

Go to page (83 pages in total): [First page] << < 42 43 44 45 46 47 48 49 50 51 52 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4601 |
PF17114 (Nod1) |
 Estimated TM-score=0.59; easy target. |
>Gef2-related medial cortical node protein Nod1 (Length=145) MVCLSTSIAENPYQSLSLESVPSKILFGSFLFYVKDRIPEKAGYIIEEQFCKRLQQIDYDAYSFSPKECNEKLKNIFSEF SQMKLKMLSVFFSIVQILLPKLSGDYQEHVQFFASISAVVAPRGYVFEIYHAVEHLSLEAREVFQ |
| 4602 |
PF05335 (DUF745) |
 Estimated TM-score=0.59; easy target. |
>Protein of unknown function (DUF745) (Length=181) GNPKQKATNIAQKAAQEATAASEAQNAAGAAAAHQVKMQLAEKAMQAAKAAEAALAGKQQIVEQLEQEVREAEAVVQEEA SSLDSAHSNVQAAIRASQQAQKQLKTLTEAVQTAQANVGNSEQAATGAQAELAEKAQLMEAAKTRVDQLLRQLSGARQDY SNTKQSAYRAACAAHEAKVNA |
| 4603 |
PF17731 (DUF5570) |
 Estimated TM-score=0.59; easy target. |
>Family of unknown function (DUF5570) (Length=123) AISWELDNVLMPSPRIWPQVTPTGRSASARSEGSTSSLWNFSAGQDVHAIVTRTCESVLSSAVYTHGCGYVRSATNITCQ SSGQQRQAARQEEENSICKAHDSREGRLGYPLSAHQPGSGGPN |
| 4604 |
PF04757 (Pex2_Pex12) |
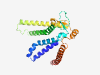 Estimated TM-score=0.59; very hard target. |
>Pex2 / Pex12 amino terminal region (Length=237) QDSLMGAVRPALQHAVKVLAESNPSRYGPLWRWFDEIYTVLDLLLQHHFLSRTSASFSENFYGLKRVVTSGLQRSAHLGL HSRQRWNSLLLLVLVPYLHAKLEKVLARQREEDDFSIQLPQTRFQRMYRAFLAAYPFVSTVWEGWTFCQQLLYVFGRTWR HSPLLWLAGVRLVHLTAHDILNMDLKPTTTSLATSQRFGEKLQQMLSTVVGGIAVSLSTSLSLGVFFLQFLEWWYSS |
| 4605 |
PF11870 (DUF3390) |
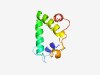 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF3390) (Length=88) RLRNEAKNPPKPGMPAMRGQEAAASKAESLAMKGFAFAASHPSIYHMGTWSATKMKALMPSNLGPWTQCRTAPKPASKTL HQLIKARK |
| 4606 |
PF18983 (DUF5717) |
 Estimated TM-score=0.59; hard target. |
>Family of unknown function (DUF5717)C-terminal (Length=308) DETMTTLSYMAFRAGKYDENILQYLCLYFKGTCKEMRDIWKAAEGFGVDVYSLGERILVQLLYSGAHVGEKTTIFRKYVE AGAKGQIEIAFLSQSAYDYFVREKLTDGYILKDMQRLIRQGEELPFVCKLAYTKYYAENKKLINEEISRWLILFLREILD KKLYFPYFREYTDHIAFMRRFEDKTMIEYHAKEGSHVEIHYLVEREDQSDGEYQKEEMQDMFHGIFVKQFILFFGERLQY YITEKDGEKEQLTESGTLSRNETDAVQKEGRFNLLNDISTGRTLHDYSTMERLLEEYFLQEQMVKEIF |
| 4607 |
PF03733 (YccF) |
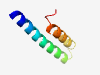 Estimated TM-score=0.59; hard target. |
>Inner membrane component domain (Length=51) IGNILWFVLAGLWMGIAYIVAGVLQCITIIGIPFGLQSFKLAGYAFWPFGR |
| 4608 |
PF12494 (DUF3695) |
 Estimated TM-score=0.59; very hard target. |
>Protein of unknown function (DUF3695) (Length=96) QYKNPTHLAQQQEPWSRLSSTPTITSMRRDAYFFDPKVPKDDLDFRLAALYNHHTGAFKNKNEILVHQETIQDTHGIVKM PEEFLPPPPPLPITSL |
| 4609 |
PF17264 (DUF5330) |
 Estimated TM-score=0.59; hard target. |
>Family of unknown function (DUF5330) (Length=64) PQVGASQAVSAATAAVSDMSQFCKRQPQACEVGGQAATVIGHRAQEGARKLYQIITDKKAPDHT |
| 4610 |
PF15190 (TMEM251) |
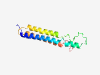 Estimated TM-score=0.59; hard target. |
>Transmembrane protein 251 (Length=128) MNFRQRMGWIGVGLYLLASAAAFYYVFEINETYNRLALEHIQQHPEEPLEGTTWTHSLKARLLSLPFWLWTIIFLVPYLQ MFLFLYSCTRADPKTVGYCIIPICLAVICNRHQAFVKASNQISRLQLI |
| 4611 |
PF06269 (DUF1029) |
 Estimated TM-score=0.59; hard target. |
>Protein of unknown function (DUF1029) (Length=53) MITNYEPFILLGIICIALLANIKLSSKVKLDIIFFVQSILFMWFIFHFVHSVF |
| 4612 |
PF07041 (DUF1327) |
 Estimated TM-score=0.59; hard target. |
>Protein of unknown function (DUF1327) (Length=112) MKKKYELVVKGINNYPDKITVTVALEIGGYPSLLLPDVAISLDRTEGATLEFYEAEAKKQAKQFFMDVAAGLCEGDGPLP ENRPVILEAQDVLITYKGKLPGRITCSLKMPP |
| 4613 |
PF03216 (Rhabdo_ncap_2) |
 Estimated TM-score=0.59; very hard target. |
>Rhabdovirus nucleoprotein (Length=307) WTNEMLKDRKVINLDAWNQVGTFDERIQIIREALTSLTTEVSVRSVGALMVCAWGLVSCENPNESVFGGLWESSKARELE EYTRLSDGILCKSSRWRSPPDDKDLILFTTTGNGLNWVRTACFYCAAVLRLATKEHDALVKAWSYLPEHYQSFYKAPLEF SLSLDHECLKCLRRLLQKSTTIRNSVAPFLLAFQELSGCSKNRAICKTLFESHLGFTGLHAYTLFISNASKLAVPHHVFR HVLRHHAAEEGLETIMEILKRYEDPSLDEGDRKSKCTWIYSRIFDSDMFGSLQTKRCVFLAALLAVI |
| 4614 |
PF05540 (Serpulina_VSP) |
 Estimated TM-score=0.59; hard target. |
