

You can:
| Name | Oxytocin receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Oxtr |
| Synonym | OT receptor OT-R OTR |
| Disease | N/A for non-human GPCRs |
| Length | 388 |
| Amino acid sequence | MEGTPAANWSVELDLGSGVPPGEEGNRTAGPPQRNEALARVEVAVLCLILFLALSGNACVLLALRTTRHKHSRLFFFMKHLSIADLVVAVFQVLPQLLWDITFRFYGPDLLCRLVKYLQVVGMFASTYLLLLMSLDRCLAICQPLRSLRRRTDRLAVLGTWLGCLVASAPQVHIFSLREVADGVFDCWAVFIQPWGPKAYVTWITLAVYIVPVIVLAACYGLISFKIWQNLRLKTAAAAAAAEGNDAAGGAGRAALARVSSVKLISKAKIRTVKMTFIIVLAFIVCWTPFFFVQMWSVWDVNAPKEASAFIIAMLLASLNSCCNPWIYMLFTGHLFHELVQRFFCCSARYLKGSRPGETSVSKKSNSSTFVLSRRSSSQRSCSQPSSA |
| UniProt | P70536 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3996 |
| IUPHAR | 369 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 243 | 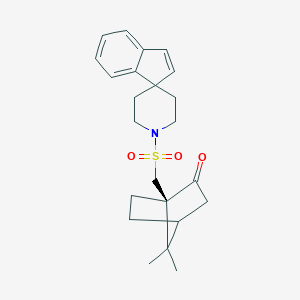 CHEMBL341267 CHEMBL341267 | C23H29NO3S | 399.549 | 4 / 0 | 3.7 | Yes |
| 244 | 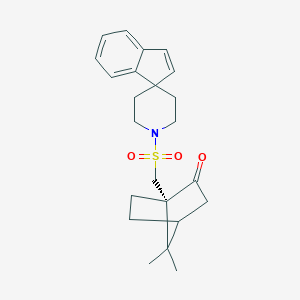 CHEMBL126962 CHEMBL126962 | C23H29NO3S | 399.549 | 4 / 0 | 3.7 | Yes |
| 246 | 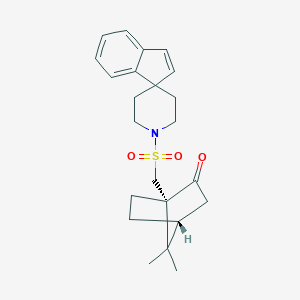 CHEMBL420144 CHEMBL420144 | C23H29NO3S | 399.549 | 4 / 0 | 3.7 | Yes |
| 1831 | 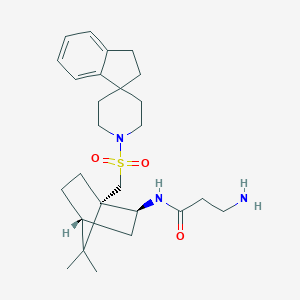 CHEMBL128328 CHEMBL128328 | C26H39N3O3S | 473.676 | 5 / 2 | 3.6 | Yes |
| 2143 | 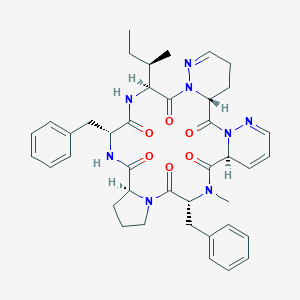 CHEMBL1790547 CHEMBL1790547 | C40H48N8O6 | 736.874 | 8 / 2 | 3.0 | No |
| 3444 | 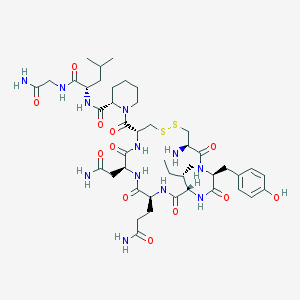 [Pip7]OT [Pip7]OT | C44H68N12O12S2 | 1021.22 | 15 / 12 | -2.2 | No |
| 8709 | 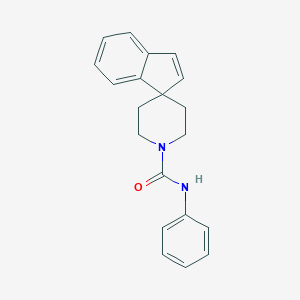 CHEMBL337976 CHEMBL337976 | C20H20N2O | 304.393 | 1 / 1 | 3.9 | Yes |
