKLSVMEVIQFSDKT AVTLYRKLKDHKHETGIEQVFWDCKQRLALLLENECGCVDCQRLASVLQRDRGVMRPPNLNPHQKHSQAATFLTWIYNQA VLNLNVAVLEDQIEVLFIKPDEFFDFAPDMRDELSLLTSCLHFCWLHFILKHYVHLELDVIQEHI |
| 4738 |
PF04109 (APG9) |
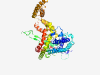 Estimated TM-score=0.58; easy target. |
>Autophagy protein Apg9 (Length=501) WNWVNVYNLDAFLQEAYYYYEGKGFYSIALSRGLNLLTVGFVIGFSTFLLGCVDYSRIRPDKVTRLSEVVVKRCVSRFSG FTLLFFVLFTAFFAWQLVSYVLSMMRLVDMYRFYTYLLHIPDEDIQTISWPEVVRRIAAIRESNPLTALSSNAAAHQSTT TAKLDAHDIANRIMRQENYLIALFNKELLDLRVPLPEALKRIPMFRVDEDGKGRTLTRALEWNLRFCLMEYLFDESGRVR KVFLKSKNRAILIEGLRRRFIFMGILNAIFAPFIVLYLLMYSFFRYFEEYHKNPSNIGGRAYTPYAQWKFREFNELPHLF KRRLNESYPTASMYIGQFPNEKVTLIVRFVAFIAGSFAAVLLLASVVDPDLFLHFEITPHRTVLFYITVFGSILAVARGM IPEENRVFDPELLMSEVVQYTHYMPDEWKGELHSKKVHAEFGELFQMKVVIFAQELLSVLLTPFVLWYSLPPCAPAIVDF FREFTVHVDGLGYVCSFAVFD |
| 4739 |
PF12283 (Protein_K) |
 Estimated TM-score=0.58; hard target. |
>Bacteriophage protein K (Length=56) MSRKIILIKQELLLLVYELNRSGLLAENEKIRPILAQLEKLLLCDLSPSTNDSVKN |
| 4740 |
PF03470 (zf-XS) |
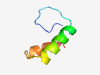 Estimated TM-score=0.58; hard target. |
>XS zinc finger domain (Length=42) CPYCQEKSGRHLFYEDLLQHASGIGKSKIRTARDRANHLALA |
| 4741 |
PF05868 (Rotavirus_VP7) |
 Estimated TM-score=0.58; trivial target. |
>Rotavirus major outer capsid protein VP7 (Length=249) MASLLLLVLAAAVTAQLNIVPSTHPEVCVLYADDHQADANKFNGNFTQIFHSYNSITLSFMSYSSSSYDVIDIISKYDLS SCNILAIDVFNASMDFNVFLQSTNNCSKYNANKVHHVKLPRGEEWFSYSKNLKFCPLSDSLIGMYCDTQLSDTYFEISTG GTYEVTDIPEFTQMGYTFHSSEEFYLCHRISSEAWLNYHLFYRDYDVSGVISKQVNWGNVWSGFKTFAQVLYKILDLFFN SKRNVEPRA |
| 4742 |
PF09441 (Abp2) |
 Estimated TM-score=0.58; hard target. |
>ARS binding protein 2 (Length=170) LPDRNVTAATIEEAYVQFVLHCNPAVPAETDTAALREAFRVPPKSGGKSFSTFTLFQLIRQLETKEIKTWAELALKLGVE PPDQDKGQSSQKIQQYAVRLKRWMHSMHVDAFFEYLMDHPHPYWTEIPDDPTPVCEAGRDGVMAEDDMALRALLPHIKPR RGRKRPEDDD |
| 4743 |
PF14009 (DUF4228) |
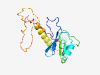 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4228) (Length=168) MGNYISCALATPLIKNSKAVRVVLPGGEVRQFRDSVKAAELMLESPSHFLANAQSLHIGRRFSALAADEELEFGNVYLMF PMKRVNSVVTAADLATFFMAANSAARRISSAKIRVLNENRNLQASEAMPRISDGNEGPRLSLEGVEGFPMHRLSVCRSRK PLLETIKE |
| 4744 |
PF15087 (DUF4551) |
