

Go to page (83 pages in total): [First page] << < 53 54 55 56 57 58 59 60 61 62 63 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 5701 |
PF11445 (DUF2894) |
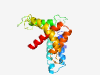 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF2894) (Length=181) LDAWRERGADRLDPVRFHFIEALDRRAAGHSGETRRILDDRLSRLLETYAGDLERAASKAADAGSAASPCKPARGALAGL IDYIASHASADGDGAAAGNRVTRPPSWPELEVLDYFRETWSKVSTERQLRQSLEQVPGNAGPLNSSSLVHRSLSLMRELS PGYLQQFLSYVDTLSWMEQMN |
| 5702 |
PF14167 (YfkD) |
 Estimated TM-score=0.51; very hard target. |
>YfkD-like protein (Length=232) PSSVLNITKENTYPNPVQDQPTLQPSELTQSLLDTSKVKIENPDLIRMLNESSVNSTPFAFGYRAIVYLGQWPLNYESTE TAPNWEYQKINTNYFDNRGGKAPYQIHYVQEAQKVVRGALTAKIPKAQDVQNMMLLKAMEKSKLPLAFETVIGAGTKKDQ VYNIPPKRLGYLYGYAPAVNEKGKVTYGEVYLMLKGSKRFLVIKNVTSQGIGAWIPVQDHISFGFMASERPR |
| 5703 |
PF17276 (DUF5341) |
 Estimated TM-score=0.51; very hard target. |
>Family of unknown function (DUF5341) (Length=158) VTDTVHMLQFDSPLGSHAAVHLNTGLDSMVNDITEDENPGSLESTLRYTDQKVLHKRYNYFDAQWISWAWDNANNDLLRI AANDRGLEEEEYDSSVYNYFRDSPAWKYCWTVTDNPHPGEGEDYNQLGAENAIHGELYFNTYGGVDGYCNDNKDGAQC |
| 5704 |
PF13051 (DUF3912) |
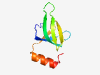 Estimated TM-score=0.51; easy target. |
>Protein of unknown function (DUF3912) (Length=71) MDFDITGQKAYVKDGPYRNRIGIVKQKEQQAEPNFIIVIDGQNIDVELKDIVLVGVDVGQFHEWCEQNGYL |
| 5705 |
PF04877 (Harpin) |
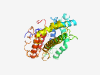 Estimated TM-score=0.51; very hard target. |
>HrpZ (Length=299) MNMISNMPGLVAGNFSNFSIGSQSNSERPQNMDQVVDKLADMLKGSDLSSSPLGQMVKDKLDDSGGLMGILKKGGMENAV KDALKDIISEKLGDNFGAAQSFGLGNGSGTGGAGGAGGGQQPDLLERVLSGLAKSSLDDLLGKQGEGTKFSNKDMPMLEE IAKFMDDPRSGIKPPDSGSWANELKEDNYLNKEETAAFRAALDQVGGALDQKASGQPSGMPGGLGMPGNAGQNMLGGEGG LGGSGTGSAAGNPLQQLNDLIQGLQGLASQLSLQNLASSAMDTANALVGAMLPNNSRTA |
| 5706 |
PF07753 (DUF1609) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF1609) (Length=226) EVEAGEEAEMPSVEVGGARRKTGKKSEGDQKRLKIHSRVLRWRKSPEKIKKEWDRGSEERWRGRSLEEIKEQKVFHDIVG VLELLKSPDADRFFMDTGKYTKGGSERQRMVAIGVLERGEKKMTGVVEVGMFKDSPSGCPVVYHLMFRVTGIEEMGNVVN PEFPEVSDVEKIDEDKEYQDAGKFVYPKGVKFETVKETDAFQIVWKNPSDTSEVLQSLTVQCRPPV |
| 5707 |
PF13726 (Na_H_antiport_2) |
 Estimated TM-score=0.51; easy target. |
>Na+-H+ antiporter family (Length=87) NPVVISVIVMSVLCLFKLNVLLSILLAAMTAGVAAGMPIKETMNTLIDGMGGNSQTALSYILLGTFAMAISKTGLATILA KKISQVV |
| 5708 |
PF01639 (v110) |
 Estimated TM-score=0.51; very hard target. |
>Viral family 110 (Length=102) LLSIPTLLYTYEIEPLERTSTPPEKELGYWCTYANHCRFCWDCQDGICRNKAFKNHSPILENDYIANCSIYRRNDFCIYY ITSIKPHKTYRTECPQHINHER |
| 5709 |
PF12734 (CYSTM) |
 Estimated TM-score=0.51; hard target. |
>Cysteine-rich TM module stress tolerance (Length=46) QPPTGYPTEKKKMKKCWPRSKPKGDRGFIEGCLFALCCCWICDICF |
| 5710 |
PF08384 (NPP) |
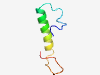 Estimated TM-score=0.51; hard target. |
>Pro-opiomelanocortin, N-terminal region (Length=44) QCWEHPSCQDLNSESSMMDCIQLCRSDLTAETPVVPGNAHLQPL |
| 5711 |
PF17330 (SWC7) |
 Estimated TM-score=0.51; hard target. |
>SWR1 chromatin-remodelling complex, sub-unit Swc7 (Length=97) DKSLDLAALLKEPIVDKEVLTQFQNHKLVKMYAPELCNVHLRLLKSLVADIFMTGTPGDETHDDTTVITLANYYYNQRIE ELTQDQLPRIRHEIAEL |
| 5712 |
PF12547 (ATXN-1_C) |
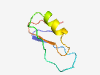 Estimated TM-score=0.51; very hard target. |
>Capicua transcriptional repressor modulator (Length=57) GICAGKPVTRTPSNSLSSTSLPFPPPPLSVESLKAAVTTLSPHTVIQTTHNATEPLS |
| 5713 |
PF16465 (DUF5046) |
 Estimated TM-score=0.51; easy target. |
>Domain of unknown function (DUF5046) (Length=267) LSNTVTGEELDGFDQVCGNGTASFRTEDGQYQLIDLASTEQSEVLATFDRSVLYHAPGAVVCWNAGNDYRYQFHDLTTGE MKPVYNVDADDKTLAVYATDGTLRVYDRTTGAILTDVNVDSVENQDSAQVWAVGEGYALVMLYPAGDHSKPTIRLYDAQG LVRQLAISASTPDGYYYFAPLTTTDGHAYFRYGYAGPNGKTLYDVLDENGNAVLKGLALCYSYYGGNGLNDLPAGAFMAR KGFYYGWMTPDGQWLYCRSIFASSTDE |
| 5714 |
PF15873 (DUF4730) |
 Estimated TM-score=0.51; easy target. |