>Serpulina hyodysenteriae variable surface protein (Length=371) KKVLLTAMALLTIASASAFGMYGDRDSWIDFLVHGNQLRARMDQLGFVLGNGTIKGTFGFRSQSAVTKIGNILSGNTGNV DLQTTISAGIGYTSEPFGIGVGYNYTYVNPRLGVHTPVLMINALNNNLRIAVPVQIAVSHDPFNDSAKFPYSSSTKDYMG ISTDIQLRYYTGIDAFNAIRVYFKYGQAGYKTADGANEAFAQSLGFEARFYFLNTPVGNVTINPFIKVAYNTALQGYNRT VRAGEAVTYDASKEYAGLGAGSFDAKDQKWDSNPYDVKAQAVLGITANSDVVSLYVEPSLGYQATYLGKHTSENIDSKVQ HSLAWGAYAELYVRPVQDLEWYFEMDVNNGGTRQPSNIPVYFATTTGITWY |
| 4615 |
PF17667 (Pkinase_fungal) |
 Estimated TM-score=0.59; hard target. |
>Fungal protein kinase (Length=409) KLDMFLASTDAVLLSGEHDWSNVLIIGEHKQNPDDDGSTKTLVQLAGYVREVFGSQPERRFVPGFTICGSIMRLWVSDRS GSYNSEKFDVHKEPERFIRVIAGYALMTDAELGLNTFVKHDSNGKYIVARDVKISLEDKPIASTKAIVCRGTTCYRGRRS DSTEWDYVVKFAWPSDKRQREGELLKLAKERGVTGIATWFNHKQIVIDGDPDTTSHFRRDMKFGRPRKLSSKASWVDGSP ESSRAIQETVVDSLAHCRNEWQRNNVDCIQEAGVDSFTDCESETYDNRVNYCLVTSPAGRPLREYHSVRELLEVLRDAIR GHRSLLEDGKILHRDISENNIIITELPAEGDPKGRLIDLDLAKELDSMPSGARHRTGTMQFMAIEVLEGNGHTYRHDLES FFYVFLWMC |
| 4616 |
PF07268 (EppA_BapA) |
 Estimated TM-score=0.59; hard target. |
>Exported protein precursor (EppA/BapA) (Length=136) EKNYAKAKGSFSKENFNLINERLNNYDFKNEYDRSKFFSYAPKIRGDLRKIGIKEKSVFLDALDVVGYLIKNRFIYVDEY EFECINSLINGSPNTIFNDLIELLDSDEIDYPEKYGEKAIKKFKESYSKDKSNTVK |
| 4617 |
PF12676 (DUF3796) |
 Estimated TM-score=0.59; hard target. |
>Protein of unknown function (DUF3796) (Length=126) PKLGLLGLFGLFGFLGFVPQFFGESGVLDVPFPLIFFCFFGFFGFYYEGKMSGTMIDERYEANVNRAAAIANRVSLTLII MAAILALSLFRIHDSYGMLKLLLAVIGFAFGLSLFLQEYLLYRFEN |
| 4618 |
PF15789 (Hyr1) |
 Estimated TM-score=0.59; hard target. |
>Hyphally regulated cell wall GPI-anchored protein 1 (Length=44) IVSQSGSSFTTLTTFPAAEEPVETEFTSTWEVTNTDGSVSTESG |
| 4619 |
PF13683 (rve_3) |
 Estimated TM-score=0.59; easy target. |
>Integrase core domain (Length=61) DIKLKLIQPGKPTQNAFIESFNGKFRDECLNEHWFCSLAEARIRIAAWRRDYNEHRPHSAI |
| 4620 |
PF09883 (DUF2110) |
 Estimated TM-score=0.59; very hard target. |
>Uncharacterized protein conserved in archaea (DUF2110) (Length=219) LATKCYIDGDARDRALDSLTSLVANDIGDLDVEYDIGVRDDDFISVTVSGDDETVARNVLREEWGEVTPHLSAGETYVGT LEHWDDDGFVLDAGIEVRIPTEELGLGRGSATQIRDRFGLVQHMPLTFVYGGDDEPSRLADEQVDRLYDWTRGSGRVNVN SATRGEVRATVNRAGHAKDIVTVERLGLLEQSIVCRDETDPPGLLASIGSYLPAELKCV |
| 4621 |
PF10107 (Endonuc_Holl) |
 Estimated TM-score=0.59; hard target. |
>Endonuclease related to archaeal Holliday junction resolvase (Length=164) LLFLAVLVLFYKYSRIQGQVERKAREIFESWRQREREDIENWKEQELKRLSDEKAKILFETWRQDEEGNIRTDAIKRSQS VTRGRVTECLIPYFPDFPYNPKDARFLGTPVDLIIFDGLSDADEVQNVVFVEIKTGKAANLSKRERAVRECIKAGRVQYF TIHQ |
| 4622 |
PF15012 (DUF4519) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF4519) (Length=56) MRQLKGKPKKETSKDKKERKQAMQEARQQVTTVVLPTLAVVVLLIVVFVYVATRPN |
| 4623 |
PF05862 (IceA2) |
 Estimated TM-score=0.59; easy target. |
>Helicobacter pylori IceA2 protein (Length=55) MAVVIKVVNGKIQEYENGIHKRTYGSNIVAADTDGHIVAAVTAKGRIEEYENGIH |
| 4624 |
PF17068 (RRG8) |
 Estimated TM-score=0.59; hard target. |
>Required for respiratory growth protein 8 mitochondrial (Length=279) MSELLVKGVKHVYGRKMRKTRPTLPNVESPLVTHFHRWAGKRIRLSFLAAELGTREGQHILPDWNLRGNLFAGLLAAPMR WDRLGNLRVPKSFLMQTKARALEAPSDGKLVKIISVVEERGPGSSSYVPQSKAALQRAKQAMFVPVALQQSDMRYFSSEE IAVDKDTLLAEYSAQLISKVKEGLEVLLSMSGTSHERVELEDWDVVVTYDPDNANDIELRKCKGIPGDPVVIILNMGCLK CTELEELVNLRLRNHEYGLVLKVLKNERLLKFMYRLITF |
| 4625 |
PF10812 (DUF2561) |
 Estimated TM-score=0.59; hard target. |
>Protein of unknown function (DUF2561) (Length=206) MVSRYSAYRRGPDVISPDVIDRILVGACAAVWLVFTGVSVAAAVALMDLGRGFHEMAGNPHTTWVLYAVIVVSALVIVGA IPVLLRARRMAEAEPATRPTGASVRGGRSIGSGHPAKRAVAESAPVQHADAFEVAAEWSSEAVDRIWLRGTVVLTSAIGI ALIAVAAATYLMAVGHDGASWISYGLAGVVTAGMPVIEWLYARQLR |
| 4626 |
PF04149 (DUF397) |
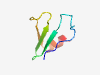 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF397) (Length=50) AWFKSSYSGTEGGECVEVAPATTLVHVRDSKTATGPMLTVSRAAWTGFID |
| 4627 |
PF00839 (Cys_rich_FGFR) |
 Estimated TM-score=0.59; hard target. |