 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF4551) (Length=586) RRNARLDSFLQRKLERRVYERIRAYEPCVVLSESINKVYMHVVLSDECVYLTEYPPRSLTAAVSFRRVRYIELVNDLPDF LSGKHQKRCQHIRIVYVTEKPAAKQRNWLKRDKKEGLPPAAPPSRRNSHCPEITSTLQACGVPILLSSSVNEAGDGANCC LVTMKCCKTDANSIKLLKHTRSASCPNPETLGLIRVPQPPPFHSPSVSPASSPSLSEKSLKSLESGQLQWRIGSVLSRLL RRGCVDSEEEREAELHLYAVSETSRLYLHLQSLWNSFSIRSTLLLDPLYRRRNCASLSAPAISWERTAHLFGQLSSELLQ DGISVESMYLLLQELRTAAHRSVTLRRLFWRSSEVCTFLLCTLEGCLHSRQSLGGLYTADQLLLSSLIVDTLAIMFRETE VEAARLSLLSAKKGALASRMLLALICDPQPQTHSGGSLMDSELQALLAEYLDTAASLLFELLLVGHVTSRCVSADNFLSV GWILRVLQPHPHLLSFIGYQVQQVVLILTGLQGSVPSPVQSVLLFQRLHLLLACLQYNSQLAQHLRSHFREEFRYSVMPS GSEVKLPPHYPISRPTLRLVEHIRTL |
| 4745 |
PF18549 (NitrOD1) |
 Estimated TM-score=0.58; hard target. |
>Nitrosopumilus output domain 1 (Length=68) SNAVLIRYKLKGDKQYTTCVVTRIQYENFKILPVVKECEIVQRYVGITGDQIEHANQTLGEAIRKEIK |
| 4746 |
PF03004 (Transposase_24) |
 Estimated TM-score=0.58; hard target. |
>Plant transposase (Ptta/En/Spm family) (Length=149) DSHIWKAMCEYWDTEEAIARSKIYSKARLSDRNGLGPHIHLSGPKSYQNIKHELEKELGRRVTVGEVFIKTHTKPDGTYV DKKAEQIVKNYEKKLQEKLSEMEAETSNVSDGESRPRELTIEDHTVIFLEVTETDTRGNYYGVGSLKDN |
| 4747 |
PF16334 (DUF4964) |
 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4964) (Length=81) VFCSCETQVEQHEKNELRAPAYPLVTIDPYTSAWSTTDNLYDSPVKHWTGKDFPLLGVAKVDGQTYRFMGTEELELRPLV K |
| 4748 |
PF10139 (Virul_Fac) |
 Estimated TM-score=0.58; hard target. |
>Putative bacterial virulence factor (Length=855) AIDWIDDVRPNVVRLNNEADSLILELRRLRNTAKRLGAVSDKPITAGFFGLSQAGKSFLISALAADEQGKLETLFDGQKL DFIKHINPPGGGKEATGLVTRFTRKETKGVAGYPLELRLFSEIEVAKILVNAYFNDFDKERVEYEITQSRINQLVKSLSG RVSANLVPGVNQDDVVDLQDYAQDSFGKSLSVLQGNYWANATALAPYLAIQERATLFSILWGEIQELTDIYIQFAHSLAR LGHPERVYAPLSAVVKETPEGGLSQADSIMNVDMLERLGTARDEQIDVRPAQGEEVGGAVTISLAELTALTAELVFPLMN PTRVPAVERVDLLDFPGYRGRLAITSLAEVKEGNPVSQLILRGKVAYLFERYTDSQEMNILIVCTPSTKQSDVNSVGPVL ERWIHKTQGESPEQRAERKPGLLWAITMFDMRISQDLTKEEELLKISWGSGGLLKQTILERFGNYDWFNNWANGNPFDNV FLVRKPGFKVPFLNVEGENEVSINQSETAQLALLKRTFCQDPDIRKHIAQPEEAWDAMLLLNDGGMKRISQYLETIALPE VKAHRLTEQLNERIHHFVENRFASWYQSDGEEEVAKKRQLANALAKELHPQNPRSLCMGELLRHLQLPEETIRSLYFSDL DDILGEEEVEQTYSPAEEFDLFSDPAPVTTTTPQRIEKVEESRFAQAVFKAWVSHLRNLTADHRLMQYLGVSVDNVENVV NELVTGATRLKLQEQLSKIALRNERSGSKRDQLAERQVFTMNTAIADFIAWLGYSDITLEQRPASRVLAGQMIFEQRPVE QHNGLPKLNDHTSNYDDNYRLDWLVAFGHFAVNNAGHSAGREINAAQNAQLGNVL |
| 4749 |
PF18917 (DUF5668) |
 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF5668) (Length=41) LVFGVILVLIGLVILLDKLGVVYLSWKKILSILLVIAGAYL |
| 4750 |
PF14162 (YozD) |
 Estimated TM-score=0.58; hard target. |
>YozD-like protein (Length=55) MKEIEVVIDTEEIAEFFYRELIRRGFVPTQTELEELADITFDYLLEKCIIDEELD |
| 4751 |
PF13038 (DUF3899) |