>Domain of unknown function (DUF4730) (Length=55) MEAVHHMIVAAILSLLLFTPGVEAKMTAGDVIALILGLVLGILGICACLGYYART |
| 5715 |
PF10492 (Nrf1_activ_bdg) |
 Estimated TM-score=0.51; hard target. |
>Nrf1 activator activation site binding domain (Length=79) SGMITVPVSTQMYQTMVANIQQLPNGDGTVCITPMQMCDLVENGETMETITVGPGMHQMMIQGPPGSEPQVLQVLSLKD |
| 5716 |
PF04640 (PLATZ) |
 Estimated TM-score=0.51; very hard target. |
>PLATZ transcription factor (Length=79) RRSSYHDVVKVSELEDILDISDVQTYVINSSRVVYLTERPQLRSCGVSNTKLSSSQTYKCEICSRTLLDDFRFCSLGCN |
| 5717 |
PF12129 (Phtf-FEM1B_bdg) |
 Estimated TM-score=0.51; hard target. |
>Male germ-cell putative homeodomain transcription factor (Length=139) KIGAYDQQIWEKSVEQREIKGLRNKPKKTGHVKPDLIDVDLVRGSAFAKAKPESPWTSLTRKGLVRVVFFPFFFRWWIQV TSKAIFFWLLFLYLLQAAATVLFFTNSQTHNVPVTEVFGPICLMLMLGTVHCQIVSTRT |
| 5718 |
PF12155 (NADHdh-2_N) |
 Estimated TM-score=0.51; easy target. |
>NADH dehydrogenase subunit 2 N-terminal (Length=88) MELTLGLIILIVLTYGLKAPTLRLAMLLAGAVGAAGLLAEPHLLCWTQAIKMLVMLSGLAILCMLDHRTSHRSSSLLILL VILGNLLL |
| 5719 |
PF10266 (Strumpellin) |
 Estimated TM-score=0.50; easy target. |
>Hereditary spastic paraplegia protein strumpellin (Length=1088) ESKADQQKYAEVIMDFSYLKIAEAQDQKIEQDPLLQDLDEEIRENYLEILTRFYLAFESTHQYAKDLQQFIDELNRGYYI QQTLETVLQDEEGKQLMCEALYIFGVILLIVDFHIPGAVRERLLISYYRYSGSKSHSDSNIDDVCMLLRSTGYISPKANN QTNNKTSKDGISNYPESYFARFKFDENFIDMVIGRLRCDDIYNQLSIFPHPDHRSTALSTQAAMLYVCLYFSPKILHSQV AQMREIVDKFFSDNWVVSIYMGITVNLVDAWDQFKAAKAALSIVETTAIKGFCLLRKEKMEKVLKQSQEILRDGILTDFF VLQNISKIINLIRQCNVLLRWYFTHASKTIYEISRGPVTQKMEQIIKLVRTELGYNELDLFKLLLNCSQLELEVKEILRS LMQEKSNRWSDFKKEALDRVKELAEAFSGSRPLSKIESNPQLKLWFDDIAKEIENLNSDNVNISGRTLIQLIQALEEVQE FHNLQSNMQVKQYLMETREYLTRMTQIINVKEDILINIQLISDLSYAWRLIDRDFTAIMQDAIKRQPKSVIRLRATFLKL ASALEIPLMRINQAKSEDLVSVSNYYSTELANYMRKVLQIIPESMFNLMARIIQLQTDVLKELPTRLEKDKLKEYAQFED RYTVAKLTHSISVFTEGILMMKKTLVGVIELDPKQLLEDGIRKELVKHLANALNVGLIFTPSGSGGKQKNPPTLELETKL DQLAKIMDGYRRSFEYIQDYLNVHGLKILQQELTRVINYNVERECNAFLRNKVQDWQSKYQSTTIPIPTFPPLQGDTTKS NNFIGRLANEILLCTDPRNTIFVELKATWYDKKSPHKEVLDSRFFCKMREAIGPAGLVGLDRLYTYMFVADLKKNIEKLQ HNIENDQMWSSTLAQLTLELESKQFPGAYTTNPLKYYAAYHQRWLKVWPTMVDWILDLGHKQILRKQLAFELKSKSKVNA KNLESALETFNRSLLNELENKDMQQKFKDNTMLVELKDYLMYTGSYEPLEEIFVLCKNSHNIALFFSLFVLHNVTNRLQY SMQIQSLLPKVNKDALDGVPLIVGLLTILQQFHKDVKTMFITYLCQFV |
| 5720 |
PF16059 (DUF4801) |
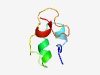 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF4801) (Length=50) QEKPPCNNNFCRLGCVCDSIKDTGDVSSEKEHCGKVDCMFGCVCTDEEKA |
| 5721 |
PF15378 (DUF4605) |
 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF4605) (Length=59) TLFGELNKCLRSIGFSQMYFGEKIVEPVVIIFFWVMLWFLGLQALGLVGVLCLVIIFIQ |
| 5722 |
PF16018 (Anillin_N) |
 Estimated TM-score=0.50; very hard target. |
>Anillin N-terminus (Length=88) VKTRMQKLAEQRRHWDNNSMTDDIPKSSLLPPMPAEEKAASPAKIPSAAASATPLGRRGRLANLAATICSWEDDVNYSSA KQNSVQEQ |
| 5723 |
PF08407 (Chitin_synth_1N) |
 Estimated TM-score=0.50; hard target. |
>Chitin synthase N-terminal (Length=72) KEVQLINGELVLECKIPTILYSFLPRRDEVEFTHMRYTAVTCDPDDFVDRGYKLRQTIGRTARETELFICVT |
| 5724 |
PF13889 (Chromosome_seg) |
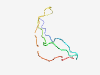 Estimated TM-score=0.50; very hard target. |
>Chromosome segregation during meiosis (Length=53) VLRYLIHLNICCPAKGRFYLYSTIRVVFANRVPDGKEKLRNEIQYPEPRYSPY |
| 5725 |
PF10870 (DUF2729) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2729) (Length=57) KLLTYCKLKLVKGFSKKFASLFCRCVVSHLSDEDDSDGDRYYQYNNNCNFIYINIVK |
| 5726 |
PF07038 (DUF1324) |
 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF1324) (Length=59) MKCTLVFQSRFCIFPLTFKSSASPRKFLTNVTGCCFATVTRIPLSNKVLTAVDRSLRCP |
| 5727 |
PF13029 (DUF3890) |
 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF3890) (Length=84) MNDSKLDTNQALSANLKTLKSENLESLQRVYEQILKLLMLDFKDLSIKEFFLYLNLLENILILKRIRIESLNDSTMFVLI YYFI |
| 5728 |
PF10284 (Luciferase_3H) |
 Estimated TM-score=0.50; trivial target. |
