

You can:
| Name | Taste receptor type 2 member 60 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | TAS2R60 |
| Synonym | T2R56 T2R60 TAS2R60 Taste receptor type 2 member 56 taste receptor, type 2, member 60 |
| Disease | N/A |
| Length | 318 |
| Amino acid sequence | MNGDHMVLGSSVTDKKAIILVTILLLLRLVAIAGNGFITAALGVEWVLRRMLLPCDKLLVSLGASRFCLQSVVMGKTIYVFLHPMAFPYNPVLQFLAFQWDFLNAATLWSSTWLSVFYCVKIATFTHPVFFWLKHKLSGWLPWMLFSSVGLSSFTTILFFIGNHRMYQNYLRNHLQPWNVTGDSIRSYCEKFYLFPLKMITWTMPTAVFFICMILLITSLGRHRKKALLTTSGFREPSVQAHIKALLALLSFAMLFISYFLSLVFSAAGIFPPLDFKFWVWESVIYLCAAVHPIILLFSNCRLRAVLKSRRSSRCGTP |
| UniProt | P59551 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P59551 |
| 3D structure model | This predicted structure model is from GPCR-EXP P59551. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3309109 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 445102 | 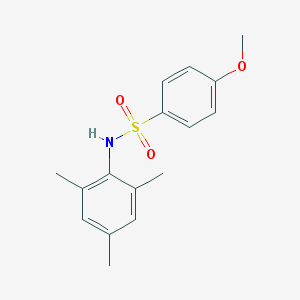 GSK137647A GSK137647A | C16H19NO3S | 305.392 | 4 / 1 | 3.5 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417