 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF3899) (Length=89) INILFYTSFIIFIVAFLILLIQEGIFDATSFGFRRLKYQLSSTKRKRMMKNDPFFNPQKVKKESYIISPWVVPTLIVNLT YIVVSIGVS |
| 4752 |
PF06683 (DUF1184) |
 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF1184) (Length=197) REMLALSPHSPYILGFDFAKQELKEEVVRLGVELSLYVAESMYLLSDDIRSMLRFCLKLWRDAKGDVLIPDSPVVERLLL AIHYVYSKDIKPKHGGVYQNDGKSVQWELIRTTWENFDAGIRDLDRLVLILRGEGTCFDGREFTSRIEEALRKVDDKLRC AKAVSEAKGFAREAMETDIGDLWKSLFDKEAKEVMKH |
| 4753 |
PF19152 (DUF5834) |
 Estimated TM-score=0.58; hard target. |
>Family of unknown function (DUF5834) (Length=159) MRIDGTSQNIKDEFQRIIQESTPLMLSNNYVTDFEVLHCEPVKYDDYHILISIFFAGKPVYMFGIINLSKNQGSLKIYNY YSFENFLFDMTDMVIKNFRNVSFKYQRKLIFNMGKELYDYLVDFNGKIRPISEEMSEDIYNNLIDIDLIEEYICYFKNL |
| 4754 |
PF09716 (ETRAMP) |
 Estimated TM-score=0.58; hard target. |
>Malarial early transcribed membrane protein (ETRAMP) (Length=85) MKLAKLFYFVAFLITIKVFVPGVNNDGFVQAKSAGADSKSLKKLDNDMLRKQRNQKIMIISTIATSLALLLGGALGGYGY YKQKK |
| 4755 |
PF15210 (SFTA2) |
 Estimated TM-score=0.58; hard target. |
>Surfactant-associated protein 2 (Length=59) AGPGMTLQLKLKDSFLANVSYDSSFLEFLEKLCLLFHLPSGTNVTLHRAGCPHHVICRV |
| 4756 |
PF03239 (FTR1) |
 Estimated TM-score=0.58; very hard target. |
>Iron permease FTR1 family (Length=313) VAVFFVVFRECLEAVIVISVLLSFLKQAIGEHDRALYRKLRIQVWIGVLLGFIICLAIGAGFIGAYYSLQKDIFGSAEDL WEGIFCMIATVMISMMGIPMLRMNKMQSKWRVKIARSLVEIPHRKRDYFKIGFLSRRYAMFILPFITVLREGLEAVVFVA GAGITTQGSHASAYPLPVVVGLICGGLVGYLLYYGASKSSLQIFLILSTSILYLISAGLFSRGAWYFENYRFNLASGGDA SEGGDGNGSYNIRKAVYHVNCCNPELDNGWDIFNALLGWQNTGYLSSMLCYNIYWLVLIIVLSLMIFEERRGH |
| 4757 |
PF07937 (DUF1686) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF1686) (Length=179) VGVSWNTALYSVAVAGIIAYQLWVLSRSYRTEGVSGMLRQDWSAIGCMVPMMVGLPHAGMIRSGVYSLTMAASGAVMLYV QDAMRSGMSLKQVCAIGGGNLVLAVACLAGSKKLFMPGACGLSQRICVGLVVFVALLVVVSYAEKGRSGGKKYSSTVGSV VFVMTLAAVTLGSWVCGRD |
| 4758 |
PF17103 (Stealth_CR4) |
 Estimated TM-score=0.58; hard target. |
>Stealth protein CR4, conserved region 4 (Length=57) DDIRKNPRKFVCLNDNIDHTHRDAQTVKAVLRDFYESMFPVPSQFELPREYRNRFLH |
| 4759 |
PF10211 (Ax_dynein_light) |
 Estimated TM-score=0.58; hard target. |
>Axonemal dynein light chain (Length=186) EDILNSILPPKEWTEDGQLWVQYVSSTPATRLDVVNLQEQLDLRLQQRQARETGICPVREELYAQCFDELIRQITINCAE RGLLLLRVRDEARMTIAAYQTLYESSIAFGMRKALMAEQKKMEAEQKIRQLETENRELVSQIDELQTRCEAVQRREEERK TNDEKKHQEEVEALRKSNDQLKASLE |
| 4760 |
PF14476 (Chloroplast_duf) |
 Estimated TM-score=0.58; very hard target. |
