

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MNGDHMVLGSSVTDKKAIILVTILLLLRLVAIAGNGFITAALGVEWVLRRMLLPCDKLLVSLGASRFCLQSVVMGKTIYVFLHPMAFPYNPVLQFLAFQWDFLNAATLWSSTWLSVFYCVKIATFTHPVFFWLKHKLSGWLPWMLFSSVGLSSFTTILFFIGNHRMYQNYLRNHLQPWNVTGDSIRSYCEKFYLFPLKMITWTMPTAVFFICMILLITSLGRHRKKALLTTSGFREPSVQAHIKALLALLSFAMLFISYFLSLVFSAAGIFPPLDFKFWVWESVIYLCAAVHPIILLFSNCRLRAVLKSRRSSRCGTP | |
| CCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCCC | |
| 998777755311157999999999999999999999999999999990787878879999999999999999998688630243214563289999999999829999999999999873441788779999999745019999999999999999999964101123445565667654200037888999999999999999999999999999999999999874779999999838999999999999999999999999999998266649999999996455977499997288089999999962306998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MNGDHMVLGSSVTDKKAIILVTILLLLRLVAIAGNGFITAALGVEWVLRRMLLPCDKLLVSLGASRFCLQSVVMGKTIYVFLHPMAFPYNPVLQFLAFQWDFLNAATLWSSTWLSVFYCVKIATFTHPVFFWLKHKLSGWLPWMLFSSVGLSSFTTILFFIGNHRMYQNYLRNHLQPWNVTGDSIRSYCEKFYLFPLKMITWTMPTAVFFICMILLITSLGRHRKKALLTTSGFREPSVQAHIKALLALLSFAMLFISYFLSLVFSAAGIFPPLDFKFWVWESVIYLCAAVHPIILLFSNCRLRAVLKSRRSSRCGTP | |
| 775543234430331231111221331233133212200310111104433031011000000112000000133310000000311343300000101113312300100000000000000223110000013304310011023122323311101203233323333434333210122233331120111012333333333313323320110022024303433533412314102200200120232333233013211111234431110201021233130100032141430130023002133548 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCCC MNGDHMVLGSSVTDKKAIILVTILLLLRLVAIAGNGFITAALGVEWVLRRMLLPCDKLLVSLGASRFCLQSVVMGKTIYVFLHPMAFPYNPVLQFLAFQWDFLNAATLWSSTWLSVFYCVKIATFTHPVFFWLKHKLSGWLPWMLFSSVGLSSFTTILFFIGNHRMYQNYLRNHLQPWNVTGDSIRSYCEKFYLFPLKMITWTMPTAVFFICMILLITSLGRHRKKALLTTSGFREPSVQAHIKALLALLSFAMLFISYFLSLVFSAAGIFPPLDFKFWVWESVIYLCAAVHPIILLFSNCRLRAVLKSRRSSRCGTP | |||||||||||||||||||||||||
| 1 | 4grvA | 0.17 | 0.22 | 0.89 | 1.33 | Download | -NSDLDV---NTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA----RKSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIC---HPFKAK--LMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGL-VCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVM--------SGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWTYFYMLTNALAYASSAINPILYNLVSANFRQV------------ | |||||||||||||||||||
| 2 | 5tjvA | 0.11 | 0.18 | 0.89 | 2.80 | Download | -NFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS--RSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLA----YKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHID-----------ETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 3 | 4n6hA | 0.10 | 0.18 | 0.92 | 2.20 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMK-TATNIYIFNLALADALATST--LPFQSAKYLMETWFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRAVVCMLQFPSP------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV-----RLLSGSK-EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-- | |||||||||||||||||||