| 9476 | 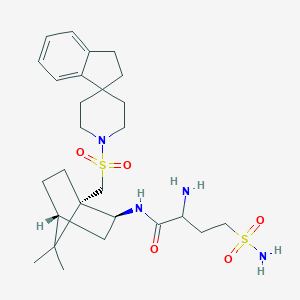 CHEMBL340639 CHEMBL340639 | C27H42N4O5S2 | 566.776 | 8 / 3 | 2.3 | No |
| 10252 | 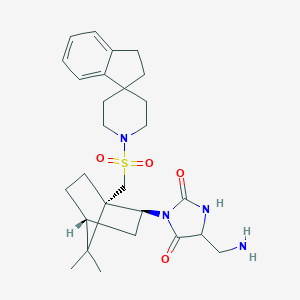 CID 10029477 CID 10029477 | C27H38N4O4S | 514.685 | 6 / 2 | 3.3 | No |
| 11564 | 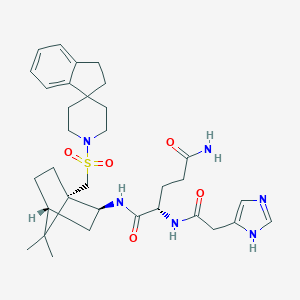 CHEMBL336082 CHEMBL336082 | C33H46N6O5S | 638.828 | 7 / 4 | 2.4 | No |
| 11607 | 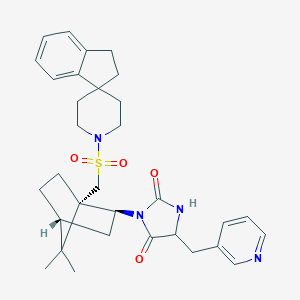 CID 10393379 CID 10393379 | C32H40N4O4S | 576.756 | 6 / 1 | 4.6 | No |
| 12064 | 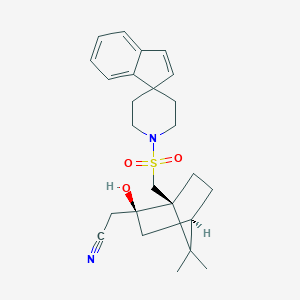 CHEMBL131499 CHEMBL131499 | C25H32N2O3S | 440.602 | 5 / 1 | 4.1 | Yes |
| 12065 | 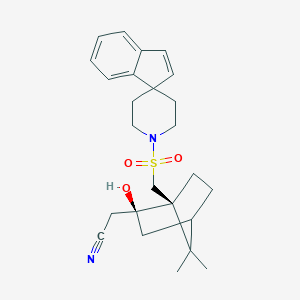 CHEMBL127049 CHEMBL127049 | C25H32N2O3S | 440.602 | 5 / 1 | 4.1 | Yes |
| 12334 | 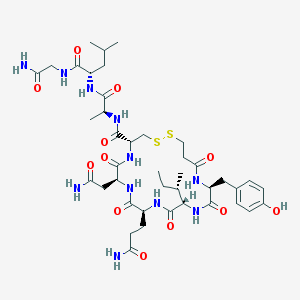 CHEMBL3277915 CHEMBL3277915 | C41H63N11O12S2 | 966.14 | 14 / 12 | -2.1 | No |
| 12880 | 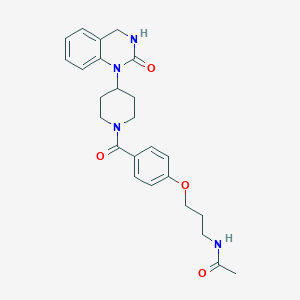 CHEMBL143341 CHEMBL143341 | C25H30N4O4 | 450.539 | 4 / 2 | 2.0 | Yes |
| 13170 | 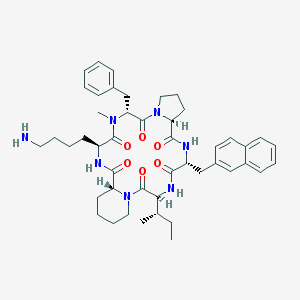 CHEMBL338018 CHEMBL338018 | C46H61N7O6 | 808.037 | 7 / 4 | 5.1 | No |
| 14871 | 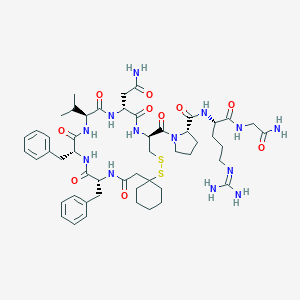 CHEMBL385062 CHEMBL385062 | C51H73N13O10S2 | 1092.35 | 13 / 11 | 0.1 | No |
| 14874 | 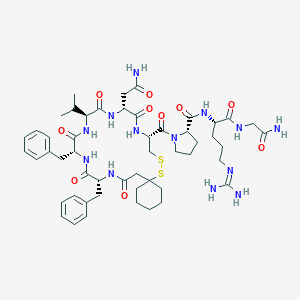 CHEMBL266643 CHEMBL266643 | C51H73N13O10S2 | 1092.35 | 13 / 11 | 0.1 | No |
| 442222 | 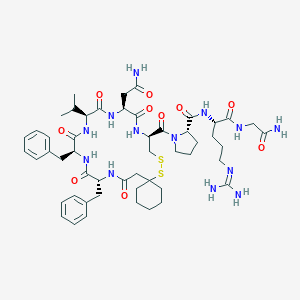 CHEMBL3350125 CHEMBL3350125 | C51H73N13O10S2 | 1092.35 | 13 / 11 | 0.1 | No |
| 17423 | 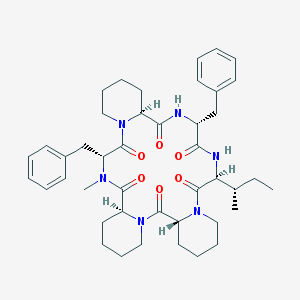 CHEMBL129089 CHEMBL129089 | C43H58N6O6 | 754.973 | 6 / 2 | 5.1 | No |