>Cysteine rich repeat (Length=58) PECHHFLWDYKKNLTKDIRFDQAAYEVCKEPLAQLPECSKLETGQGLVISCLMDHKGN |
| 4628 |
PF17012 (DUF5091) |
 Estimated TM-score=0.59; very hard target. |
>Domain of unknown function (DUF5091) (Length=145) LIFKLNNAIQKPTLEAFLNIFEHDGYVRLNMVYISPISLKHLRQKLESFDLIEMFFTTNGLAFIFKTFRLNFFGNMNVVV KNKIYCIEINFDPTIDENCGIPIMSEIEHYLKGCDALFFMTYFVDIDKEILHNSLSESVDFLYKR |
| 4629 |
PF07950 (DUF1691) |
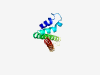 Estimated TM-score=0.59; easy target. |
>Protein of unknown function (DUF1691) (Length=113) LLASHVAMNRVLPLVVDGDSSNIGLGYVAHGFARHAPSAYLAYALLLSVGAGHMVWGMARWLDFAPPANWKKITFDKPTR SRRRRAWWAVNAAAASLAALWAAGGLGVVARAG |
| 4630 |
PF03025 (Papilloma_E5) |
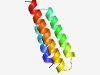 Estimated TM-score=0.59; hard target. |
>Papillomavirus E5 (Length=71) LSLIFLFCFCVCLYVCCHVPLLPSVCMCAYAWVLVFVYIVVITSPATAFTVYVFCFLLPMLLLHIHAILSS |
| 4631 |
PF11055 (Gsf2) |
 Estimated TM-score=0.59; very hard target. |
>Glucose signalling factor 2 (Length=374) MEIYVRLNNDLENDNAFQISREDTIKDKLVHIFPRDKNPKSLSDVMVLRPTVFHNYAPDYFCKSVHQGYLSEGGCLLFHY DAADDKYLTKLDEDKPLFDQLWPGQLIVPKWSYNKWNLFTYTAIILLWLYTDLPDVVSPTPGICVTNQLSRMIIPILEYM ERPTLASKLREEIQVNYSSFWAQWAFFLLHVIKVSVLTLFLRLGIANPITFNPIKVYQFQNGNAVSSDHVKAILKSIGWI GSRRGTYDQYQSNFYSYTIKKYGGPVQAYRAGVIKTAANPGVELNAGEGFQTPLDQRFTNSTFETMESQGKFVLSEEYFI ELEDNLKENLNKCNGDIGKMNEEIRRFRKFGIYEPSEKLAKMVQKRKQVAKEQK |
| 4632 |
PF05353 (Atracotoxin) |
 Estimated TM-score=0.59; trivial target. |
>Delta Atracotoxin (Length=42) CAKKRNWCGKNEDCCCPMKCIYAWYNQQGSCQTTITGLFKKC |
| 4633 |
PF01573 (Bromo_MP) |
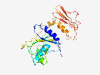 Estimated TM-score=0.59; very hard target. |
>Bromovirus movement protein (Length=276) SSSSFSVSYAEEMLLADEVSKTNSMSILGPNQLKLCTQLVLSNGAAPVVLSLVSKEKKSILNRMLPKIGQRMYVHHSAIY LLYMPNILKSSSGSITLKLFNEATGELVDVDTDHDATQACIFAGRYPRSILAKDAAKGHDLKLVVHAVASTNMNSAVGVL YPIWEDELSRKQILERGADFLKFPIAKTEPVRDLLNAGKLTDFVLNRTRLGVGSKSDPSPVLLEPRAKIAGKAKTLFIPE GPSVPSTTINGMAPTVRIDAGSPKGLGVPKGFTYES |
| 4634 |
PF04932 (Wzy_C) |
 Estimated TM-score=0.59; very hard target. |
>O-Antigen ligase (Length=159) ASIIFAGLVASIARSNWISFVVYSAVLFIYCLKKKKYIKRYMMCLGIFVVVFLLLNLLTDSSIASRLNSIGLDAKKITMK DNDTAGSSRLYIWKRGITFIPEHPLLGSGPDTFGVVFLEKYFDELKYMQDITGGIVDKAHNEYLQLAVTSGIPALLIYL |
| 4635 |
PF15596 (Imm57) |
 Estimated TM-score=0.59; hard target. |
>Immunity protein 57 (Length=109) MADKAILWALISASTKEGRKACSLSYFACKAAEAELGLAYMAANDNKEFLTSLSNIMRYKIDAGLSESYTCYLLSKGKII RPYLKNLNPLQLAADCIETVNKIKDKNKK |
| 4636 |
PF04718 (ATP-synt_G) |
 Estimated TM-score=0.59; easy target. |
>Mitochondrial ATP synthase g subunit (Length=92) KTPALVNAAVTYSKPQLATFWYYAKVELIPPTPAEIPRAIQSLKEIVNSAQTGSFKQLTVKEAVLNGLVATEVLMWFYVG EIIGKRGIIGYD |
| 4637 |
PF14072 (DndB) |
 Estimated TM-score=0.59; hard target. |
>DNA-sulfur modification-associated (Length=336) FPAIRGIQAGREFYVSMCPLRLLPKIFTFDDEELIPELRAQRVLNKARIPEMARYIISNMNDYVFSAITASIDAKVRFEA LGQSGEANRVGSLHIPMDARFIINDGQHRRAAIELALREKPDLADETIAVVFFLDKGLNRCQQMFADLNRYAIRPSKSLG ILYDYRDDMALLAKSVVLKSPVFKDVVEMEKSTLSPRARKLFTLSSIYFATNELLESIDGSQEEKTNMAINYWVEVAKYI KEWNLVRARKMTSAEVRREFIHSHGIALQSIGKVGNHLLRNNPANWKRTLKKIENIDWSRSNSQLWEGRCMIGGRVSKSS HNVTLTTNAIKAALSL |
| 4638 |
PF07384 (DUF1497) |
 Estimated TM-score=0.59; hard target. |
>Protein of unknown function (DUF1497) (Length=59) MGYYDTRNEARRISKLASQNISSEQTKKEFELDRQSKFNQEMQAEFHEKIKKLGEKNGS |
| 4639 |
PF06461 (DUF1086) |
 Estimated TM-score=0.59; easy target. |
>Domain of Unknown Function (DUF1086) (Length=136) RAKRRPDQKSERDRPLPPLLARVGGNIEVLGFNARQRKAFLNAIMRFGMPPQDAFNSQWLVRDLRGKSEKNFKAYVSLFM RHLCEPGADNAENFADGVPREGLSRQHVLTRIGVMSLIRKKVQEFEHINGYYSIPE |
| 4640 |
PF08756 (YfkB) |
 Estimated TM-score=0.59; hard target. |
>YfkB-like domain (Length=151) MYPSDFAANLEVLSLDEIREAIHHLLDIRDESVWMLFGTLPFYPCSTNPEDQALLKRLYESKNVTLRNDPDGRSRLNVNI FTGDVIVTDFGDTPPLGNIQHDRLPDVFEKWLETPIAKSLNCHCPQVKCLGPNILVKNAYYKNIDFTQRKA |
| 4641 |
PF12813 (XPG_I_2) |
 Estimated TM-score=0.59; easy target. |