>Luciferase helical bundle domain (Length=67) CEKTGLELSGTAKGGALSAAHVEHLDSEAFREGLHQPKFHTDGLHMPHTSGEKTYETGFHYLLEVHD |
| 5729 |
PF12288 (CsoS2_M) |
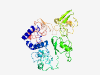 Estimated TM-score=0.50; very hard target. |
>Carboxysome shell peptide mid-region (Length=425) PSRALVLARREAQSKHGKTAANQPTSAASVARQGDPDLSSREISQRVRELRSKSGSTDKKRSGACRPCGPNRNGSKQAVA ADAHWKVGLSETNTGQFVTGTQANRSSKTTGNEASTCRSITGTQYLGSEVFDAFCQSSPQPGQPLKVAVTNTSHGNRVTG NEVGRSEKVTGDEPGTCKNLTGTEYISANQANQYCGVSQPSPRKVGQSITEDGRKVSGVMVGRSERVTGDEAGSNRQLTG DQYLGADPLPEGRPAEKVGSFHTLRGAGVTGTNVARSEYVTGNEPGSCKRVTGDEYVGTQQYKTFCGGKPNPEAAKVGLS LTNKSQTVSGTLTGRSELVTGDEPGTCKAVTGTPYSGVEQASGWCDTNSVREIQDRTPKLLGTPGAVMTGLQPGVGGVMT GAEKGACEPLTGTPYVGGDQLVQAC |
| 5730 |
PF15195 (TMEM210) |
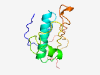 Estimated TM-score=0.50; very hard target. |
>TMEM210 family (Length=113) TYCDCSLGLSREALIALIVVLAGVSASCFCALMVVALGVLRAKGEPSTRHVDNRLVGHFGVQEDRMDLHAVHVESHLMDP ELEVSMMPSLEDHGLMTIPMEATLEEPPPPPPP |
| 5731 |
PF10191 (COG7) |
 Estimated TM-score=0.50; easy target. |
>Golgi complex component 7 (COG7) (Length=765) DFSKFLADDFDVKDWINAAFRAGPKDGAAGKADGHAATLVMKLQLFIQEVNHAVEETSLQALQNMPKVLRDVEALKQEAS FLKEQMILVKEDIKKFEQDTSQSMQVLVEIDQVKSRMQLAAESLQEADKWSTLSADIEETFKTQDIAVISAKLTGMQSSL MMLVDTPDYSEKCVHLEALKNRLEALASPQIVAAFTSQSVDQSKVFVKVFTEIDRMPQLLAYYYKCHKVQLLATWQELCQ RDLPLDRQLTGLYHALLGAWHTQTQWATQVFKNPYEVVTVLLIQTLGALVPSLPMCLSEAVERAGPELELTRLLEFYDTT AHFAKGLEMALLPHLQDHNLVKVVELVDAVYGPYKPYQLKYGDLEESNLLIQISAVPLEHGEVIDCVQELSQSVHKLFGL ASAAVDRCAKFTNGLGTCGLLTALKSLFTKYVSHFTNALQSIRKKCKLDDIPPNSLFQEDWTAFQNSVRIIATCGELLRQ CGDFEQQLANRILSTAGKYLSDSYSPRSLAGFQDSILTDKKSPAKNPWQEYNYLQKDNPAEYANLMEILYTLKEKGSSNH NLLSVSRTALTRLNQQAHQLAFDSVFLRIKQQLLLVSRMDSWNTAGIGETLTDDLPAFSLTPLEYISNIGQYIMSLPLNL EPFVTQEDSALELALHAGKLPFPPEQGEELPELDNMADNWLGSIARATMQTYCDGILQIPAVTPHSTKQLATDIDYLINV MDALGLQPSRTLQNIATLLKAKPEEYRQVSKGLPRRLAATVATMR |
| 5732 |
PF15172 (Prolactin_RP) |
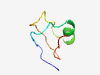 Estimated TM-score=0.50; very hard target. |
>Prolactin-releasing peptide (Length=44) QGHRHSMEIRTPDIDPAWYTGRGIRPVGRFGRRRAALGDAMKPS |
| 5733 |
PF12360 (Pax7) |
 Estimated TM-score=0.50; very hard target. |
>Paired box protein 7 (Length=44) QDGSSAYCLSSGRHGFSGYSDSFVPPPGHSNPVNPSISNGLSPQ |
| 5734 |
PF10152 (CCDC53) |
 Estimated TM-score=0.50; hard target. |
>Subunit CCDC53 of WASH complex (Length=151) LNQFVVHTVRFLNRFSAVCEEKLSNISLRIQQIETTLCILEAKLSSIPGLEDVTVDGLNQQQSVQTNGPVVASQNQPAGV TAAVAPPSQPDQGSPEAAPAQEKAEAVSENIMTVAKDARYARYLKMVQVGVPVMAIRNKMIMEGLDPNLLD |
| 5735 |
PF05318 (Tombus_movement) |
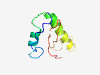 Estimated TM-score=0.50; very hard target. |
>Tombusvirus movement protein (Length=71) MDNQSESDNSVKRRSGKKVNKQVRGSEKSATKRSVASHAARSFSQRPSGDSAGGNWVLVADKIEVSISFNF |
| 5736 |
PF17305 (DUF5354) |
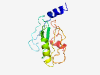 Estimated TM-score=0.50; hard target. |
>Family of unknown function (DUF5354) (Length=128) NTVQDAQEQLTKKIRCWEPIEENIAGNNFSGKHTLSEPIYDLCSYMPDPKNWNKGYVNGVDMDSDDYTNVFNFFQVTHPR LAMLNVCLQEAFQFHVAGRPSQTSIRCLCKRDGCNLPKEFTTFLDFNK |
| 5737 |
PF12601 (Rubi_NSP_C) |
 Estimated TM-score=0.50; very hard target. |
>Rubivirus non-structural protein (Length=66) VCAVGGGPRRVSDRPHLWLAVPLSRGGGTCAATDEGLAQAYYDDLEVRRLGDDAMARAALASVQRP |
| 5738 |
PF02990 (EMP70) |
 Estimated TM-score=0.50; very hard target. |
>Endomembrane protein 70 (Length=539) PYSYYSLPFCKPDTIVDSAENLGEVLRGDRIENSPYVFEMREPKMCQIVCKATISDKQAKELKEKIEDEYRVNMILDNLP LVVPIARPDRDDVVFQGGYHVGVKGQYAGSKDEKYFIHNHLIFLVKYHKDENSDLSRIVGFEVKPFSVKHQFEEKWNDAN TRLSTCHPHANKIVINSYTPQEVEAGKDIIFTYDVGFEESDIKWASRWDTYLLMTDDQIHWFSIVNSLMIVLFLSGMVAM IMLRTLYRDISRYNQLETEEEAQEETGWKLVHGDVFRPPTNSDLLCVYVGTGVQFFGMLLVTMMFAVLGFLSPSNRGGLM TAMLLIWVLMGLFAGYASSRLYKMFKGSEWKSITLKTAFLFPGIAFGIFFVLNALIWGEKSSGAVPFSTMFALVLLWFGI SVPLVFVGSYLGFKKPAIEAPVKTNKIPRQVPEQAWYMNPAFTILIGGILPFGAVFIELFFILTSIWLHQFYYIFGFLFL VFIILIITCAEIAIVLCYFQLCSEDYMWWWRSYLTSGSSAIYLFLYAGFYFFTKLQITK |
| 5739 |
PF16020 (Deltameth_res) |