| 18635 |  CHEMBL1790550 CHEMBL1790550 | C40H52N8O5 | 724.907 | 8 / 2 | 3.0 | No |
| 19940 |  CHEMBL339493 CHEMBL339493 | C36H52N6O6 | 664.848 | 6 / 2 | 3.2 | No |
| 21634 |  [Mpa1, D-Tic7]OT [Mpa1, D-Tic7]OT | C48H67N11O12S2 | 1054.25 | 14 / 11 | -0.7 | No |
| 21637 |  [Mpa1, L-Tic7]OT [Mpa1, L-Tic7]OT | C48H67N11O12S2 | 1054.25 | 14 / 11 | -0.7 | No |
| 22596 | 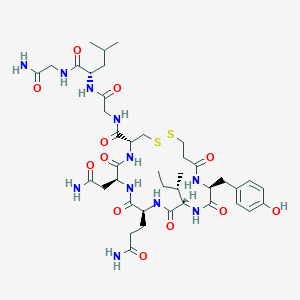 CHEMBL3277916 CHEMBL3277916 | C40H61N11O12S2 | 952.113 | 14 / 12 | -2.5 | No |
| 22762 | 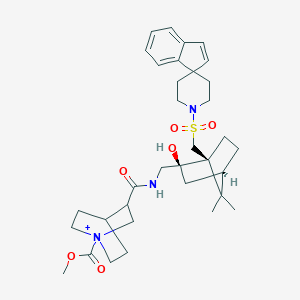 CHEMBL335886 CHEMBL335886 | C34H48N3O6S+ | 626.833 | 7 / 2 | 3.9 | No |
| 24394 | 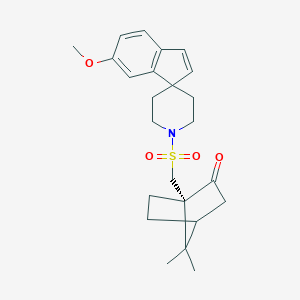 CHEMBL340531 CHEMBL340531 | C24H31NO4S | 429.575 | 5 / 0 | 3.7 | Yes |
| 24468 | 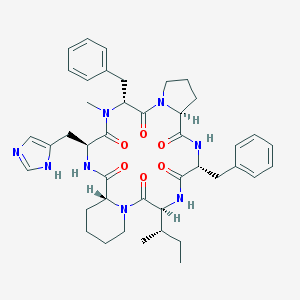 CHEMBL339450 CHEMBL339450 | C42H54N8O6 | 766.944 | 7 / 4 | 3.9 | No |
| 24491 | 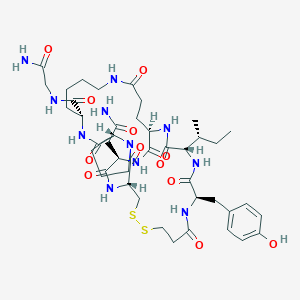 CHEMBL1790201 CHEMBL1790201 | C43H63N11O12S2 | 990.162 | 14 / 11 | -2.2 | No |
| 26351 | 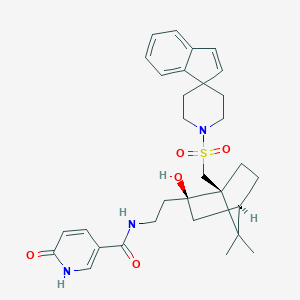 BDBM50043061 BDBM50043061 | C31H39N3O5S | 565.729 | 6 / 3 | 3.7 | No |
| 28254 | 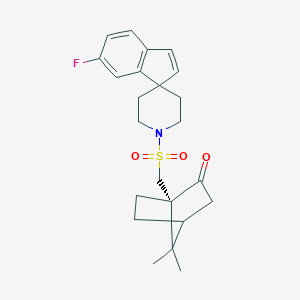 CHEMBL129498 CHEMBL129498 | C23H28FNO3S | 417.539 | 5 / 0 | 3.8 | Yes |
| 30078 | 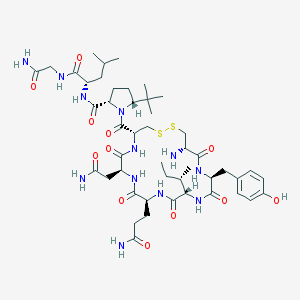 CHEMBL429161 CHEMBL429161 | C47H74N12O12S2 | 1063.3 | 15 / 12 | -0.8 | No |
| 32105 | 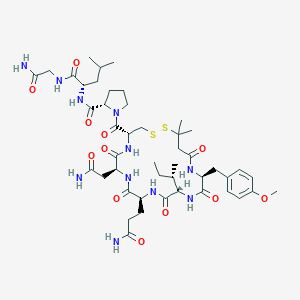 CHEMBL3229638 CHEMBL3229638 | C46H71N11O12S2 | 1034.26 | 14 / 10 | -0.8 | No |
| 32199 | 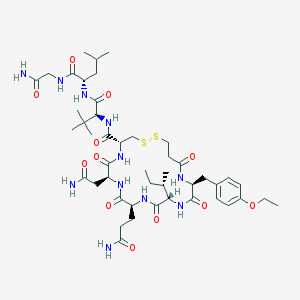 CHEMBL441128 CHEMBL441128 | C46H73N11O12S2 | 1036.28 | 14 / 11 | 0.0 | No |