>XPG domain containing (Length=241) PAFLVPAVLDALRGSKYGPITEVMGGEADGYCAVHVRKSGGLVLTSDSDLLVHDLGENGGVIFFTDIDLDSENSKLVAPQ FRHAEICRKLSIKPDVGFSYMAFEISADPHLTLEQAAERSRRGEAVMYSREEYDSFIKTYLLPETAPKTVATCGLQLDPR ISELVLRYLQITTTASKEKDASLEMFLPFLLDCPSRTSAWEVSKPIRKLAYSLLQSGQKTSLKTVSEMRRLQTLSSGSQV D |
| 4642 |
PF16032 (DUF4788) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF4788) (Length=226) IVTQLRLNKYEFDEPQNLMVRAKFNKIGLAITSSRINVTEFRAGRDIEFKIEPQMLRNNLKKIGMLVTVQYGSISLGTGT LMFPQSFIDKIDYGMSDLVHADTCSIQSRNTEVGFVELLCRLIIKCEDETLGLEACQKRCEHLDEEINQADIMFLVDKPE LDAIPCEPCSDELKQEEDDAQLRWDLSRYRSFNQRVTQSPEDTSAMQACSQLKNLTAKCGEIIDSV |
| 4643 |
PF03072 (DUF237) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF237) (Length=138) LRWELINRANIQLQLQNVKVRENNFEVNGWGVPVSINWNDHNNGINYRATTPWTYGFEITMNYKGSYGLKGIYWTLANWG LGGIPPEWSGDMELKFQIDGKLANWITQKQDYPGSLFQFQNDKLLFTLHVVQRITVKD |
| 4644 |
PF05774 (Herpes_heli_pri) |
 Estimated TM-score=0.59; very hard target. |
>Herpesvirus helicase-primase complex component (Length=127) WLESVQFGLNILLCTKCNQSICDVLETLLQLYFKNRHNAKFWLVPQHFLTSKPQKPNIPSDCVAPTLFFFTTDGPLCWHQ KLNVPLNINYEEYIHKAITVFEKVPSLTINKDMYIKIESWKNILCLL |
| 4645 |
PF13935 (Ead_Ea22) |
 Estimated TM-score=0.59; hard target. |
>Ead/Ea22-like protein (Length=128) SNIDKQALREAAEKATPGNWHRSSSRFNGITATPFSLCGEEVMLAHTVEKRDAEFIAAANPATMLALLDENLQLQREKDA IEAVALALRDDMRQAREQLEAAEKRNAEQREYYEGVIADGSKRIAELE |
| 4646 |
PF19164 (DUF5846) |
 Estimated TM-score=0.59; hard target. |
>Family of unknown function (DUF5846) (Length=115) MYNYYFIHSTTDYNNLKDILESGIIYPGKYLRKDQQKLTIDSEFVYANLYFEDLNNIPNLPNFGLLIHPKILYENGMYFH KGWTGPKEGDIHIKATNTPKQISKKLEKIREFVKN |
| 4647 |
PF17023 (DUF5098) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF5098) (Length=468) LHVLKEKIEWIKAFLEHIELVSKRSGELLFRATQIPRKSSWKDPFILELLEYSDKQRVIYDELSHLYTITIMNDVNELTS QINEEFAAINEEREKFDKNTKELYTVLEAKRNSYIEALKGNKDDPWLKLADLRSAFNKYYIEKNRSNELFRAQSFESEKN HDYYKKKYENIVKNFFKIQSSHFMNLSSTIDYDFENKLSELKDEIYYDFPSQHEEKSTESENEIERFEKDFKEILEQIYS SFGKKEDQISIKKPVIIKCGIYKQKKGKNDWKPVFIVLSNKLFIHAFDMDVIMRKSPNLRARFSLLYKKLNDGSYKKFIS FFDKPKSRGLNFNEELELKELSTAIISETDLISYHLPNISMNLNHWIVKLDKEKFEMIFSEKRVNTFTSIFINNTLRIKS FALKDLYELYFGIFDHNKILIDNSNIEISDCNVVANESVEKEKEENIEVPKPTNVPDIWKSLDDHNPW |
| 4648 |
PF12345 (DUF3641) |
 Estimated TM-score=0.59; hard target. |
>Protein of unknown function (DUF3641) (Length=136) LPPAQQALESSYKTHLKEHYDIDFNRLYTITNMPIKRFGSTLISKGSFDDYMQLLVDSYNPDTLEQVMCINTISVDWQGY LYDCDFNQMLDLPLADNQGKMHIADLVATDLQSDKIATRQHCYGCTAGQGSSCGGA |
| 4649 |
PF06919 (Phage_T4_Gp30_7) |
 Estimated TM-score=0.59; hard target. |
>Phage Gp30.7 protein (Length=121) MNYINFERKYVSNGIAGSIDTICLWKHQNGSVCEIEQYMTPNYVYMRFENGITVSITMKGSNFKIALDDDFRQRDLGTHP CWNGANRKLLVKTWIRHILSNRAKPEHLEAIFDVVLNEFDI |
| 4650 |
PF18825 (LPD13) |
 Estimated TM-score=0.59; very hard target. |
>Large polyvalent protein-associated domain 13 (Length=137) DGTLKVDKVNSINELTDADFENPTRNVELPAIPENVDKAIGANGKPVVIKKNIFEKNKKSHNFSNRQSRDILTNALYNTD LVGQTQPTKRPKHWVAIQLDEKSPITVLEVNENKDNVEVVGWYTLDNRNLERIKRQA |
| 4651 |
PF13339 (AATF-Che1) |
 Estimated TM-score=0.59; hard target. |
>Apoptosis antagonizing transcription factor (Length=145) EVEKGKAVKNQLALWDQLLEGRIKIQKALVTANQLPQPQTFPEFKRRGGGEFTAALKNTHKALKALQRSLLELHDQLLHQ NADTRSIDTGEDEEITSDDEKEGSAQQARAPKRKLEMAEYPDFMAKRFAAFQPYCNATLQKWHDK |
| 4652 |
PF10758 (DUF2586) |
 Estimated TM-score=0.59; easy target. |
>Protein of unknown function (DUF2586) (Length=369) FPSVQINALNQLSGETKEIERHALFVGVGTTNQGKLLALTPDSDFDKVFGETDTELKKQVRAAMLNAGQNWFAHVYIAQE DGYDFVECVKKANQTASFEYCVNTRYLGVDKASIGKLQECYAELLAKFGRRTFFIQAVQGINHDQSDGETWDQYVQKLTT LQQTIVADHVCLVPLLFGNETGVLAGRLANRAVTVADSPARVQTGALVSLGSANKPQDKDGNELTLAHLKSLETARYSVP MWYPDYDGYYWADGRTLDVEGGDYQVIENVRVVDKVARKVRLLAIAKIADRSFNSTTSSTEYHKNYFAKPLRDMSKSATI NGKDFPGECMPPKDDAITIVWQSKTKVTIYIKVRPYDCPKEITANIFLD |
| 4653 |
PF18729 (HEPN_STY4199) |
 Estimated TM-score=0.59; hard target. |