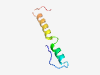 Estimated TM-score=0.50; hard target. |
>Deltamethrin resistance (Length=48) DDLPVPSGSWQARYDANQRRYNGILLFGIAFSVGSIIAVRYSSTLHCH |
| 5740 |
PF04328 (Sel_put) |
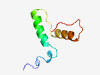 Estimated TM-score=0.50; hard target. |
>Selenoprotein, putative (Length=62) ITRAVRYLGQSMRLMVGVPEYSTYVAHMKTTHPDQPVMSYREFFRERQEARYGGNGKVTRCC |
| 5741 |
PF18189 (RIF5_SNase_2) |
 Estimated TM-score=0.50; easy target. |
>TbRIF5 SNase domain 2 (Length=154) DVMRGDVYRVSWMSADKCDDAVTLPPLLILDATVCQLPSTVKGFAAMRWAQTSLLHRELLVHVHAVFPSDKDIAYNTQLE MYEYLANVRGTAWVLCESTGEESHGTDVGEVLVSLGLGWIRRVADAGVPRSSNLLSLCRSVGISATQALSRSFA |
| 5742 |
PF13438 (DUF4113) |
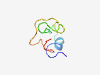 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF4113) (Length=50) RLAAAIDAINRKNGHNTIKVAVQGTTDKSWHLKCEHISKQYTTNLDDVIL |
| 5743 |
PF14070 (YjfB_motility) |
 Estimated TM-score=0.50; hard target. |
>Putative motility protein (Length=55) IASLSMVMSQSKVQTAASIAVMKKAMDTNKENAAQMTEMMRNVAVDPNLGNHIDV |
| 5744 |
PF17584 (ComS) |
 Estimated TM-score=0.50; hard target. |
>Bacillus competence protein S (Length=44) MNRSGKHLISSIILYPRPSGECISSISLDKQTQATTSPLYFCWR |
| 5745 |
PF19224 (pATOM36) |
 Estimated TM-score=0.50; hard target. |
>pATOM36 family (Length=274) MSSTLLATANQFYLLQQKKGRSFPHHGFTPRAIATKRFAENTTTLSLSVGFFLPTMTQVHQQLEQLTVERDDIPMEKVVT TVVEAIKDNLVLFGDKLARLAVQTAIACGLAKKNKPYMNSVVYLWNVYPGLHDHYVGETLRYLRLPRFLSSLFGPHALKS DVDVFMQDISAVAAAEDSMKRAVLDMVADSVVDMVRVLLPPWLLRGPALSFKRRLKLFGHDILMRFTVAVTKVTGAGVGR AIAGHRGEYWGEIIGVATASFLFERISAMFLDYK |
| 5746 |
PF03617 (IBV_3A) |
 Estimated TM-score=0.50; hard target. |
>IBV 3A protein (Length=57) MIQSPTSFLIVLILLWCKLVLSCFREFIIALQQLIQVLLQIINSNLQSRLTLWHSLD |
| 5747 |
PF15722 (FAM153) |
 Estimated TM-score=0.50; very hard target. |
>FAM153 family (Length=114) MVDKDTERDIEMKRQLRRLRELHLYSTWKKYQEAMKTSLGVPQRGDLEDLEEHLPGQTVSEEATGVHMMQVDPATPAKKL EDSTITGSHQQMSASPSSAPAEEATEKTKVEEEV |
| 5748 |
PF12043 (DUF3527) |
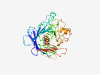 Estimated TM-score=0.50; easy target. |
>Domain of unknown function (DUF3527) (Length=355) TVSQGMLQCTWKGGIPHFVFSVDDQREVYVANQWKAELPDDKALDCMYLFHSRTGGQKEHKIPDNKADLVGKMKVSSSFT LCPNDSKIMETEFVFFGGRENYASKMQTSSSIIRKNKGLSKKVVEVFRTSHSSKQRTTSKFGGTTAILENDSWEQCQDTC NNPSALGRTNLLENDLPPNLELAAIVVKDHLPGKRQEAEVGGWGLKFLKKVGLKQNSASLEASLPSECCLRSTGDCSTSM DILVPAGIHGGPRTRNGGPSSLIERWRSGGHCDCGGWDIGCPLTVLKTRATKEEFSHHTDIQEEYKSFDLFAQGSELSAP ILRVVNVHDGLYFIHFRSMLSALQSFSIAVAIIHT |
| 5749 |
PF15214 (PXT1) |
 Estimated TM-score=0.50; hard target. |
>Peroxisomal testis-specific protein 1 (Length=51) MQLRHIGDSIDHRMVQEDLQQDGRDALDHFVFFFFRRVQVLLHFFWNNHLL |
| 5750 |
PF14910 (MMS22L_N) |
 Estimated TM-score=0.50; hard target. |
>S-phase genomic integrity recombination mediator, N-terminal (Length=708) PPCFSCALDNRDGGRNFSVESHLASGSLKRLLLRLDPSPTDYEEDTVEIFGFHWVTEIALVESCRLLFGLLRQQINKLEN LVQMSSSDFGQAANLHSEAGNIRHQCIAFLHYVKVFIFRYLESHKTEDDGLLHPYEELEVQLPSVLVEELHALTLHIGHL YELPSSILGAFTIQHQAKLFPPSWHLLHLHLDIHWLVLEILHVLGEKMLRQIVYAHQFMNLTGENLTNISLFEAHCGNLF CDLISLSINKYTKVRPSEALTSHHYPCSCIKELWVLLIHLLDHRNKGSHTESFWSWVNKLLKKVFERPSTAEGMSVSEPV QCKDPLSFSWWIITHLASLYQFDRNGNVEEKKQVESNWNFVEELLKRSTDAQAAVLEEQIRMHLQCCLTLCSFWDLNLPT VTILWEYYSKNLNSSFTIPWLGLKGLASVSKTPLSMLELVKNCCSDQQSPDLYKSSNSYLIFLCILAQMMKEAMESNVTH PWKQIKGRIYSKFHQKRMRELTEVGLQNFFNLFLVLAAVAETEDIASRVLDLLDFLSPTSTSTSQRALIWRGYFAFLLMY IEKNMDISVLAEKLSHAFCEKAKEFLVTKNDHTHKQNLWTPLSTYIDGVQEVFETSCYLNLSQEKLLNDGFSMLLPACRG TELNMVLNFLQVVLSRLRSVHKRMIQGPQLGKTGPNTQLPLVAKEHHLAVASALWRNFFPFLKSQRMS |
| 5751 |
PF01998 (DUF131) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function DUF131 (Length=59) GIIKSSKKERNENQREETKTEYGGVIFIGPIPIVFGSNKDMAKMALIIGVIMVVLIIAF |
| 5752 |
PF17079 (SOTI) |
 Estimated TM-score=0.50; very hard target. |
>Male-specific protein scotti (Length=100) ENPQVAMLLDAPHEPPIELHHMLEPVNVPQRPRKKRSFLTISKPFHVQPERCALISNGWRAVQCVQPEKRGEYFANYLIK HMNSRNYPNGEGLPNRWGQF |