| 36182 | 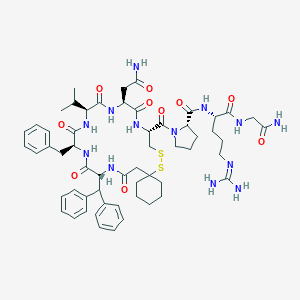 CHEMBL558950 CHEMBL558950 | C57H77N13O10S2 | 1168.44 | 13 / 11 | 1.5 | No |
| 36184 | 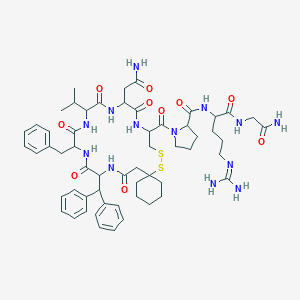 CHEMBL558950 CHEMBL558950 | C57H77N13O10S2 | 1168.44 | 13 / 11 | 1.5 | No |
| 37056 | 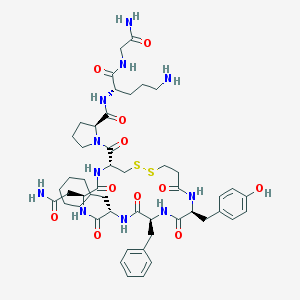 d[Cha4,Orn8]VP d[Cha4,Orn8]VP | C49H69N11O11S2 | 1052.28 | 14 / 11 | 0.5 | No |
| 38952 | 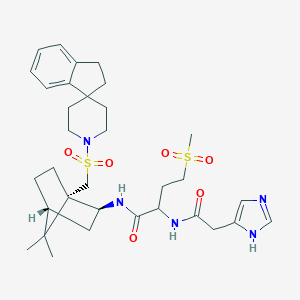 CHEMBL131477 CHEMBL131477 | C33H47N5O6S2 | 673.888 | 8 / 3 | 3.0 | No |
| 39034 | 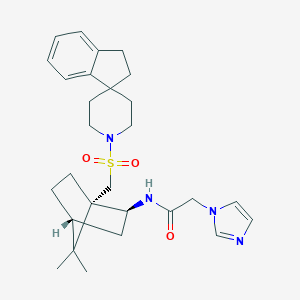 CHEMBL128398 CHEMBL128398 | C28H38N4O3S | 510.697 | 5 / 1 | 4.0 | No |
| 39221 | 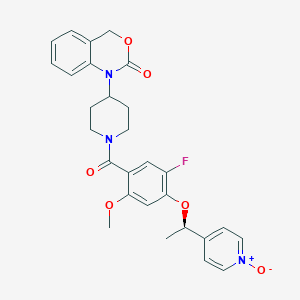 CHEMBL105514 CHEMBL105514 | C28H28FN3O6 | 521.545 | 7 / 0 | 2.8 | No |
| 39653 | 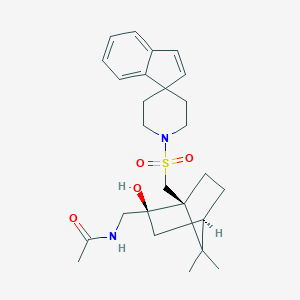 CHEMBL339151 CHEMBL339151 | C26H36N2O4S | 472.644 | 5 / 2 | 3.2 | Yes |
| 40891 | 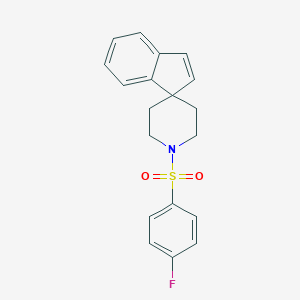 CHEMBL341108 CHEMBL341108 | C19H18FNO2S | 343.416 | 4 / 0 | 3.9 | Yes |
| 42484 | 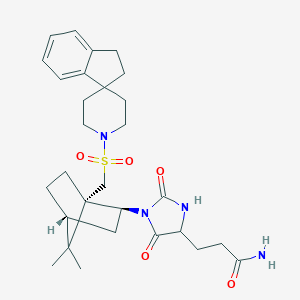 CID 10415322 CID 10415322 | C29H40N4O5S | 556.722 | 6 / 2 | 2.9 | No |
| 43066 | 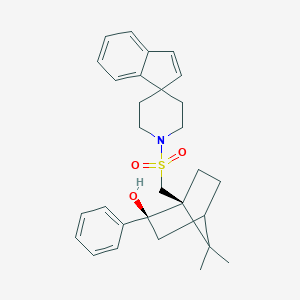 CHEMBL340153 CHEMBL340153 | C29H35NO3S | 477.663 | 4 / 1 | 5.3 | No |
| 43958 | 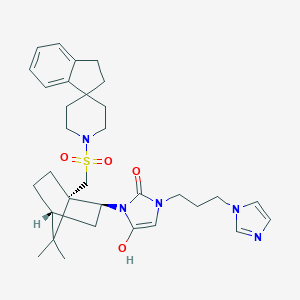 BDBM50043148 BDBM50043148 | C32H43N5O4S | 593.787 | 6 / 1 | 4.0 | No |
| 44503 | 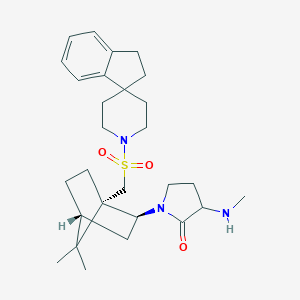 CHEMBL128388 CHEMBL128388 | C28H41N3O3S | 499.714 | 5 / 1 | 4.0 | Yes |
| 44555 | 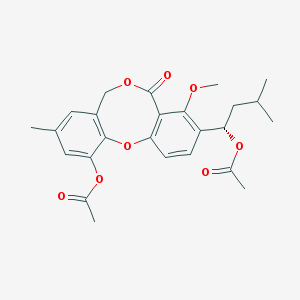 CHEMBL115277 CHEMBL115277 | C25H28O8 | 456.491 | 8 / 0 | 4.4 | Yes |