| 4 | 4djh | 0.11 | 0.22 | 0.91 | 1.57 | Download | ------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKAL-DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWDLFMKI--CVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFTPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEAGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDCFP------ | |||||||||||||||||||
| 5 | 5glh | 0.14 | 0.19 | 0.89 | 1.19 | Download | --PPCQ-GPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYK--------NGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVAS---W-----------WTAVEIVLIWVVSVVLAVPEAIGFDIITMDYK---GSYLRICLLHPVQKFMQFYDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNEGLRLTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARIKLTLYNNVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL------- | |||||||||||||||||||
| 6 | 4grvA | 0.16 | 0.22 | 0.88 | 1.49 | Download | ----NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA----RKSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAICH----PFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMG-LQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMSGS------------VQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISHYFYMLTNALAYASSAINPILYNLVSANFRQV------------ | |||||||||||||||||||
| 7 | 3uon | 0.11 | 0.22 | 0.88 | 1.72 | Download | ---------------EVVFIVLVAGSLSLVTIIGNILVMVSIKV---NRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMA----GMMIAAAWVLSFILWAPAILFWQFIVGVEDGECYIQ-----FFS----NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEADAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------ | |||||||||||||||||||
| 8 | 4buoA | 0.16 | 0.24 | 0.92 | 2.38 | Download | -NSD---LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARK---KSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICH-------PFKAKTRSRTKKFISAIWLASALLAIP-MLFTMGLQNLSGDGTHPGGLVCTPIVDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQ-----PGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYILFDFYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------- | |||||||||||||||||||
| 9 | 4djhA | 0.11 | 0.18 | 0.88 | 1.46 | Download | ------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKA-LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE-DVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSASSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----- | |||||||||||||||||||
| 10 | 5c1mA | 0.13 | 0.17 | 0.90 | 1.45 | Download | --GSHSLPQTGSPSVTAITIMALYSIVCVVGLFGNFLVMYVIVRY---TKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVC---HPVK-ALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCT----LTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKS------VRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
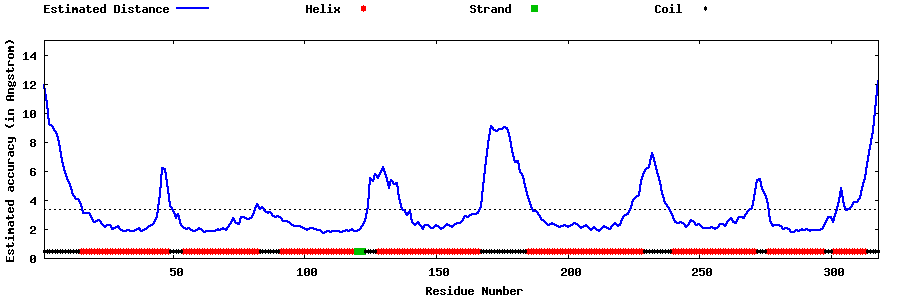 | |||
|
|
|||
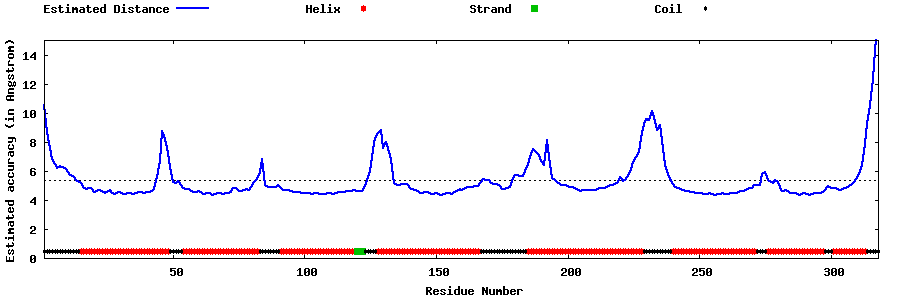 | |||
|
| |||
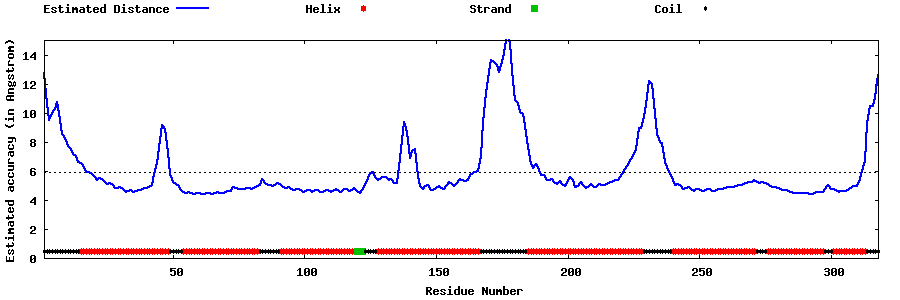 | |||
|
| |||
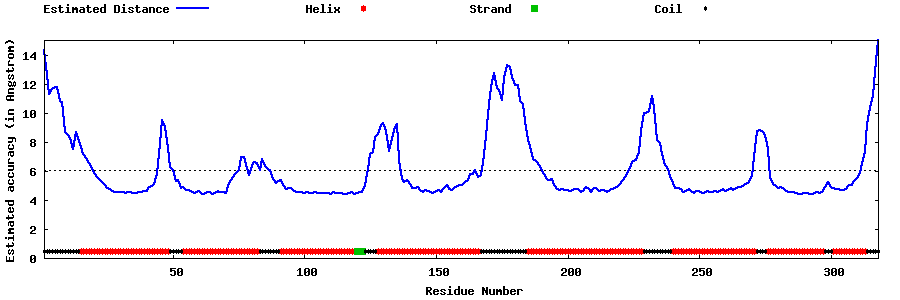 | |||
|
| |||
 | |||
|
| |||