| 5753 |
PF03430 (TATR) |
 Estimated TM-score=0.50; hard target. |
>Trans-activating transcriptional regulator (Length=575) MTQINFNASYTSASTPSRASFDNSYSEFCDKQSNDYLSYYNHPTPDNGADTVISDSETAAASNFLASVNSLTDNDLVECL LKTTDHLEEAVSSAYYSESLEQPIVEQPSTSSTYHAESSDGVNQPSATGTKRKLDEYLDNSQCIVGQFNKNKLRPKYKKS TIQSSATLEQTINHNTNICTVASTQEITHYFTNDFAPYLMRFDDNDYNSNRFSDHMSETGYYMFVVKKSEVKPFEIIFAK YVSNVVYEYTNNYYMVDNRVFVVTFDKIRFMISYNLVKETGIEIPHSQDVCNDETAAQNCKKCHFVDVHHTFKAALTSYF NLDMYYAQTTFVTLLQSLGERKCGFLLSKLYEMYQDKNLFTLPIMISRKESNEIETASNNFFVSPYVSQILKYSESVQFP DNAPNKYVVDNLNLIVNKKSTLTYKYSSVANLLFNNYKYHDNIASNNNAENLKKVKKEDGSMHIVEQYLTQNVDNVKGHN FIVLSFKNEERLTIAKKNKDFYWISGEIKDVDVSQVIQKYNRFKHHMFVIGKVNRRESTTLHNNLLKLLALILQGLVPLS DAITFAEQKLNCKYK |
| 5754 |
PF14738 (CFAP91) |
 Estimated TM-score=0.50; hard target. |
>Cilia- and flagella-associated protein 91 (Length=161) TQTLYRESSAQTLAYLPNIEDKDIDEHVELFKLPSALPGDKPPGLYEVEVLERSRRRWAFNEALTVNLKKQLDEARELAM QTKYKSILEAFEWESWIEREEYIQECQRMRLEIVIKMFDKREKEMHSASKTRMEMACERIEKRRQRGLHKNEIEYQRGKR R |
| 5755 |
PF19171 (DUF5853) |
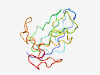 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5853) (Length=123) RNYSAPIGPLPRQNMSGSAGLFGQRPFDQQINYSAPIGPVQGPGLGMSYDQNRNYPQPIGPLPRQNMSGSAGLFRNRPLN RKINYSQPIGPEPLSEDRPRNANFIPTASFLGQNTDESVGASP |
| 5756 |
PF10733 (DUF2525) |
 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF2525) (Length=60) DVEALLEAIQDRSSGEVKEFMDNDRAHKVRVDGREYRSYTELADAFELDIRDFTVSEVNR |
| 5757 |
PF05358 (DicB) |
 Estimated TM-score=0.50; hard target. |
>DicB protein (Length=62) MKTLLPNVNTSEGCFEIGVTISNPVFTEDAINKRKQERELLNKICIVSMLARLRLMPKGCAQ |
| 5758 |
PF06075 (DUF936) |
 Estimated TM-score=0.50; very hard target. |
>Plant protein of unknown function (DUF936) (Length=714) LVPGVLLKLLQHMNTDVKVAGEHRSSLLQVVSIVPALAGSDLFSNRGFYLKVSDSSHATYVSLPDEHDDLILSDKIQLGQ FIYVDRLEAASPVPILRGVRPVPGRHPCVGHPEDIVATSSLGFLGTEKLRLSNGSKDDAKLSSDKEKNKLAKQNGTIRTE EVEKKKVVLGKSSSVLLKQAADGNLEKKEAIGVRSRSMNSRSIPSSPTSVYSLPASFEKFSNDLKQQAKVKGIEKPPSSR LGLLERTTSVLKVTAAGRKPAAGNSTRNLVLGIESSPKALRRSWEGSMETKGRVHSNSKAGKLKTKSETQSTSVPRMKPP LNGKSLSKEDNKVQTPAKKGTANASSDDSDRSNKQRSSIIKKTQEASNNLNLANLVKFVSNSKRWTDGSVSWASFPSSLV KLGKEVLKYRDAAQLAAIQALQEASAAESLIRCLSMYVEVTSTAKEDNPQPAVEEFLALHASLSRASQVTDSLANTAVTS PDQPPGGNPIPEDALKISADRRRRATSWVRAALATDLSPFSLYQHKPHPPPLSAAAASTSVAVVLDGPSKTAAASSKAKP RSSPPPIPASSGSGKPSPATTSQGKARGAAAPPSPPPEWARGGGLEEGSELARALREEARRWFLGFVERFLDADVAAPGP SDGEQVAGMLSQLKRVNDWLDTIGRQREGEAAEAATAEDRVGEGGGVPEETIERLRKKIYEYLLAHVESAAVAL |
| 5759 |
PF04347 (FliO) |
 Estimated TM-score=0.50; hard target. |
>Flagellar biosynthesis protein, FliO (Length=84) WLVRRLGLQNRQGGRLLKVVSSLMVGQRERVVLVEIGDTWLLLGVGAGQVNTLHTMAAQHAPDASPKVDDTDPDTFANRL RQSL |
| 5760 |
PF17446 (LtuA) |
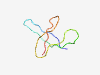 Estimated TM-score=0.50; hard target. |
>Late transcription unit A (Length=46) MFFIRARFIGFLDVHGYLAAKKGQQVMRSGSSIWVGSHGPIFYKVF |
| 5761 |
PF13142 (DUF3960) |
 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF3960) (Length=89) MKAVQRDPNWNLVTDTYIEPNNFAELFSLLVPCHPKGEGKERTILVWKEKEFYKEENLAAFIVYGMNKAKNLPQFHKDEI PTLVRILRL |
| 5762 |
PF14073 (Cep57_CLD) |
 Estimated TM-score=0.50; easy target. |
>Centrosome localisation domain of Cep57 (Length=178) AILSALKNLQEKIRKLELERTQAEDNLRRLSQETSEYKKVLDKEIRQKDSSKTEVARQNRELSTQLAAAESRCTLLEKQL EYMRKMVKNAESERTTVLQKQASIGRERSLDHLELQTKLEKLDMLEQEYLKLTATQNIAQNKICLLEQKLREEEHQRKLV QDKAAQLQTGLETNRILL |
| 5763 |
PF18951 (DUF5695) |
 Estimated TM-score=0.50; easy target. |