>STY4199-like HEPN (Length=282) MILSQSHQIREQFERCLGIIRQASVEILLLLKVHVAEGKDPRWFLEQLDTARLGLGGWAAVAKRLNLNDAQLSQFTLQLR LLQQRVPQYESGQEVSDNQLIAATRFVTSLEHLRLQQPLLTYSTELSDSDQAQQQQAQKQVRALEMMIKGLIIQAWPDPV RLNNHLKTLFSADKVRRWLRQGERNDVLSGMLFSELALMLVDKKEFSRSYAPLFNDPSMLTLLVEPRKTLQTFLDDLRVI RNKIIAQQPLSSAQTLLLENYYQQIAGPVQRAFEEGRTRVNP |
| 4654 |
PF12030 (DUF3517) |
 Estimated TM-score=0.59; hard target. |
>Domain of unknown function (DUF3517) (Length=351) VFDFMLSFVKLGRHELAAFLGHPDFLRWLLWIVLADANTEALLPNQFAKLASVISRRMPNRPPSYESIIQLLDFLLAHMR CGYTPSGQPTGVTDSTKRLDPDGGLDQPFALTKAEADLLHQMGPRTFPVNIFIDRLISIGQNPAASDSIIASLMKQSRRM ENSVFRTLLHRITGQIAHNVGPYLHVASAVFCRVASDAGLISDLIKHVSQQCLILQNSEGKAFLDFMRETFDAPRERSGE TRHQVIMAGLDSVPDWAPGLLGYFDTSVIDETERFLRDKIFQPRISQSPSGNESEEDREKAEKVRQTARALGTRCLWYLR DNYVARNTEVTERSVGGLQRVIKQCSKYFNL |
| 4655 |
PF12962 (DUF3851) |
 Estimated TM-score=0.59; very hard target. |
>Protein of unknown function (DUF3851) (Length=125) MNPNILNKNPLMFFDRAVNAQRSQLLTVMADAVSECRTAADQAAELNETGQVGLLRLAEIWSAIRAKEGMGGLVLEGTEA KILSDVVAQFYAYLSGCMFNDPVGMAIYAELHYMMSSLMLGEWFE |
| 4656 |
PF10497 (zf-4CXXC_R1) |
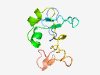 Estimated TM-score=0.59; easy target. |
>Zinc-finger domain of monoamine-oxidase A repressor R1 (Length=99) YDKVLGNTCHQCRQKTIDTKTVCRNQSCGGVRGQFCGPCLRNRYGEDVRSALLDPDWMCPPCRGICNCSYCRKRDGRCAT GILIHLAKFYGYDNVKEYL |
| 4657 |
PF05840 (Phage_GPA) |
 Estimated TM-score=0.59; hard target. |
>Bacteriophage replication gene A protein (GPA) (Length=324) HQIQTLATMTAAMFSGTFEKLCDGFGATDGELTMDVTLKAYQMLARMALHLHAMPPHYDALTTDKDRRNEPDTELLPGAI LRLTCAEWWKRKLWLLRCEWREEQLRAACLVSRKTSPYLSQDALSEFRAQREKTRDFLKSFMLENEDGFTIDLETVYYAG VSNPIHRKAEMMATMKGLELLAEARGDRAVFLTVTCPSKYHATTENGHPNPKWNGATMRDSSDYLVNTFFAAVRKKLNRD GLRWYGIRTVEPHHDGTVHWHMMVFAHPEEIDTIVSHTRDIAIQEDRHELGNDITPRFKVEYVDGSKGTPTSYIATYIGK NLDS |
| 4658 |
PF15729 (ALS2CR11) |
 Estimated TM-score=0.59; hard target. |
>Amyotrophic lateral sclerosis 2 candidate 11 (Length=419) KGSDSEELEITQDTPNLVPFGDVVGCLGIHIKNCRHFTPKISVQHFANLFIRISINKAAKCTKMCSLLSKNNEKNTVIKF DEVKYFSVQVPRRYDDERNNILLELIQYDNREKRAFLLGSVQIHLYEVIQKGCFIEEVQVLHRNIFVCRLEVEFMFSYGN FGYGFSHQLKPLQKITEPSMFMNLAPPPERTDPVTKVITPQTVEYPAFLSPDLNVTVGTPAVQSSNQPSVVRLEKLQQQP RERLEKMKKEYRNLNTWIDKANYLESILMPKLEHKDSEETNMDEASENLKSNQPEEELENIVGVDIPLVNEEAETTANEL LDNDSEKGLTIPTLNQLDQDNSTADASKSDKSTPSPTEVHSLCTISNQETIKAGRIPPLDERQSESMPDRKMKNVFFPSE VKLKDNYPSILKADSSLSE |
| 4659 |
PF13821 (DUF4187) |
 Estimated TM-score=0.59; easy target. |
>Domain of unknown function (DUF4187) (Length=53) EDEEEEELEPSEKLEHLTMYLRRTHQYCIWCGTKFDDDRDMADNCPGDTAQDH |
| 4660 |
PF12421 (DUF3672) |
 Estimated TM-score=0.59; hard target. |
>Fibronectin type III protein (Length=142) LTPDGKLTAKNADISGSVNANSGTLSNVTIAENCTINGTLRAEKIVGDIVKAASAAFPRQRESSVDWPSGTRTVTVTDDH PFDRQIVVLPLTFRGSKRTVSGRTTYSMCYLKVLMNGAVIYDGAANEAVQVFSRIVDMPAGR |
| 4661 |
PF07305 (DUF1454) |
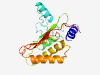 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF1454) (Length=187) LLLSLALTFPVWAEDAPPQDRSDHLPAAPYLLPGAPTFDMTIAQFREKYNAANPDLPLMEYRAIDNQDDKSNLTRAASKI SETLYASTALEQGTGKIKTLQITWLPVPGPDEKNAQEKAMAYMTALLRFFSPGLSPEDGRKKLDSLLSAGKGSRYFSRNE GALRFVVADNGDKGLTFAVEPIKLALA |
| 4662 |
PF18550 (NitrOD2) |
 Estimated TM-score=0.58; easy target. |
>Nitrososphaera output domain 2 (Length=104) MVDAETLAEAILDSLKEIFGPPVFHSLMELIAEDYLGEMDARTAIIERPDLFERAFVGLLGEAGKKILADICEGLCAEFL LDENAADLKTGDLAECMAIIIPKS |
| 4663 |
PF11134 (Phage_stabilise) |
 Estimated TM-score=0.58; hard target. |
>Phage stabilisation protein (Length=469) QQLPMMKGMGKDFKNADYIDYLPVNMLATPKEILNSSGYLRSFPGITKRYDMNGVSRGVEYNTAQNAVYRVCGGKLYKGE SEVGDVAGSGRVSMAHGRTSQAVGVNGQLVEYRYDGTVKTVSNWPADSGFTQYELGSVRDITRLRGRYAWSKDGTDSWFI TDLEDESHPDRYSAQYRAESQPDGIIGIGTWRDFIVCFGSSTIEYFSLTGATTAGAALYVAQPSLMVQKGIAGTYCKTPF ADSYAFISHPATGAPSVYIIGSGQASPIATASIEKIIRSYTAEEMATGVMETLRFDSHELLIIHLPRHVLVYDASSSQNG PQWCVLKTGLYDDVYRGVDFMYEGNQITCGDKSEAVVGQLQFDISSQYDKQQEHLLFTPLFKADNARCFDLEVESSTGVA QYADRLFLSATTDGINYGREQMIEQNEPFVYDKRVLWKRVGRIRKNIGFKLRVITKSPVTLSGCQIRIE |
| 4664 |