| 44754 | 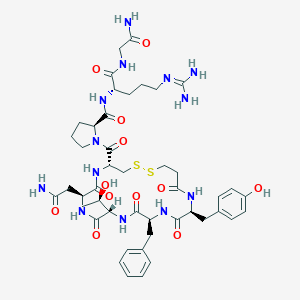 CHEMBL2369831 CHEMBL2369831 | C45H63N13O12S2 | 1042.2 | 15 / 13 | -2.9 | No |
| 44763 | 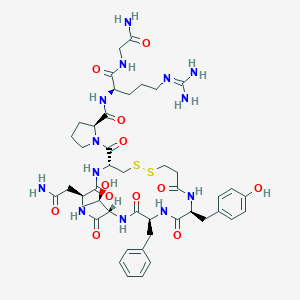 CHEMBL3277498 CHEMBL3277498 | C45H63N13O12S2 | 1042.2 | 15 / 13 | -2.9 | No |
| 46798 | 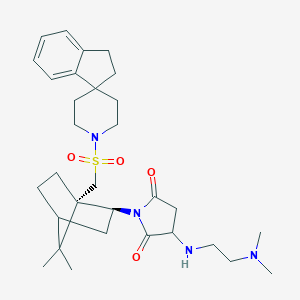 CHEMBL417458 CHEMBL417458 | C31H46N4O4S | 570.793 | 7 / 1 | 3.4 | No |
| 51467 | 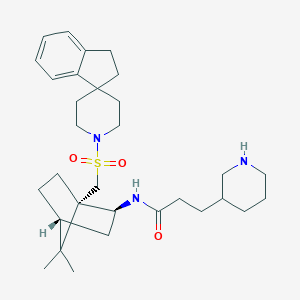 CHEMBL339155 CHEMBL339155 | C31H47N3O3S | 541.795 | 5 / 2 | 4.7 | No |
| 51474 | 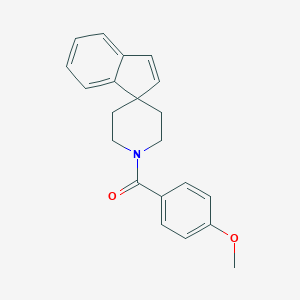 CHEMBL129647 CHEMBL129647 | C21H21NO2 | 319.404 | 2 / 0 | 4.1 | Yes |
| 51906 | 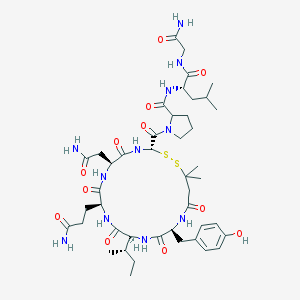 CHEMBL291005 CHEMBL291005 | C44H67N11O12S2 | 1006.21 | 14 / 11 | -1.0 | No |
| 51907 | 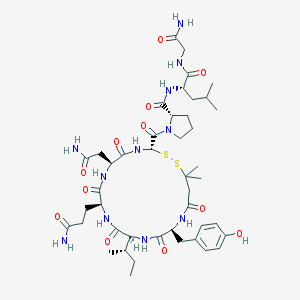 CHEMBL2372251 CHEMBL2372251 | C44H67N11O12S2 | 1006.21 | 14 / 11 | -1.0 | No |
| 52720 | 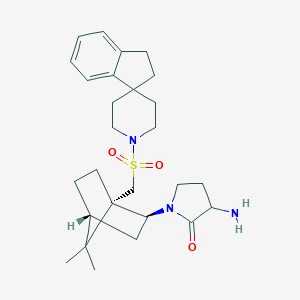 CHEMBL336195 CHEMBL336195 | C27H39N3O3S | 485.687 | 5 / 1 | 3.5 | Yes |
| 53307 | 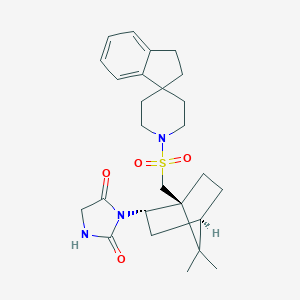 CID 10051081 CID 10051081 | C26H35N3O4S | 485.643 | 5 / 1 | 3.7 | Yes |
| 53745 | 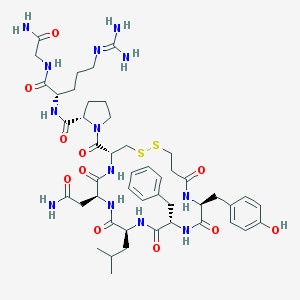 d[Leu4]AVP d[Leu4]AVP | C47H67N13O11S2 | 1054.25 | 14 / 12 | -1.5 | No |
| 54085 | 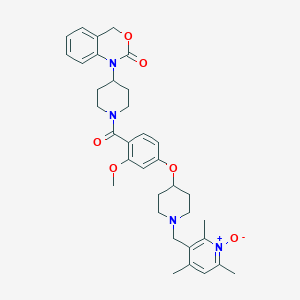 CHEMBL306010 CHEMBL306010 | C35H42N4O6 | 614.743 | 7 / 0 | 4.1 | No |
| 54423 | 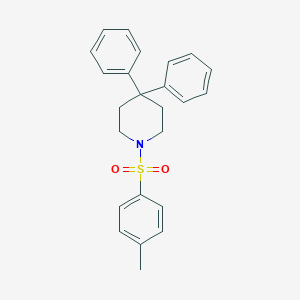 CHEMBL419603 CHEMBL419603 | C24H25NO2S | 391.529 | 3 / 0 | 5.3 | No |
| 54583 | 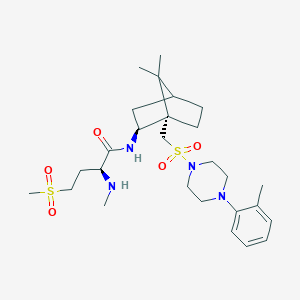 CHEMBL2112898 CHEMBL2112898 | C27H44N4O5S2 | 568.792 | 8 / 2 | 2.6 | No |
| 54589 |  CHEMBL2391300 CHEMBL2391300 | C27H44N4O5S2 | 568.792 | 8 / 2 | 2.6 | No |