>Petal formation-expressed (Length=318) KLYAIADGVADRVEMHANIGEQRNNWNNLLLNSINAITLTAATMAGLSMIPTGTNFVLVMKITSTLLYSAATALLLVVNK IQPSQLAEEQRNASRLFRQLHEQIRTTLTLGTPTLKDVDEAMEKMLALDKAYPLPLLPGMLDKFPATVEPTRWWPAERKQ RQPQQLLSKQKGMNGWNQDLEEEMRGVLGVLKRKDTADYMRLSKLVLKINRILATSGPLLTGLAALGTALAGSSSYGSWA GLVGVAGGALATIVNTFEHGGQVGMVFEMYRNCAGFYRFLEERIESNLIERDFHKRENGEMFEMKMAIELGRSIQELR |
| 4761 |
PF14936 (p53-inducible11) |
 Estimated TM-score=0.58; hard target. |
>Tumour protein p53-inducible protein 11 (Length=179) MKKHSQTDLVSRLKTRKILGVGGEDDDGEVHRSKISQVLGNEIKFAVREPLGLRVWQLVSAVMFSGVAIMALAFPDQLYD TVFDEESASSKTPIRLYGGALLSIALIMWNALYTAEKVIIRWTLLTEACYFTVQFLVTTVSLVESSRIATGAVLLLVSRA LFILISIYYYYQVGRRPKK |
| 4762 |
PF17271 (Usher_TcfC) |
 Estimated TM-score=0.58; trivial target. |
>TcfC Usher-like barrel domain (Length=433) DGHMQWFIQAGETADNKVVSSTDDEDSNSTSRKPVVQAGARLPVFAELTATAGVANTDNENYGEAGLQWTHGFTGKVIDG VLDMQANVFSGTDGSKGNVEQISYNDGFSMSIYRNAAYGKSCTSADAGQYDYADIGCYESVSGTLSVPVKGWSASLGYTY NKNTSLSPDYSSYDPSKPFEDNMINNTESQSTSKTLQLSLNKSFNWKQMTITTRFGGYRRQSSSASDNDKGIYMGFSLSR STPKNSQLRSSNSSFSTDYQGSQNGDAQMTYNASQNWDWGSNNDRELGIDVGGTDTDNANASVHGRLNGQYGEGELTISD SYDNEQKTHQGAVTGSYSSSVAVSKSGFFWGPGGTGSPGAAVAVKVKGKDEDADAADDALVDVSVQGGGATKMKQNSKAL FPVTGFEEGKVSVDESRDVSNGSQASITQGAGS |
| 4763 |
PF10777 (YlaC) |
 Estimated TM-score=0.58; very hard target. |
>Inner membrane protein YlaC (Length=153) MNTIKQILIQDLERINIEEQRDGKPRFNRQFISNHPYLCLAMFVSYLFLAALIWYTPYFSLLSLLAFTVLFIIMSAVLLF EIKPVYRFDDIGVLDLRVCYNGEWFVSEKISDCAIKELCSHPHVSLTIKDEIKRIIQTKGEIHFYDVYIMAFP |
| 4764 |
PF14445 (Prok-RING_2) |
 Estimated TM-score=0.58; easy target. |
>Prokaryotic RING finger family 2 (Length=56) DPHSFAQFTCDLCNTAHPIAELRQCVLCGRWACNACWKDEYYTCRSCAGLIKIHQL |
| 4765 |
PF17140 (DUF5113) |
 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF5113) (Length=162) YNDLYQIAGAYVSIGKYMNEHGRYTEALDTLAKALDCVNQHHMLYYHHAADTLDKLHVFVEGDTTYTGVPWIMQEDVRTV PEWISRIREQLSVSYAGLGMKYASDYNRNIYLDILNYTRQDKELESRYLSLEADSRQMTLVLSLVIVGLVLVVILWWFFN KR |
| 4766 |
PF05398 (PufQ) |
 Estimated TM-score=0.58; hard target. |
>PufQ cytochrome subunit (Length=74) MTDMTSNTPVEARSHKPPKTEFMVYFAIIFVATLPLATLTWALAAIKSGSFTEKGPVARAWSQARIITPMIFTA |
| 4767 |
PF12398 (DUF3660) |
 Estimated TM-score=0.58; very hard target. |
>Receptor serine/threonine kinase (Length=42) ETPFVDQVRSQDLLMNEVVISSRRHISRENKTEDLELPLMEF |
| 4768 |
PF17540 (DUF5457) |
 Estimated TM-score=0.58; easy target. |
>Family of unknown function (DUF5457) (Length=90) EEIPLHEVVLELMFEANKEKPAELSANEIFWKISDSSLIERNISEVLNWLVSKRRAEEYLEKYTLSRNEFLDRRNIDRGI SVSKEEVSEA |