PF16292 (DUF4938) |
 Estimated TM-score=0.58; easy target. |
>Domain of unknown function (DUF4938) (Length=302) PLRIRHLAALYGPNIYRPQPGVVLRVGAAADYSDRLRTALKGGALAIGLVIAQLHVEAKPDNDDMLMSAFFTTPEPAIGA ELCRYVVDGINAELGHDESWDPDEPLLALRDRRQAQALPVAALQLMANARHRDIPTTILDDGRLLIGQGERGWIVAPADL RAAPDLQPPWSRLGRILIIAVTGERYRQQAIAEQAARHQGIILDQANRASVITALADPACTALIVGFDTDSLIREGLPFD RCDLAIITDRAGSRPPTAADDDEWLRALGLPMLCSVAPTLINLTDPALHPLLSYAPNGVIGW |
| 4665 |
PF10234 (Cluap1) |
 Estimated TM-score=0.58; hard target. |
>Clusterin-associated protein-1 (Length=269) RALGYPRLISMENFRTPNFPLVAEVLIWLVKRYEPNADLPMDVDTEQDRVIFIKSVAQFMATKAHIKLNTKKLYQADGYA VKELLKVTAMLYNAMKTNTGQQEEEQEEETTAMSFDISSRIADLKASRQLASEITTKGASLYDLLGREVELREQRSVAIN KPLEINEIEKHLKVSIRAVEQEIKKTEHMLNNVASDEANLDAKISKKKLELERNRKRLQTLQSVRPAFMDEYERLEEDLQ KQYESYLQKFRNLGYLEQQLEEHNRAEQD |
| 4666 |
PF13824 (zf-Mss51) |
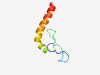 Estimated TM-score=0.58; easy target. |
>Zinc-finger of mitochondrial splicing suppressor 51 (Length=59) CPHHDHQPGGGMAPAHVDFECPDCGIPVYCSKEHWADDYEAHLQICDTLRQVNEDDHDL |
| 4667 |
PF12156 (ATPase-cat_bd) |
 Estimated TM-score=0.58; easy target. |
>Putative metal-binding domain of cation transport ATPase (Length=84) SCYHCGLPVPSGSEFYAAILGQRRRMCCHGCQAVAESIVDNGLEDYYQHRTELPQTAEELVPEELRQLELYDHPEVQKSF VHDE |
| 4668 |
PF18870 (HEPN_RES_NTD1) |
 Estimated TM-score=0.58; very hard target. |
>HEPN/RES N-terminal domain 1 (Length=126) NYQDGYCDYCGKELKVVPLESLMEFIMDGISNFYEDAANFMSYNSREGGYLGEIYTPDELIQEHIELNAEPFEVIEDVVN SIEDIAWAQPDLYYDNIKDDLEFQWNYFKEIIKHKSRYLFSSGDSS |
| 4669 |
PF07149 (Pes-10) |
 Estimated TM-score=0.58; hard target. |
>Pes-10 (Length=416) MNPTEMQYQVLAMMIKARVDSEVITKSINLLSNHPISTDAFDRCNLPCLIMRYASHIGAARDLLLIYRIKKTSEMKEDQP QLLETFYKANMTIFEGAEPPAVLNEFLRKMLEDEDEEYVRLAYRLLTDIDFHMEFYLNIIARARRDRLRIFEAGWLVDKV EKILRYEEIVDEDMDSDAEEDEEIAGPALPPMHIVDENLEEEGAPEEDSDADDSFDWVSEDEEDDESDSDESGYEDESDE MVEERLDEEEEEFNQFGAAIGEVFMRNLARSLKSGNERKIESAIEAISHFQLHPSVYRKYEISRLIFELSFHNFNAMVLF IQLERREEEVYSQQKLEVFHEFLNYLNSDNIDSVNVYKVMAEYLTDKRFEKQVVKVFLNGSVTREQFEKWGIKEYLLNCS EKSDDVKELIEKIHKL |
| 4670 |
PF05112 (Baculo_p47) |
 Estimated TM-score=0.58; very hard target. |
>Baculovirus P47 protein (Length=308) EHTTQFLPQACKLEDEVLVLYAIYLNGFDYTLPRFVKREVQVNADGFVRFDYNIKIFDFARFTNMTQATPEDIDDYINLT RIDSLSDHDLKMLHLVCRDRWYKGDVARLRRILQQKDVDDLVKFVCNVMWERAYEDHYTLGQQLSIRITTKLIQSGLDFK HQPDTTAPVSVRGWEDATFEKYLQSITSISEVIKRHVFSKKYICLEVAASYWSDAVELLQQENFKIILNSKTPYVLLIEM DDDKNSMMYLRKLAHLLENKIVNLLFVTDVEFYFKNGNFIFYLYNSLKFYYYCLKNKFAFENVDKEIF |
| 4671 |
PF15562 (Imm17) |
 Estimated TM-score=0.58; hard target. |
>Immunity protein 17 (Length=62) LLFTFFLGIYFSAILLNKDWVLFPGDGKYNFDYFIKIFGRTIVRIFIGTLTLIGIGITFSLF |
| 4672 |
PF07580 (Peptidase_M26_C) |
 Estimated TM-score=0.58; hard target. |
>M26 IgA1-specific Metallo-endopeptidase C-terminal region (Length=757) NLAEYNLGNTGLLYTPNQFLYDRDSIVKEVLPELQKLDYQSDAIRKTLGISPEVKLTELYLEDQFSKTKQNLGDSLKKLL SADAGLASDNSVTRGYLVDKIKNNKEALLLGLTYLERWYNFNYGQVNVKDLVMYHPDFFGKGNTSPLDTLIELGKSGFNN LLAKNNVDTYGISLASQHGATDLFSTLEHYRKVFLPNTSNNDWFKSETKAYIVEEKSTIEEVKTKQGLAGTKYSIGVYDR ITSATWKYRNMVLPLLTLPERSVFVISTMSSLGFGAYDRYRSSDHKAGKALNDFVEENARETAKRQRDHYDYWYRILDEQ SREKLYRTILLYDAYKFGDDTTSGKATAEAKFDSSNPAMKNFFGPVGNKVVHNQHGAYATGDGVYYMSYRMLDKDGAITY THEMTHDSDQDIYLGGYGRRNGLGPEFFAKGLLQAPDQPSDATITINSILKHSKSDSTEGSRLQVLDPTERFQNAADLQN YVHNMFDLIYMMEYLEGQSIVNKLSVYQKMAALRKIENKYVKDPADGNEVYATNVVKELTEAEARNLNSFESLIDHNILS AREYQSGDYERNGYYTIKLFAPIYSALSSEKGTPGDLMGRRIAYELLAAKGFKDGMVPYISNQYEEDAKQQGQTINLYGK ERGLVTDELVLKKVFDGKYKTWAEFKTAMYQERVDQFGNLKQVTFKDPTKPWPSYGTKTINNVDELQALMDQAVLKDAEG PRWSNYDPEIDSAVHKLKRAIFKAYLDQTNDFRSSIF |
| 4673 |
PF17282 (DUF5347) |
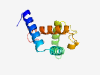 Estimated TM-score=0.58; hard target. |
>Family of unknown function (DUF5347) (Length=102) VQLSAGQRVNGLNHIAAIRTKLWGDNCGNELKRFMADMRDKRDTQYEQNKRALGAIFFLANIRSERHDVEFDELTSDEKY ALISAMNHIRAVVSLFPKKLTL |
| 4674 |
PF01666 (DX) |