>Family of unknown function (DUF5695) (Length=848) QLVKASQTVAALRPNSNKKFDFTPGDRLKVRSSDGMYQLGDIELRLKSAGDADWKGYSTAAKRAAVNPMPNKSKTVLAAA DLSATLPADIPLNIQRYWKVADGHLVLQFKLKNKSDKVVEIGSLGIPMIFNNIMQGKSLEETHAENVFYDPYIGKDAGYL QVTRLNGHGPALVVVPFGKTPFEAYNPLNNDPTPRGIDFEGFYEWMVYSKAYAENEWKNAEPWNTPTSVKLNPGETISFG VKFLLADSIKGIEHKLAENKRPVTVGIPGYVLPQDVNAKLFLNYPKEVKSIQVEPEGALAITPASSSESKWKTYDIKGVT WGRARLTITYADGLCQTINYKVIKPETDVLKDFGNFLTTKQWYENKSDVFGRDQSVITYDYDKKQQVTQDNRAWIAGLSD EGGAGSWLGAMMKELVQPDKAEVDKLERFVDHTLWGNIQHSEGEQKYGVKKSVFYYQPDSLPKGTYSDSIRWNTWSAWNH VAANDIGRSYNYPHVASAHWVMYRLARYHTGLVSGQTWQWYLDNAYQTTMAMIDKAPYYVQFGQMEGDVFFKILMDLKAE GWNDKADLLEKAMKVRADHWRTLMYPFGSEMPWDSTGQEEVYLWSEYFGYTDKAEITIDAILGYMPTIPNWGYNGSARRY WDFLYGGKLQRIERQLHHYGSGINAIPVLSEYRAKPNDLYLLRVGYGGVMGSISNITQDGFGPAAFHSYPSTLKIDGLSG DYGSNFFGYALNTATYITHDKELGWLVFGGNMTIKPDAVNVEITTAAKSRIYIAPAGLWLTLDAGTFKSVTFNPETGKVN VLLNPADQYTPVAYLHIEHPANGASASTYKLVSPSKMERGAYVINLTN |
| 5764 |
PF05436 (MF_alpha_N) |
 Estimated TM-score=0.50; very hard target. |
>Mating factor alpha precursor N-terminus (Length=88) MKFSTILSTAAFVSSSVMAAPVNVTETEQENLAIPSEALIGYLDLKGDDDISILPVSNSTNSGLLFLNSTILTQAMNENT EAALSKRE |
| 5765 |
PF12020 (TAFA) |
 Estimated TM-score=0.50; very hard target. |
>TAFA family (Length=90) AGTCEIVTLDRDSSQPRRTIARQTARCACKKGQIAGTTRARPACVDARIVKTKQWCDMVPCLEDEGCDLLVNKSGWTCTQ PSGRVKTTTV |
| 5766 |
PF19226 (DisA) |
 Estimated TM-score=0.50; very hard target. |
>DisA glycoprotein (Length=359) LFTFKQFSTIYRSDIFNDFKIYHANAWNEFVGISCPEHHVPYYLVHHIFSAIYVCLPVPHVLEPLYTAYELILGETNFWI FCGLTGSSSEFAKFTPKDYTDYLILVDSNNKYHLSSKYCVGHMYQVGPDNKEIMTLNYTVNDHSVTLDLPSEHCKILPLP ISKLDFTLLPIEVDVKSSVDVLDCLRVERSSNLHPEKPFFELALDDTTLLTYNTSTNGSHNFAYIWFSSHQFPTKTFHIS SAVNVNPLSWIITGLMSFFYPLLDMLLSSILYIFESVLDIFESHTFLDLFDRLLNVVITILTRIFQFFVSTIFPRIVDLL NRIPTRYKFLFGLMFVLYLKTTKLLFSFCITVLVHFCIK |
| 5767 |
PF10168 (Nup88) |
 Estimated TM-score=0.50; easy target. |
>Nuclear pore component (Length=713) RWRRELASHQIFLRLQENVRGEAESTAKAPAKNLLFNLDGDLFLWDGDNSVFYTTNLRQLNSEQPSPAAPYQTLLCINPP LFEVHQVLLSPTQHHVVLIGSKGLMVLELPQRWGKRSEFEGGKDTVNCRTISIAERFFTSSTSLTLRHAAWYPSETEEPH LLLLTSDNSLRIYSLKEPQTPLKVLSLSQSEEESVLLPRGRSYTAPMGEVAVAFDFGPLVAVSSHLVGQKGKEEVQAFPL YILYENGETFLMYLFLSHGIGSFGKLLGPLPMHPAAEDNYGYDACAVLCLPCVPNILVIATESGMLYHCVVLEAEEEEDQ TSSESWDPRSEMVPSLYVFECVELELALKLPSGEEEEPMESDFSCPISLHRDPLCQSRYHCTHEAGVHSVGLTWLNKFQK FLASDEEDKDGLQELAAEQKCIVEHILCTRPLPCSQSAPIKGFLIICDLSLGASMICITNAFECITRPLLSTIRPASPPL LCSKAESELAASPLRTFAVDSFEKHIRNILKRTSTNPVFLKASEKETSPPPQECLQLLSRATQVFREEYIIKQDLAREEI QRRVKLLRGQKEKQLEDLEHCIEERKNLREMAERLADKYEEAKEKQDIIMNRMKKVLRSFSSQLPVLSDSEKDMKKELNT INGQLRHLGNTIKQVKMKMEYQNRQTEKGASPRKSTLSLSDYQKKCIYNVLKEDGEHIAEIVKQLNDIRSHVD |
| 5768 |
PF15369 (KIAA1328) |
 Estimated TM-score=0.50; very hard target. |
>Uncharacterised protein KIAA1328 (Length=324) DLCLEDKRRIANLIKELARVSEEKEVTEERLKAEQESFEKKIRQLEEQNELIIKEREALQLQYRECQELLSLYQKYLSEQ QEKLTMSLSELGAARMQEQQVSNRKSTLRSSSVELDGSYLSVAKPQTYYQTKQRPKSANQDPASESLVEFRNNSLKPAAL HYPKEDLDRVQSETRTCNYGSPGRKLVDAVPTEKAPPEELKMKECQHLKPAPSSQCCDHRLSENADHVHDSHPTNLAPQY SKTHPESCSYCELSWASLMHGRGALQPSETDVKKQLSEDRRQQLMLQKMELEIEKERLQHLLAQQETKLLLKQQQLHQSR LDYN |
| 5769 |
PF10837 (DUF2575) |
 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF2575) (Length=71) MQHSLRSDGAGFYQLACCEYSLSMRKIALSGGFWGGVCRMAMKSIFFMFHQGNRRLTLTAVQGILLRFSLF |
| 5770 |
PF10840 (DUF2645) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2645) (Length=98) FVLCCIWFIVAFLWITITSALDKEWMIDGREINNVCDVLMYLEEDDTRDVGVIMTLPLFFPFLWFALWRKKRGWFMYATA LAIFGYWLWQFFLRYQFC |
| 5771 |
PF15201 (Rod_cone_degen) |
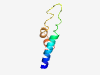 Estimated TM-score=0.50; very hard target. |
>Progressive rod-cone degeneration (Length=54) MCTTLFLLSTLAMLWRRRFANRVQPEPSDVDGAARGSSLDADPQSSGREKEPLK |
| 5772 |
PF11448 (DUF3005) |
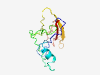 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF3005) (Length=126) QTGRAHPRSVELDNDDTHDSTVDTDGKHREASRLAGAGPISPDQITRSNASLVNAMPEAGDGFAGFDSRPDGNHPAFALR AGYRVIEKGFDTPPADDAPFGPVHRMYGSAYWPGHGRRPERVIELT |
| 5773 |
PF04764 (DUF613) |
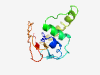 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF613) (Length=118) MYIYRKPYEQNYWNNFILLSLYVNNKRVGVNKNITKLEYILLDIFLHGPLHTEFINYLEITNYINNRTYLYEKMLKEKSV NAINIILPPVMHTASFEYNYEIIEDLDSNKILNIYILN |
| 5774 |
PF12726 (SEN1_N) |
 Estimated TM-score=0.50; hard target. |