| 4769 |
PF12005 (DUF3499) |
 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function (DUF3499) (Length=123) MRRCCRPGCKNPAVATLTYVYSDSTAVVGPLATVAEPHSWDLCETHASRITAPKGWELVRHEGGFSSSTPDDDDLTALAE AVREAGMRRRTPEPEQRRYPDQPPAPPRPSRTGRRGHLRVLPD |
| 4770 |
PF07096 (DUF1358) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF1358) (Length=111) SHRTEMSDEERKFRYRASAFLAGVAGIAAVAGFSKTIMMAKKSDPQFFERGVQGSIAMQETGASLAMRALGWGTLYAFLG TGTICYGIWKLTGASNMQEFRESIGNIFPRI |
| 4771 |
PF10036 (RLL) |
 Estimated TM-score=0.58; hard target. |
>RNA transcription, translation and transport factor protein (Length=249) FERYLTALQFPRVGQLNIDDPKDFRSFVMWLEDQKIRFLPIEERQPLRKINAPDEWNRAYEKYKKDIKVPEHLKTKHEEV AWLLLYAIRLEYTDNAEQYRPLTGARKLQQESKKTNAPEVQSSNPFDSFDFTSADFEEGSWKLAERLGISYHPDHLVSLQ AAAKVIKGAYNKEALKEPPPIAGQASFPIAQGSGMGLEKDQDLEQCARILRLLQIQNLRTLQTTINETIVSVQNVTADPR TDTKLGKVG |
| 4772 |
PF06074 (DUF935) |
 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF935) (Length=511) ILDIHGNPFRFEDRLQTENESRLAALQHHYSEHPASGLTPMKAARILRNAEQGDLVAQAELAEDMEEKDAHLQSELGKRR GAMLNVEWQIVPPANATPEEQRDAEMIEEILRDAVWFDDCIFDATDAILKGFSCQEIEWETHGNMTLIKGVHWRDPAWFM TPQFERNSLRLRDGSAHGVELQPFGWVKHIAKAKTGYLSRIGLVRTLVWPFIFKNYSLRDFAEFLEIYGLPLRLGKYPEG AGEKEKQTLLRAVMSIGHNAGGIIPRGMEIEFQKAADGSDATFMAMIDWAEKSMSKAILGGTLTSQADGATSTNALGNVH NDVRLEVRNADLKRLAATLTRDIVYPLYALNCKSFNDARRIPRLEFDIAESEDLNAFAEGLNKLVDIGFKIPKQWAHDKL QVPIAGENEEVLARNQPNPTAYLNSQSHQKMAVLSVQRDPDDLLDELEPSVEAYQAIIDPMLKPVVNALAEGGYEFAQEK LASLYADMDDSELEKILTRAIFVSDLLGRAN |
| 4773 |
PF14118 (YfzA) |
 Estimated TM-score=0.58; hard target. |
>YfzA-like protein (Length=90) KERPSRIRNWLKTIGAFFVMQLIYIILDMNSWIPNFKEGGVGDRLVNSEFFTEWFAPYKTKQFNVLTAVMAILLFLNVVT SAIKDAFSRK |
| 4774 |
PF15641 (Tox-MPTase5) |
 Estimated TM-score=0.58; easy target. |
>Metallopeptidase toxin 5 (Length=110) ELSKATGKTTASRNRAIDAIISEDFPDLNLTHRPVYSYGVNSGMAKPNVGTHIGFKRFASRALLRNTVVHEELHHRWFAR GLRNHHPRDGSGTSERFYGIIDRYMSIRGW |
| 4775 |
PF17109 (Goodbye) |
 Estimated TM-score=0.58; hard target. |
>fungal STAND N-terminal Goodbye domain (Length=121) WDLALKRYTAAVGRQLDDPSLPHPSSVEELLNRLDSQNGHFSQFRENKRSLFNILTCICNPIERFGSMAASIASNAFPAS SVCFSAITYLIDAAKDVSASYNAVIELFFTLKDFLVRLAIY |
| 4776 |
PF05959 (DUF884) |
 Estimated TM-score=0.58; very hard target. |
>Nucleopolyhedrovirus protein of unknown function (DUF884) (Length=188) MNVNLYCPNGEHDNNISFIMPSKTNAVIVYLFKLDIEHPPTYNVNAKTRLVSGYENSRPININVRSITPSLNGTRGAYVI SCIRAPRLYRDLFAYNKYTAPLGFVVTRTHAELQVWHILSVRKTFEAKSTRSVTGMLAHTDSGPDKFYAKDLIIMSGNVS VHFINNLQKCRAHHKDIDIFKHLCPELQ |