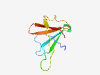 Estimated TM-score=0.58; very hard target. |
>DX module (Length=75) FQTNMDCNANEEVDQKFDFMFCHNDTGNLWVMGQYNVNGDEVIKHWTHCNTNNDCGEGLVCVKEDLCRYRCYDDP |
| 4675 |
PF11296 (DUF3097) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF3097) (Length=258) WKRSAPVPEVPAEPDLVVEEVASGFCGAVIRCEKTAQGPTVTLEDRFGKHRVFPMEPRGFLLEGKVVTLVRPSAGGPARP TRTASGSVAVPGARARVARAGRIYVEGRHDAELVERVWGDDLRIEGVVVEYLEGVDDLPAIVAEFAPGPDARLGVLVDHL VPGSKESRIAAEVTGDHVLVVGHPYIDVWEAVKPSSVGIDAWPVVPRGQDWKTGVCRALGWPENTGAAWQRILSTVRSYK DLEPQLLGRVEELIDFVT |
| 4676 |
PF03086 (DUF240) |
 Estimated TM-score=0.58; very hard target. |
>Domain of unknown function (DUF240) (Length=121) MKFHTVYVGKNTKIDLDFALQAQTNNFSSLEELRESFTNSGQTLSTQLFWKPVIDKLITDEGNDLTTIARTAIGENLFDL KVNLTDSVIDGTVLTKARKSFEERILNPFIEQRKEAKRIHD |
| 4677 |
PF14402 (7TM_transglut) |
 Estimated TM-score=0.58; hard target. |
>7 transmembrane helices usually fused to an inactive transglutaminase (Length=246) SLLDVEGGENSNVSFSIIESTQPANSRTLVEKANAKAALVDFSIYSLPIEQQNIFKSILLVPIGALIVVLMRIIVGIRTS GTFMPILIAIAFIETSLVTGLAIFLIIVSIGLFIRYYLSHLNLLLVARISAVVIVVIGIMAGMSILSYKLGFSEVMKVTF FPMIILAWTIERMSILWEEEGPKEVFIQGGGSLLVATLAYLSMTNTYIEHLTFNFPELLLALLGGILMLGQYTGYRLTEL RRFQPL |
| 4678 |
PF06166 (DUF979) |
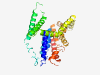 Estimated TM-score=0.58; easy target. |
>Protein of unknown function (DUF979) (Length=312) EYIYYIAGIFLSIIAFLSFKDKSNSKRLLTGTFWGLFAISFIFGKVIPPMYMGVLVIIIALIAGFGGVRMGSYHQTTEEE RKASAKKIGNKLFIPALLIPLVTIIGTVVIKDMKIGNLFLLDNKNVTLVSLGVACVVSLIVAMMLTKEKGIQPVKESRRL LDAIGWAAVLPQMLATLGGLFAVAGVGKVVSDLISSAIPVDNRFFVVLAYVLGMAIFTMIMGNAFAAFPVITAGIGLPLL VHMHGADPAIMGAIGMFSGYCGTLLTPMAANFNIVPTALLDLSDKYAVIKAQAPTAIILLTVNIFLMYFLVF |
| 4679 |
PF03935 (SKN1) |
 Estimated TM-score=0.58; easy target. |
>Beta-glucan synthesis-associated protein (SKN1) (Length=504) FPLHIDEKEPDDYLHNPDPVQDALYDKHRFWNDLKNADRRSAWGLVGFIILITAAVGVFIILPVLTFTGVEVHRTPQEYE VLTDYQYPMLSAIRQTLVDPDTPEDALTITAKDGSQWVLVFSDEFNAEGRTFYEGDDQFFTAPDLNYAATRDLEWYDPDA ATTADGTLQLRMDAFNNHDLFYRSGMLQSWNKFCFTQGRIVMSARLPNYGTVSGLWPGMWTMGNLGRPGYLASTEGVWPY SYDTCDAGITPNQSSTDGISYLPGQRLNSCTCKGESHPNPGVGRGAPEIDVIEAAIGTDPTGKTPNIGVASQSLQLAPFD IWYMPDYSYIEIYNSSVTTMNTYAGGPFQQALSGTTTLNSSWYEFGPFEHNFQRYGFEYLNDDDKGYLTWFVGDDPTFTV YPQSLHPNGNVGWRHLSKEPMSIIMNLGISNSWAYIDWPSITFPVTFRIDYVRVYQPKDQINVGCDPPDYPTYDYIQQHL NIFENVNLTLFEMGGYTFPKNKLT |
| 4680 |
PF19133 (DUF5816) |
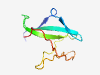 Estimated TM-score=0.58; hard target. |
>Family of unknown function (DUF5816) (Length=72) DGETVYVAREEDERGSKGPFFVVYRGEDRERRYGYFCGNCERIDNAMDAMGRIECNVCGNIRKPTEWDAAHE |
| 4681 |
PF13701 (DDE_Tnp_1_4) |
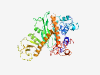 Estimated TM-score=0.58; easy target. |
>Transposase DDE domain group 1 (Length=414) QLSFDGGHLTQYGGLIFFQELFSQLKLKERISKYLATNDQRRYCRYSDSDILVQFLFQLLTGYGTDYACKELSADAYFPK LLEGGQLASQPTLSRFLSRTDEETVHSLRCLNLELVEFFLQFHQLNQLIVDIDSTHFTTYGKQEGVAYNAHYRAHGYHPL YAFEGKTGYCFNAQLRPGNRYCSEEADSFITPVLERFNQLLFRMDSGFATPKLYDLIEKTGQYYLIKLKKNTVLSRLGDL SLPCPQDEDLTILPHSAYSETLYQAGSWSHKRRVCQFSERKEGNLFYDVISLVTNMTSGTSQDQFQLYRGRGQAENFIKE MKEGFFGDKTDSSTLIKNEVRMMMSCIAYNLYLFLKHLAGGDFQTLTIKRFRHLFLHVVGKCVRTGRKQLLKLSSLYAYS ELFSALYSRIRKVN |
| 4682 |
PF04148 (Erv26) |
 Estimated TM-score=0.58; hard target. |
>Transmembrane adaptor Erv26 (Length=205) MWILPLLGYLGSVVGFCFMTLAIASGLYYLSELVEEHTVMAKRFLTRLIYSIMALQLLLCVVDRFPFLPTLLGIFSHFVY LGNMRRFPFVKLTDPLFLASCVLVLLNHYVWFKHFSHHQERAYQNMTSYYDTPDDIPSFTEIASYFGLCVWLVPFSLFVS LSASDNVLPTMGSEPSTSAAAGMVDTGNARRRGQGLVKALVDSVR |
| 4683 |
PF14210 (DUF4322) |
 Estimated TM-score=0.58; hard target. |
>Domain of unknown function (DUF4322) (Length=66) VIPNLPHQNNVQRIGYKLLSMLDFKGKKAEDVGKTLISACLWNSSIENKSSAYNVSPQTVRNYVEE |
| 4684 |
PF07046 (CRA_rpt) |
 Estimated TM-score=0.58; easy target. |
>Cytoplasmic repetitive antigen (CRA) like repeat (Length=42) EAEKRKAAEATKVAEAEKQKAAEATKVAEAEKQKAAEATKVA |
| 4685 |
PF06771 (Desmo_N) |
 Estimated TM-score=0.58; hard target. |