>SEN1 N terminal (Length=768) WIQRTESFLVKCDGCVRAWHMGRKALRSRLLKEFDEETTNIMEGRLHDYDTARIDRGLRQAVAILEQHGPMSSAKLVAID ATAVLAFYEALCCVEYLSLPENRRLFNIVFQKTQEKKPLKIASAVIPTMTVFLFQADPLRNRFARTAWERRPAQSLTPEQ FEWAVSDHLADAIDAVSKPDAADAAVQLFWDGFLLLLDAMSEDVVLHCLRGMQIPMDVYQLAVLHLDRPADGVLAGSLRA LCALMDKSAKAFWDALSRMNPPQLPMFLCRSPAFAPFLARSFEEAMQLVPDSTSTRARMPLLVAFVRSLMRSLNRNQRSD VCDYLLSVFLTNTADPAVTATREGHSTCIVAGLTALQMTLDGYLDARYRLTSESALFVTTVINSAVKHHDYVVRSANLHP SDRYNIGLSQAAVGVVESALALDTRAIQDEWRSLQQNARLPPGNERRSAQLWHGFRTALGPGRLDLARVMLTATLSLRGI QRFSPKKRQVQVEDGPRRRFNDELEPAAEAIAQTIERVGESLSCADLKTLCSDTQSRSMDAVASTLIHAEDAIREAGFLL VGAIVDDDGGSRSEIIGKVVSELPVPFLTSFADAIDHLTGGEGRLGSASPFEPMRHIYRCSKDVLDGLCDSSAGLLRSRT LTAEERKAVKRWWTMEWQFVDHSFLRARAWSEDVEKSGMENYCRDVMEFARLLMAQDGLIASALAENGGSTSMEPFAHSS TIAAPPSSSSSTKNDSAMREVLDAPQKHSMGLFEMIQLRDQYLVGIIV |
| 5775 |
PF05559 (DUF763) |
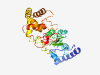 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF763) (Length=331) GVANLPLHGGHAPKWLFKRMVKLTKGIVDVMLYEYDADEFLRRISDPYWFQAFSCVLGFDWHSSGTTTTTCGALKLAIDP EEHGILVAGGKGKASRKTPSDIEKAGDIFSLSTGKIDEMVHASKISAKIDNSCIQDGYQLYHHSFILTEKGEWAVVQQGM NEETRYARRYHWLSESVSEFVEEPHKGICCDKPELQTLDMTAHQSNEARDVSVDLICDNPEHLKKYFKDRTPLVEKSQTH LEDYFNEFRMPRHHPVLDMDLSDREFDVLKRAWEIQPSDYEELVSLEGIGPKKVRALALISDLVYGSEPSWKDPVKYSFT HGGKDGYPYPV |
| 5776 |
PF12564 (TypeIII_RM_meth) |
 Estimated TM-score=0.50; hard target. |
>Type III restriction/modification enzyme methylation subunit (Length=56) NNELLSLLLSNEQIKERFFENVDGTLVFDKQKFAWFIESKEFLPDSYTRYTNKIGL |
| 5777 |
PF00713 (Hirudin) |
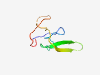 Estimated TM-score=0.50; trivial target. |
>Hirudin (Length=64) VVYTDCTESGQNLCLCEGSNVCGQGNKCILGSDGEKNQCVTGEGTPKPQSHNDGDFEEIPEEYL |
| 5778 |
PF17687 (DUF5535) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5535) (Length=105) MTVLEAVMEIQTIADVGLLWRQSHPGRTKLMLGPNPTRKDLLGWLLSHGVPREQVDRQPTKVLLELYIKEAKRSRGHPNY GLNEEQPPPPPYSDQACGEEQPVRH |
| 5779 |
PF11278 (DUF3079) |
 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF3079) (Length=50) MAKKFPLHPVHPERICWGCDHYCPADAMRCGNGSSRTQHPIELLGDDWYE |
| 5780 |
PF01307 (Plant_vir_prot) |
 Estimated TM-score=0.50; very hard target. |
>Plant viral movement protein (Length=101) AQPPDYSKSVFPIAVGVAVAVVLFTLTRSTLPQVGDNIHNLPHGGNYQDGTKRISYCGPKNSFPSSSLISSGTPMIIGII IFLIFAIYVSEKWSRSGNRRC |
| 5781 |
PF07911 (DUF1677) |
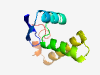 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1677) (Length=87) VEDAKCECCGMSEECTLEYIRRVREKFSGKWICGLCTEAVKEEVDKNGGGEEEALSAHMSACVRFNRIGKAYPALYQADA MREILKK |
| 5782 |
PF15317 (Lbh) |
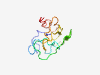 Estimated TM-score=0.50; very hard target. |
>Cardiac transcription factor regulator, Developmental protein (Length=101) MDLVPESSEDGPWPRDSPGSFQHPESARLTNPLWKDRGEIDRVEDHQDIQVVSQKTHLPSIVVEASEISEESGELRWPHE ELLVLTDDEEEAEGFFQDQSE |
| 5783 |
PF09740 (DUF2043) |
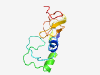 Estimated TM-score=0.50; hard target. |
>Uncharacterized conserved protein (DUF2043) (Length=105) EIPILSFGLDLKYWGEDDVVPAEVPRNNADCHRFWRPSDESCCTSRDSAEVYRARVITFVGPGLKATRRCRAPLKNGELC SRMDFRRCPLHGNIIDRDEMGYPID |
| 5784 |
PF10164 (DUF2367) |
 Estimated TM-score=0.50; very hard target. |
>Uncharacterized conserved protein (DUF2367) (Length=107) PSYGTIPAAQVGFQPPPPYPYPAAPGFSGGPPPASVQPPYSSTYTIIQQPATTSVVVVGGCPACRVGVLEDDFTCLGILC AIVFFPIGILFCLALRQRRCPNCGATF |
| 5785 |
PF13994 (PgaD) |
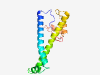 Estimated TM-score=0.50; hard target. |
>PgaD-like protein (Length=138) LIIEKPELQSHAHRFGWSSITLFFWGLYIYLWLPFITLLAWWIGVKLFHLQIVELHGYAGVIGKLGLYSAIVVILSVTLI GWANIERFRFKDSARRVDNTPVSVREIAKLFKLQENQLIQLRQKQSVIVRFSDKGHLT |
| 5786 |
PF05390 (KRE9) |
 Estimated TM-score=0.50; very hard target. |
>Yeast cell wall synthesis protein KRE9/KNH1 (Length=101) DSRSFTVPYTKQTGKARYAPMQMQPGTKLTAKSWTRKYATSAVTFYTSMRKSLQQVTTITPGWSYAISSDVNYATHAPFP SDNGGWYDPSKRLSLTTRKIN |
| 5787 |