| 55576 |  CHEMBL394411 CHEMBL394411 | C45H70N12O12S2 | 1035.25 | 15 / 11 | -1.9 | No |
| 55591 |  CHEMBL128652 CHEMBL128652 | C33H46N6O5S | 638.828 | 7 / 4 | 2.4 | No |
| 55835 |  CHEMBL2372955 CHEMBL2372955 | C40H54N8O6 | 742.922 | 8 / 4 | 2.7 | No |
| 55911 | 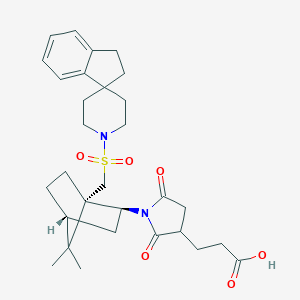 CHEMBL338503 CHEMBL338503 | C30H40N2O6S | 556.718 | 7 / 1 | 3.6 | No |
| 57488 | 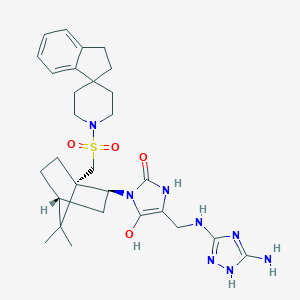 BDBM50043141 BDBM50043141 | C29H40N8O4S | 596.751 | 9 / 5 | 3.3 | No |
| 57666 | 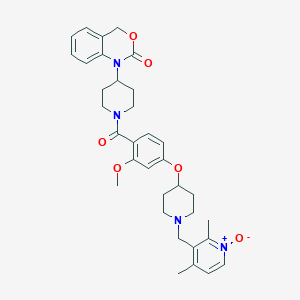 CHEMBL302297 CHEMBL302297 | C34H40N4O6 | 600.716 | 7 / 0 | 3.7 | No |
| 60339 | 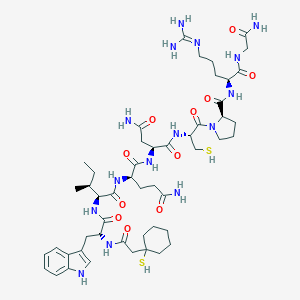 CHEMBL262157 CHEMBL262157 | C50H77N15O11S2 | 1128.38 | 14 / 15 | -2.1 | No |
| 60747 | 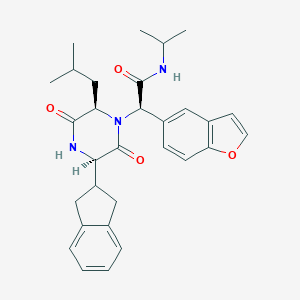 CHEMBL208787 CHEMBL208787 | C30H35N3O4 | 501.627 | 4 / 2 | 5.1 | No |
| 62558 | 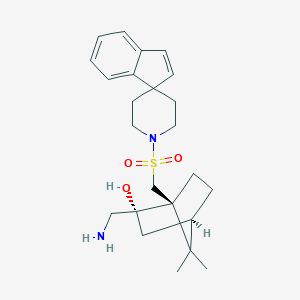 CHEMBL274955 CHEMBL274955 | C24H34N2O3S | 430.607 | 5 / 2 | 2.9 | Yes |
| 62559 | 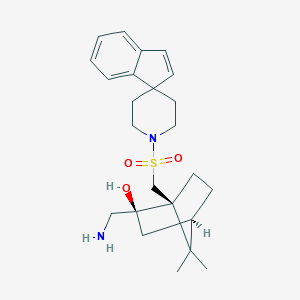 CHEMBL128124 CHEMBL128124 | C24H34N2O3S | 430.607 | 5 / 2 | 2.9 | Yes |
| 63103 | 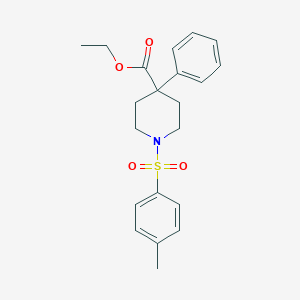 CHEMBL126883 CHEMBL126883 | C21H25NO4S | 387.494 | 5 / 0 | 3.6 | Yes |
| 63820 | 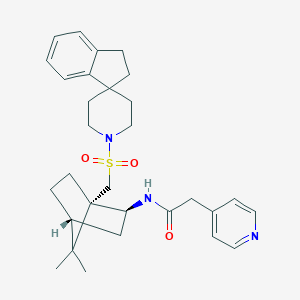 CHEMBL131269 CHEMBL131269 | C30H39N3O3S | 521.72 | 5 / 1 | 4.6 | No |
| 63938 | 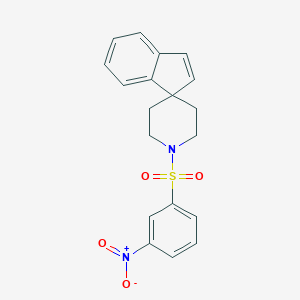 CHEMBL129708 CHEMBL129708 | C19H18N2O4S | 370.423 | 5 / 0 | 3.7 | Yes |
| 65998 | 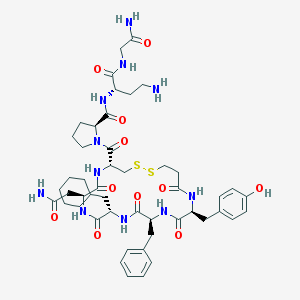 d[Cha4,Dab8]VP d[Cha4,Dab8]VP | C48H67N11O11S2 | 1038.25 | 14 / 11 | 0.1 | No |