>Viral Desmoplakin N-terminus (Length=90) KYGGTDVNTRTVHDLLNTINTMSARIKTLERYEHALREIHKVVVILKPSANTHSFEPDALPALIMQFLSDFAGRDINTLT HNINYKYDYN |
| 4686 |
PF14768 (RPA_interact_C) |
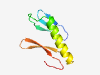 Estimated TM-score=0.58; hard target. |
>Replication protein A interacting C-terminal (Length=79) ICPLCERNSLLENKGVIVCKCGLRINTEQDGITLKNVQESLEAGANEHSAVCGSKPVFSLDEKFGTSTLTMSCQDCDFL |
| 4687 |
PF11083 (Streptin-Immun) |
 Estimated TM-score=0.58; hard target. |
>Lantibiotic streptin immunity protein (Length=93) IAQLDIELTNLQEKIATLNKMAEVVVNLQSDNQDTKQLAKYECSKMNLSQNVTIGQIESEIEIIQNQLDNKIEEYENAVR KLDEYVRVLNMDK |
| 4688 |
PF16357 (PepSY_TM_like_2) |
 Estimated TM-score=0.58; hard target. |
>Putative PepSY_TM-like (Length=200) YWLKTLHQWHWISSAVCLLGMLLFSVTGITLNHASQIESRPAVTARELQLPPELKALVAPDASSPSSPRAPLPARLADWV DTQLAVDVRGRDAEWSDEELYVSLPRPGGDAWLRIDRESGAAEYERTDRGWISYLNDLHKGRHTGVAWSWFIDVFAVACL VFCLTGLFILKLHAANRPTTWPIVGLGLVIPLLLALLFIH |
| 4689 |
PF08081 (RBM1CTR) |
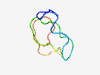 Estimated TM-score=0.58; hard target. |
>RBM1CTR (NUC064) family (Length=45) SNSGMGGRGSPSRGRGNYRGPSHGMSVSSWRRDRMSPKHGGYATK |
| 4690 |
PF13746 (Fer4_18) |
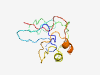 Estimated TM-score=0.58; easy target. |
>4Fe-4S dicluster domain (Length=114) AREQVCIYMCPYARFQSVMFDKETLLVSYDAKRGEGEAGRKPLKQGLMTLEERHDQGVGDCIDCKLCVQVCPTGIDIRDG LQLECISCGLCIDACDQIMTKRGWPTGLIRYASE |
| 4691 |
PF16104 (FPRL1_inhibitor) |
 Estimated TM-score=0.58; easy target. |
>Formyl peptide receptor-like 1 inhibitory protein (Length=105) FFSYEWKGLEIAKNLADQAKKDDERIDKLMKESDKNLTPYKAETVNDLYLIVKKLSQGDVKKAVVRIKDGGPRDYYTFDL TRPLEENRKNIKVVKNGEIDSITWY |
| 4692 |
PF13542 (HTH_Tnp_ISL3) |
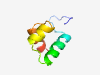 Estimated TM-score=0.58; trivial target. |
>Helix-turn-helix domain of transposase family ISL3 (Length=50) ETSIVEKNHQISNLVRQKVAQLLTEKVSLTDIARRLRVSTSTVYRKLDQF |
| 4693 |
PF06260 (DUF1024) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF1024) (Length=82) MNNREQIEQSVISASAYNGNDTEGLLKEIEDVYKKAQAFDEILEGMTNAIQHSVKEGIELDEAIGIMVSQVIYEYKEELE NE |
| 4694 |
PF05715 (zf-piccolo) |
 Estimated TM-score=0.58; easy target. |
>Piccolo Zn-finger (Length=57) LCPVCKNTELNLKSKDSLNHNTCTQCKSEVCNMCGFSPPDATAKEWLCLNCQMQRAL |
| 4695 |
PF10762 (DUF2583) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2583) (Length=88) MKRKNASLLGNVLMGLGLVVMVVGVGYSILNQLPQLNLPQFFAHGAILSIFVGAVLWLAGARIGGHEQVSDRYWWLRHYD KRCRRNQH |
| 4696 |
PF17242 (DUF5315) |
 Estimated TM-score=0.58; hard target. |
>Disordered region of unknown function (DUF5315) (Length=74) KTAGSTRSTDRTDALWAEMQATLEEVELSASGGTHVFGPDHDRKLEELRTAQIALAQAWARSEADEAIEMTGQR |
| 4697 |
PF10929 (DUF2811) |
 Estimated TM-score=0.58; hard target. |
>Protein of unknown function (DUF2811) (Length=57) SILTEIPEQLHESLKRYLETHPDWDQDRVFTAALSLFLLQNGNSDRSAARVYLETLF |
| 4698 |
PF17102 (Stealth_CR3) |
 Estimated TM-score=0.58; hard target. |
>Stealth protein CR3, conserved region 3 (Length=48) SHMPHFIQKRLFRELKARYAAEFNATSAHRFRHPEDMQFGFSYMHYVM |
| 4699 |
PF14999 (Shadoo) |
 Estimated TM-score=0.58; very hard target. |
>Shadow of prion protein, neuroprotective (Length=133) CDSSTAKGGRGGARGSARGGLRSGARGTSRVRVRPAPRYSAAGSSLRVAAAGAAAGAAAGAATGLVAGAGWRRVTGPGEG GLEDDEDGLSDRNGTSRSVYSYWTWTSGTGCLQCMPLCLLIGSTLGALELQQP |
| 4700 |
PF09338 (Gly_reductase) |
 Estimated TM-score=0.58; very hard target. |
>Glycine/sarcosine/betaine reductase component B subunits (Length=426) MRLELGNIYIKDVQFGNETKVENGVLYVNKEELISIISEDEHLKSVDVELARPGESVRIMPVKDVIEPRVKVEGKGGIFP GVISKVETVGSGRTNVLKGAAVVTTGKIVGFQEGIIDMTGPGAEYTPFSKLNNVVLVCEPVDGLKQHEHEKAVRMAGLKA AEYLGKAAKNVEPDDVETYEFLPLLKSIEKYPNLPKVAYVLMLQSQGLLHDTYVYGVDAKQILPTLISPTELMDGAIISG NCVSACDKNTTYHHLNNPIIHDLYKKHGKELNFVGVIITNENVYLADKERSSNWTAKLAEYLGVDGVIISQEGFGNPDTD LIMNCKKIEQKGIKTVIVTDEYAGRDGASQSLADADPLANAVVTGGNANEVIELPPMDKVIGHIEPANKIAGGFDGSLKE DGSIVVELQAITGATNELGFNKLTAK |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218