PF09779 (Ima1_N) |
 Estimated TM-score=0.50; hard target. |
>Ima1 N-terminal domain (Length=126) VNCWFCNQDTVVPYGNRNCWDCPNCEQYNGFQENGDYNKPIPAQYMEHLNHVVSGSPTFCDPTKPQQWVSSQILLCKKCN NHQTMKIKQLASFSPREEGKYDEEIEVYKHHLEQTYKLCRPCQAAV |
| 5788 |
PF19188 (AGRB_N) |
 Estimated TM-score=0.50; hard target. |
>Adhesion GPCR B N-terminal region (Length=176) DTCTTLVQGKFFGYFSSSSVFPYSNGGASASPCSWIIQNPDPRRYTLYMKVSKPTSCCNPRLIKTFQYDSFLETTRTYLG MESFDEVLKLCDTSTRHAFLQSGKQFLQMKKLVSRDEPSSALAGNGEFMAEYLVVGNRNPSLPACQMLCKWLENCLVSST VARPCGIMQTPCQCWD |
| 5789 |
PF08455 (SNF2_assoc) |
 Estimated TM-score=0.50; very hard target. |
>Bacterial SNF2 helicase associated (Length=379) KKVLLSTPYLLRIFDALKNRNFYAKVLDYGEKLVSIAEEDLPLDFSLSRKNNEVLLHLKESRSFVPLTEKGVYFYYNGRI YRPSEKQQKYYIPFISRLLGGKTDTLTFSAAQTERFVSEILPHIHKIGRVSIDKEIEDKLVKEELVAKIYFDKYKEGISA VINFHYGQLVVNPFSGQNYISDPDKIVVRDTESERKIIEIFEAAEFLVDKNIIYLTDENKLFEFLRNDLPRLQELAEVYY SDAFKNISVRFPPSFSGGIRINQASNMLEFSFDYGDIDKSELEHIFSSIKEKKKFYRLKDGSFLSLDLPELKSMSNLVEQ LDIKVKDLSKKVIELPKYRAMYIDSLLREANMNGIERSLDFKHMVQNIKEPGDMDFEIP |
| 5790 |
PF03905 (Corona_NS4) |
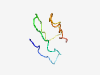 Estimated TM-score=0.50; very hard target. |
>Coronavirus non-structural protein NS4 (Length=45) MQMATTIDDTDYTNIMPSTVSTTVYLGGSIGIDTSTTGFNWFSWY |
| 5791 |
PF03491 (5HT_transport_N) |
 Estimated TM-score=0.50; hard target. |
>Serotonin (5-HT) neurotransmitter transporter, N-terminus (Length=41) NGVLQKVVPTPGDKVESGQISNGYSAVPSPGAGDDTRHSIP |
| 5792 |
PF05616 (Neisseria_TspB) |
 Estimated TM-score=0.50; very hard target. |
>Neisseria meningitidis TspB protein (Length=507) YAERFKYPIGNSDVRLDIDHKKSVVTDFRVDGQRFSGRIIEPSIIEHVPTGARSLEKVPVKFTASVSRAAVLSGVGKLAR LGAKLSTRAVPYVGTALLAHDVYETFKEDIQAQGYQYDPETDKFVKGYEYSNCLWYEDKRRINRTYGCYGVDSSIMRLMS DDSRFPEVKELMESQMYRLARPFWNWHKEELNKLSSLDWNNFVLNRCTFNWNGGDCLVNKGDDFRNGADFSLIRNSKYKE EMDAKKLEEILSLKVDANPDKYIKATGYPGYSEKVEVAPGTKVNMGPVTDRNGNPVQVVATFGRDSQGNTTVDVQVIPRP DLTPGSAEAPNAQPLPEVSPAENPANNPNPNENPGTSPNPEPDPDLNPDANPDTDGQPGTRPDSPAVPGRTNGRDGKDGK DGKDGGLLCKFFPDILACDRLPESNPAEDLNLPSETVNVEFQKSGIFQDSAQCPAPVTFTVTVLDSSRQFAFSFENACTI AERLRYMLLALAWAVAAFFCIRTVSRE |
| 5793 |
PF17669 (DUF5529) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5529) (Length=186) SQSPTMSQRPAPPLYFPSLYDRGISSSPLSDFNIWKKLFVPLKAGGAPAGGAPAAGGRSLPQGPSAPAPPPPPGLGPPSE RPCPPPWPSGLASIPYEPLRFFYSPPSGPEAAASPLAPGPMTSRLASASHPEELCELEIRIKELELLTITGDGFDSQRYK FLKALKDEKLQGLKTRQPGKKSASLS |
| 5794 |
PF12752 (SUZ) |
 Estimated TM-score=0.50; hard target. |
>SUZ domain (Length=52) QEPKIMILRRPQGKPDATANGMVSLKQKQPLKTLEQRQQEYAEARLRILGDA |
| 5795 |
PF14901 (Jiv90) |
 Estimated TM-score=0.50; hard target. |
>Cleavage inducing molecular chaperone (Length=92) ARNTMHCDCGSKHARVPVESIRASEARYCKKCRVRHPAKHNDIWAETRCGGFWWVYYACLDGVVYDITQWATCPNNRLKH MKANSHAVQYRL |
| 5796 |
PF05957 (DUF883) |
 Estimated TM-score=0.50; hard target. |
>DUF883 N-terminal domain (Length=93) DLLADFQTLVNDTEKLLQDTASLAGEQADELRVQIHESLKRARDTLRDTEQNLRERGQAAVQATEDYVQEHPWQSVGLAA GIGFLLGLLAARR |
| 5797 |
PF14886 (FAM183) |
 Estimated TM-score=0.50; very hard target. |
>FAM183A and FAM183B related (Length=103) ILEETIKKEQRHQKLYTTYSVNPFRKMYTLTGKPNSTHDSADGEEDDNFLKIIHRAHQEPVKKYTFPQTEAQEIGWMSKP LIPQDREDRRLHFPRQNSEITKY |
| 5798 |
PF02088 (Ornatin) |
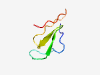 Estimated TM-score=0.50; easy target. |
>Ornatin (Length=41) LLYCGEFRELGQPDKKCRCDGKPCTVGRCNFARGDNDDKCI |
| 5799 |
PF11928 (DUF3446) |
 Estimated TM-score=0.50; hard target. |
>Early growth response N-terminal domain (Length=87) GRFTLEPAANCSNSLWAEPLFSLVSGLVGIANPPSTSAPSLSSSGPTQASAASHSPPLSCSVQPSEPNPIYSAAPTYSSA SPDVFPE |
| 5800 |
PF13494 (DUF4119) |
 Estimated TM-score=0.50; hard target. |
>Domain of unknown function, B. Theta Gene description (DUF4119) (Length=91) AKTRVKNNAQNRKKSGGRCKKAEIISEEELEKRGGLTGDETALFTVYLQKFDKGDVHMKYSKKLKDLAKHIHEKEHLYVP KQGGYKLMEVS |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417