| 4777 |
PF11046 (HycA_repressor) |
 Estimated TM-score=0.58; hard target. |
>Transcriptional repressor of hyc and hyp operons (Length=145) MTIWEISEKADYIAQRHRRLQDQWHIYCNSLVQGITLSKARLHHAMSCAPDKELCFVLFEHFRIYITLADGFNSHTIEYY VETKDGEDKQRIAQAQLSIDGMIDGKVNIRDREQVLEHYLEKIAGVYDSLYTAIENNVPVNLSQL |
| 4778 |
PF16029 (DUF4787) |
 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4787) (Length=62) DMTFREFESACEQSSACSHLSGLIKTRCIRECVSPSCYRELYQADPLEEGEIDVRLNSFKGC |
| 4779 |
PF12955 (DUF3844) |
 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF3844) (Length=113) CFLSLDACKEATNDCSGHGTCVNKFGTGNATSSDARACFACKCMATVVRRGDEPGTSGKKTVHWGGNMCQKKDVSVPFWL ITGFTITIVGAVTFAIGLLFSVGQEQLPGVIGA |
| 4780 |
PF04759 (DUF617) |
 Estimated TM-score=0.58; very hard target. |
>Protein of unknown function, DUF617 (Length=170) VIGTLFGHRKGHVHFAFQKNPTSQPTFMIELQTPISGLVKEMASGLVRIALECDKDDDKNKKGGNSKRLLEEPLWRTYCN GKKCGFGIKRECGEKEWKVLKAVEPISMGAGVLPAEKPSDDEKDGVVNDGEIMYMRAKFERVVGSRDSEAFYMMNPDSNG APELSIYLLR |
| 4781 |
PF18303 (Saf_2TM) |
 Estimated TM-score=0.58; very hard target. |
>SAVED-fused 2TM effector domain (Length=153) WNTLDFVVRSLVSWFVRARHAESWIFRGAVAVLVAIHGANWVFKYSGTIGGESGSIEFGTAGAVPDSILWIITVVCIALM LGSAMWAWIRYANEQKRLARKKVFVIEGRGLRDDDGSSLKSAVPESITGTRIDYMLDLRQRKDGVIVEPEDLL |
| 4782 |
PF04753 (Corona_NS12-7) |
 Estimated TM-score=0.58; hard target. |
>Coronavirus non-structural protein NS12.7 family (Length=105) IWHVSDAWLRRTRDFGVIRLEDFCFQFNYSQPRVGYCRVPLKAWCSNQGKFAAQFTLKSCEKPGHEKFITSFTAYGRTVQ QAVSKLVEEAVDFILFRATQLERNV |
| 4783 |
PF03048 (Herpes_UL92) |
 Estimated TM-score=0.58; very hard target. |
>UL92 family (Length=190) CQFNKLNTVVCQYHKVFCLYQCCQCKKYHICDGSKTCILINTGESLVCTLTGNCIFNNFQENTFLKQDNVKESKQQFDFT LFANILESLKQDLENYFIKSYGLEEVSKAILDNNSLKPEICDLIRCTFQHCCHIFGEIQYGYDIICSMYIHIIISIYSTK TVYGNLLFKCTKNKKYDYILKRMRETWMST |
| 4784 |
PF14855 (PapJ) |
 Estimated TM-score=0.58; hard target. |
>Pilus-assembly fibrillin subunit, chaperone (Length=187) MNRATIGFYLAVVLGSCSMNGVSQADELLTRDDFFVADESRHQWVTEHNGRTGALNVKGALVSSPCILDTPEVNFPLQKD KGRYVLNLKVSRCGDGHSDIPERQSTEHGNIVVKQSAVLKTGKDQILLSAWRNGGLSRKNLQHGDNQITYYINEAQYQKI AKTQMPVTAHKTSDSNRGVMHLNIMYE |
| 4785 |
PF09557 (DUF2382) |
 Estimated TM-score=0.58; very hard target. |
>Domain of unknown function (DUF2382) (Length=111) PLCEEEVTAEKYTRPLGEVRIHKRIVTTQKQITVPVMHEEVRVERVPAIDRNAMEYSDAMFVESTQVIPLHDEEVEIHKF PVIREGVRLRKASFQEDRTATADLRKEELDV |
| 4786 |
PF19004 (DUF5733) |
