

You can:
| Name | ACRIVASTINE |
|---|---|
| Molecular formula | C22H24N2O2 |
| IUPAC name | (E)-3-[6-[(E)-1-(4-methylphenyl)-3-pyrrolidin-1-ylprop-1-enyl]pyridin-2-yl]prop-2-enoic acid |
| Molecular weight | 348.446 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 1 |
| XlogP | 1.6 |
| Synonyms | CHEBI:83168 DSSTox_CID_2555 L917 Semprex (E)-3-[6-[(E)-1-(4-methylphenyl)-3-pyrrolidin-1-ylprop-1-enyl]pyridin-2-yl]prop-2-enoic acid [ Show all ] |
| Inchi Key | PWACSDKDOHSSQD-IUTFFREVSA-N |
| Inchi ID | InChI=1S/C22H24N2O2/c1-17-7-9-18(10-8-17)20(13-16-24-14-2-3-15-24)21-6-4-5-19(23-21)11-12-22(25)26/h4-13H,2-3,14-16H2,1H3,(H,25,26)/b12-11+,20-13+ |
| PubChem CID | 5284514 |
| ChEMBL | CHEMBL1224 |
| IUPHAR | N/A |
| BindingDB | N/A |
| DrugBank | DB09488 |
Structure | 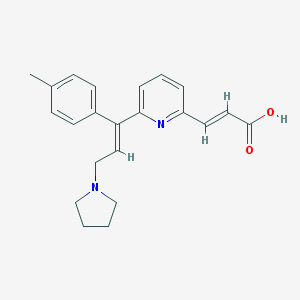 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
You can:
| GLASS ID | Name | UniProt | Gene | Species | Length |
|---|---|---|---|---|---|
| 270296 | Histamine H1 receptor | P31389 | HRH1 | Cavia porcellus (Guinea pig) | 488 |
| 270297 | Histamine H1 receptor | P70174 | Hrh1 | Mus musculus (Mouse) | 488 |
| 551254 | Histamine H1 receptor | P35367 | HRH1 | Homo sapiens (Human) | 487 |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417