| 69265 | 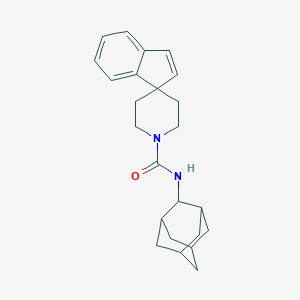 CHEMBL127119 CHEMBL127119 | C24H30N2O | 362.517 | 1 / 1 | 4.7 | Yes |
| 69341 | 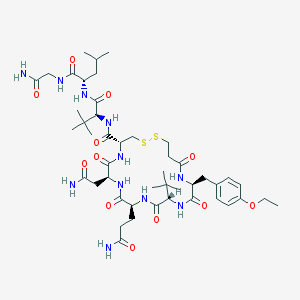 CHEMBL232501 CHEMBL232501 | C46H73N11O12S2 | 1036.28 | 14 / 11 | 0.0 | No |
| 444452 | 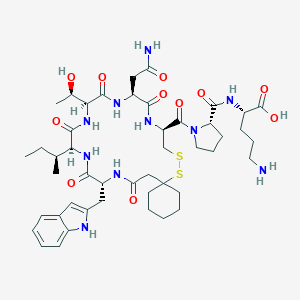 CHEMBL3349464 CHEMBL3349464 | C46H68N10O11S2 | 1001.23 | 14 / 11 | -1.4 | No |
| 444456 | 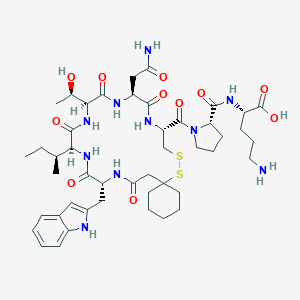 CHEMBL3349466 CHEMBL3349466 | C46H68N10O11S2 | 1001.23 | 14 / 11 | -1.4 | No |
| 70863 | 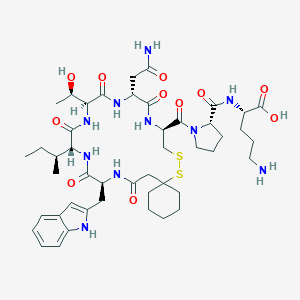 CHEMBL1790307 CHEMBL1790307 | C46H68N10O11S2 | 1001.23 | 14 / 11 | -1.4 | No |
| 70867 | 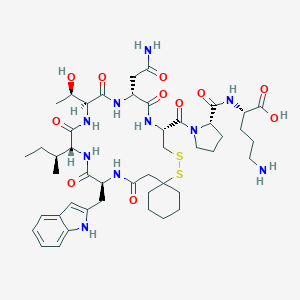 CHEMBL1790312 CHEMBL1790312 | C46H68N10O11S2 | 1001.23 | 14 / 11 | -1.4 | No |
| 71354 | 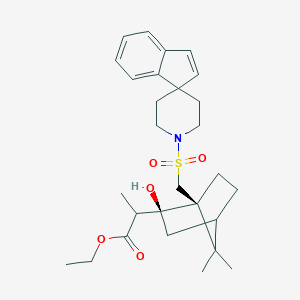 CHEMBL338409 CHEMBL338409 | C28H39NO5S | 501.682 | 6 / 1 | 5.2 | No |
| 71834 | 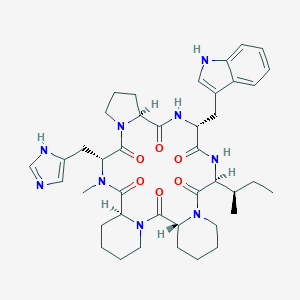 CHEMBL1790938 CHEMBL1790938 | C41H55N9O6 | 769.948 | 7 / 4 | 3.1 | No |
| 72468 | 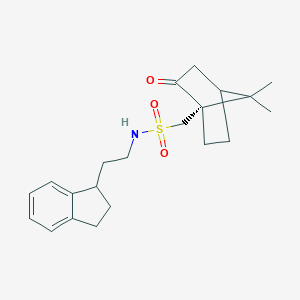 CHEMBL339093 CHEMBL339093 | C21H29NO3S | 375.527 | 4 / 1 | 3.4 | Yes |
| 74001 | 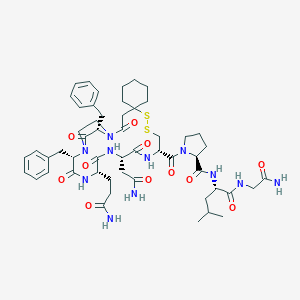 CHEMBL406533 CHEMBL406533 | C53H73N11O11S2 | 1104.35 | 13 / 8 | 0.6 | No |
| 74045 | 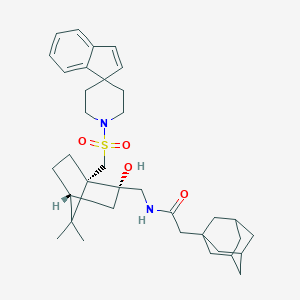 CHEMBL339464 CHEMBL339464 | C36H50N2O4S | 606.866 | 5 / 2 | 6.6 | No |
| 444712 | 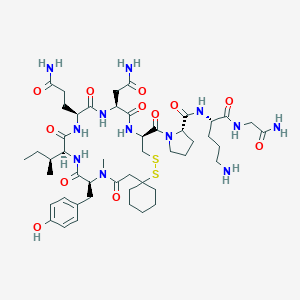 CHEMBL3350126 CHEMBL3350126 | C48H74N12O12S2 | 1075.31 | 15 / 11 | -1.9 | No |
| 76268 | 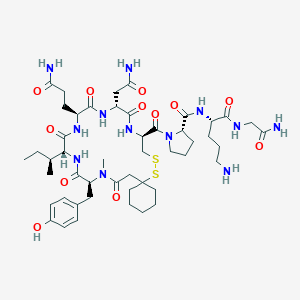 CHEMBL261914 CHEMBL261914 | C48H74N12O12S2 | 1075.31 | 15 / 11 | -1.9 | No |