 Estimated TM-score=0.58; easy target. |
>Family of unknown function (DUF5733) (Length=115) ECQLRKVIKDKKKENYTVDEAKNIYHSVQESKSGFPIYMASIYPEKVTFKPTEKGKKMRPVEYTTIGQIIQIPEDLQSFL LHVPKSKKAKSFTALFAWLDPQMSKRFEKIVHQAN |
| 4787 |
PF14006 (YqzL) |
 Estimated TM-score=0.57; hard target. |
>YqzL-like protein (Length=44) DFTWKVFSQTGNIDTYLLFKELEKDTQEMPGNQEEELAEIDFPI |
| 4788 |
PF12069 (DUF3549) |
 Estimated TM-score=0.57; hard target. |
>Protein of unknown function (DUF3549) (Length=338) TIHTLSELLKNSGCQYQVFDLGRRIQAIDTAQFEQVELGHQPYPYPLKRTAHLAIVYWNESNQPWIWFLKFTLDERGLLA QADVGQFIKYVIEAMGTRLNQEMSDEQQQKLANNPYTFKPADDKMAVFHSQIRAQLGLPCSQYYEHAQHYFTGDLGWENW QTVGLQGITDICARLGQEQNGVLLRKALNHLPPQPLYALLGALEHIALPEKLAARLQEHAEAQIAAHEPDLFLLSALVRA LSGAPAAIQNQVLDAVLNSPRLSHPEVLIGIAGRSWSMLADSGRAQRFLLRLAQSGNQSLFNQLFADLVMLPELRLVLLP LLHTSPSPELAQALMNLQ |
| 4789 |
PF04956 (TrbC) |
 Estimated TM-score=0.57; hard target. |
>TrbC/VIRB2 pilin (Length=96) SSVTTARSIASVALAAAIVSVAMVQPAFAQATGGIESVLQNIVDMLTGNVAKLLATIAVIIVGIAWMFGYLDLRKTAYVV LGIGIIFGASEIVSTI |
| 4790 |
PF09742 (Dymeclin) |
 Estimated TM-score=0.57; hard target. |
>Dyggve-Melchior-Clausen syndrome protein (Length=603) MGNADTKLNFRKAVVQLTSKSQPIDSSDDSFWDQFWSESVTNVQDVFALVPGAEIRALREESPNNLATLVYKAVEKLVKT VDSSCRTQREQQTALNCARLLTRVLPYMLEEPEWHGFFWSSLPAASENESVPLAQSLINAVCDLLFCPDFTVASTKRSGP VSIDLLLNRFLIYALISYHMGKCGQPQLTVSIPRRSYLTSSENRHALPLFTSLLNTVCSYDPVGLGLPYNHLLFADTLEP LVEVALQILIVTLDHDTSNTVNEETDESLPDNLFINYLSRIHRDEDFQFALRGVTRLLNNPLAQTYLPNSAKKVNLHQEL LVLFWKMCDYNKKFMYYVLKSSDVLEVLVPILYHLNDSRADQSRVGLMHIGVFILLLLSGERNFGVRLNKPYTATIPMDI PVFTGTHADLLVVVFHKIITTGHQRLQPLFDCLLTILVNVSPYLKTLSMVASTKLLHLLEAFSTPWFLFSQPHHHHLVFF LLEMFNNMIQYQFDGNSNLVYTIIRKRSVFHSLANLPHEHAAIARSLSAARARPPPPALFNYEQPKQPAIVLCRGLTDES EILRFLQHGTLVGLLPVPHPILIRKYQANAGTAAWFRTYMWGV |
| 4791 |
PF18174 (HU-CCDC81_bac_1) |
 Estimated TM-score=0.57; easy target. |
>CCDC81-like prokaryotic HU domain 1 (Length=59) MIELERHIEILLLSNDCVIVPGFGGFMAHHVDARYDEQENLFLPPQRTIGFNPQLTLND |
| 4792 |
PF13123 (DUF3978) |
 Estimated TM-score=0.57; very hard target. |
>Protein of unknown function (DUF3978) (Length=144) MEAYKMHDFINTNVESHQNETVFNLHICETNEFDVSLTKSTTLSFIVSKKNIKIVTKKWINSNQESMIGKSYIIPTKAFH YFLPIISETEDELNIQVQSFGLHGELLLNERLLIDKNNKHDAKITNFFETLDENVNKVLRGLQI |
| 4793 |
PF10532 (Plant_all_beta) |
 Estimated TM-score=0.57; very hard target. |
>Plant specific N-all beta domain (Length=119) TKLGLDANKVKLHMRYNPRLFGVEEEMNVCDDEDVFIYATSAKNNRRSVLVVEEISKPPEQEQLPEQLSRVGKSSVGKNY TELNSEEDEMRVDDGALIVLLEEEQGTQHQLEAIVEDHG |
| 4794 |
PF14293 (YWFCY) |
 Estimated TM-score=0.57; hard target. |
>YWFCY protein (Length=60) QQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVDIGVVDRILMNFNRTAGLF |
| 4795 |
PF13891 (zf-C3Hc3H) |
 Estimated TM-score=0.57; hard target. |
>Potential DNA-binding domain (Length=63) CNYSHRICMQNRLEGYEYCSKHLLEDKNAPYKQCNYMSNKSGKRCPNAAPKSDRKDGYCADHA |
| 4796 |
PF06210 (DUF1003) |
 Estimated TM-score=0.57; hard target. |
>Protein of unknown function (DUF1003) (Length=103) MGTANFLLYMTVFVIVWIAINVVGLFGLHWDPYPFILLNLFFSTQASYAAPLILLAQNRQDDRDRVQIEQDRSRNERNLA DTEYLTREVAALRITLREVATRD |
| 4797 |
PF19096 (DUF5784) |
 Estimated TM-score=0.57; hard target. |
>Family of unknown function (DUF5784) (Length=327) ARPLRFRHAPGRWTVGRVRNDVFQPLDGNLGAEMTAPWFRSPDGYDARRFEMDNGDIALFAWNDDEAYWLGNTETPSSLW RTEKYGFSEVPYPIARWAERELLAQLHEESPWLEPYPHVSWFFLPVFLSKDGRHTTREFFHDHAAGFPDATRDEALDFYE RFLHTGALDEYRELMAGKLGTSERIDLVRMAASMGEFNVAKVLHDAGYDLTPEIEVTTGHSLDYRADRNGDGGTLVEVTR PLPPNKRSANTPVAAVRDTAETKTNGQLEHHAGGVTLFVDCSSFPDDDWFAVRGEQPEVRHRPAVVFRARPDGHIEGYSK GSVPLDL |
| 4798 |
PF06341 (DUF1056) |
 Estimated TM-score=0.57; easy target. |
>Protein of unknown function (DUF1056) (Length=63) MIFRRIIGYLWQLSDVLLFISAMIVLDYTAFRINATLGWFVISLILFVLGWLVEVISERKRGD |
| 4799 |
PF12450 (vWF_A) |
 Estimated TM-score=0.57; very hard target. |
>von Willebrand factor (Length=92) NTENYDFIKENSFLDAQKNPLSTFSIDVDNASYSNVRRFLNQGQRPPADAVRIEEMVNYFNYDYPQPAGDAPFSVTTELS ACPWNKQNQLLH |
| 4800 |
PF18934 (DUF5682) |
 Estimated TM-score=0.57; hard target. |
>Family of unknown function (DUF5682) (Length=788) VRAALDAARPRVVLIEGPPEADALIPLAADEDMRPPVALLGHAVDEPGRSAFWPLAEFSPEWVAIRWALEHEVPARFIDL PATHTLAWGKDDAEGELTGEEQPPGPTAEPPTAETPAAGTPSSESPSASDPSGPDPSDPGTDIRVDPLAVLAEAAGYDDP ERWWEDVVEHRGVGERDAFAPFTALEEAMGALRETYGVGGHERDLVREAYMRLQVRAAQREFEDDVAVVCGAWHVPALRQ KASVAADRALLKGLPKVKADLTWVPWTHRRLSRASGYGAGIDSPGWYGHLFSAPDRPVERWMTKVAGLFREEDRIVSPAH VIEAVRLAETLAVMRGRPLPGLGETTDAVRAVMCEGSDIPLALVHDRLVVGDVLGEVPPGAPAVPLQRDLDRSQRRLRIK PEALERELELDLRKETDAGRSRLLHRLRLLGVEWGEPTASRGSTGTFRETWRLRWEPELAVRVAEAGVWGTTVLAAATAK AEADAVGAQSLADVTALAEHCLLAELPDALPVVMKVLADRAALDTDVGHLAQALPALVRSLRYGDVRGTDTGALTEVAAG LAERVFVGLPPACAALDADAAEEMRRHVDAVHGAVALLGDASGTGHGDTSEPGQSDGHGPGPGHGGLRDRWRAVLRTLSA RDTVPGVIRGRAVRLLLDEGELGPDEAARLMGLVLSPGTPPADAAAWIEGFVGGGGGLLLVHDERLLGLVDAWLTGVPAE AFTDVLPLLRRTFSSYEPGVRRTLGELVRRGPGKQVGTAATAAGIPGFGAELDAGRADAVVPVLRLLL |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218