| 76270 | 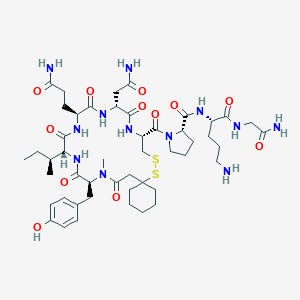 CHEMBL263521 CHEMBL263521 | C48H74N12O12S2 | 1075.31 | 15 / 11 | -1.9 | No |
| 76404 | 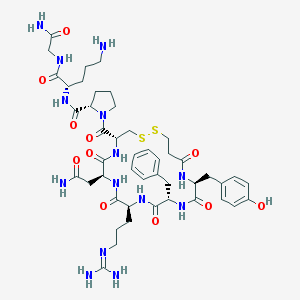 d[Arg4,Orn8]VP d[Arg4,Orn8]VP | C46H66N14O11S2 | 1055.24 | 15 / 13 | -3.4 | No |
| 76519 | 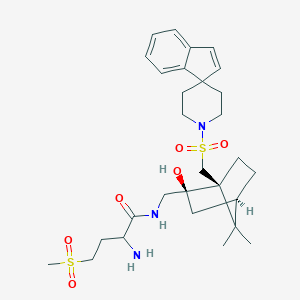 CHEMBL131388 CHEMBL131388 | C29H43N3O6S2 | 593.798 | 8 / 3 | 2.0 | No |
| 76615 | 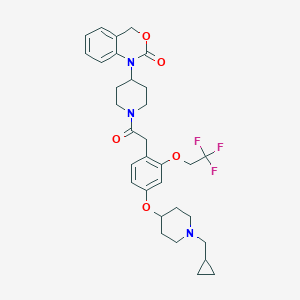 CHEMBL24456 CHEMBL24456 | C32H38F3N3O5 | 601.667 | 9 / 0 | 5.3 | No |
| 76679 | 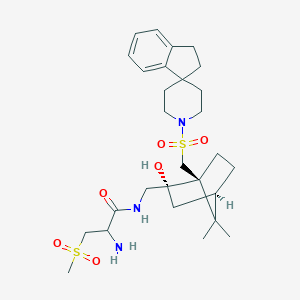 CHEMBL2112663 CHEMBL2112663 | C28H43N3O6S2 | 581.787 | 8 / 3 | 1.7 | No |
| 76754 | 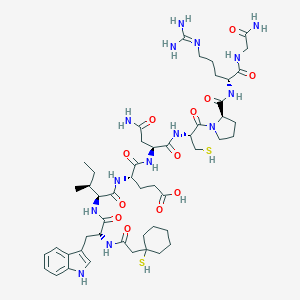 CHEMBL263153 CHEMBL263153 | C50H76N14O12S2 | 1129.36 | 15 / 15 | -1.4 | No |
| 76755 | 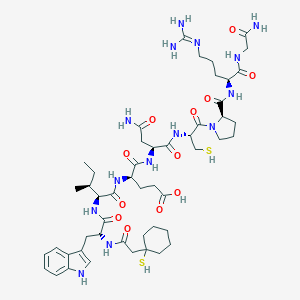 CHEMBL274635 CHEMBL274635 | C50H76N14O12S2 | 1129.36 | 15 / 15 | -1.4 | No |
| 76756 | 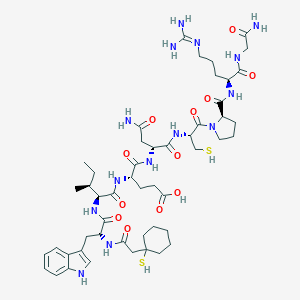 CHEMBL414373 CHEMBL414373 | C50H76N14O12S2 | 1129.36 | 15 / 15 | -1.4 | No |
| 76757 | 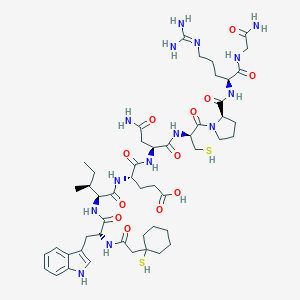 CHEMBL386547 CHEMBL386547 | C50H76N14O12S2 | 1129.36 | 15 / 15 | -1.4 | No |
| 76758 | 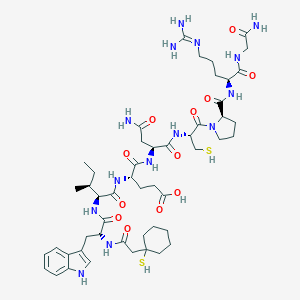 CHEMBL268779 CHEMBL268779 | C50H76N14O12S2 | 1129.36 | 15 / 15 | -1.4 | No |
| 76759 | 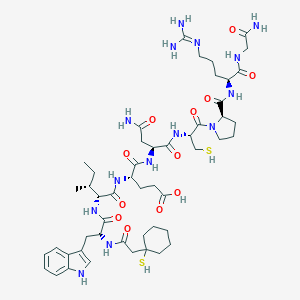 CHEMBL268778 CHEMBL268778 | C50H76N14O12S2 | 1129.36 | 15 / 15 | -1.4 | No |
| 76760 | 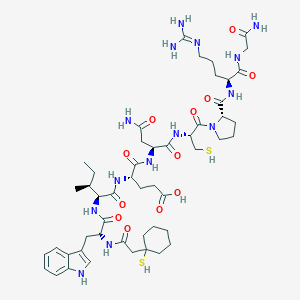 CHEMBL439482 CHEMBL439482 | C50H76N14O12S2 | 1129.36 | 15 / 15 | -1.4 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417