

You can:
| Name | Neuropeptide Y receptor type 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | NPY1R |
| Synonym | neuropeptide Y receptor type 1 NPY-Y1 receptor NPY1-R FC5 Y1 receptor [ Show all ] |
| Disease | Hypertension; Obesity; Heart disease Obesity Eating disorders reduction in food intake obesity anxiety Eating disorders reduction in food intake Eating disorder [ Show all ] |
| Length | 384 |
| Amino acid sequence | MNSTLFSQVENHSVHSNFSEKNAQLLAFENDDCHLPLAMIFTLALAYGAVIILGVSGNLALIIIILKQKEMRNVTNILIVNLSFSDLLVAIMCLPFTFVYTLMDHWVFGEAMCKLNPFVQCVSITVSIFSLVLIAVERHQLIINPRGWRPNNRHAYVGIAVIWVLAVASSLPFLIYQVMTDEPFQNVTLDAYKDKYVCFDQFPSDSHRLSYTTLLLVLQYFGPLCFIFICYFKIYIRLKRRNNMMDKMRDNKYRSSETKRINIMLLSIVVAFAVCWLPLTIFNTVFDWNHQIIATCNHNLLFLLCHLTAMISTCVNPIFYGFLNKNFQRDLQFFFNFCDFRSRDDDYETIAMSTMHTDVSKTSLKQASPVAFKKINNNDDNEKI |
| UniProt | P25929 |
| Protein Data Bank | 5zbh |
| GPCR-HGmod model | P25929 |
| 3D structure model | This structure is from PDB ID 5zbh. |
| BioLiP | BL0411971 |
| Therapeutic Target Database | T89213 |
| ChEMBL | CHEMBL4777 |
| IUPHAR | 305 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 337 | 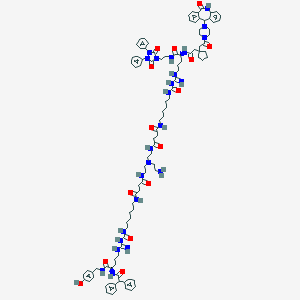 BDBM50442547 BDBM50442547 | C104H138N24O15 | 1964.4 | 20 / 19 | 6.9 | No |
| 601 | 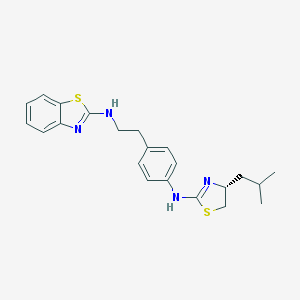 CHEMBL1076276 CHEMBL1076276 | C22H26N4S2 | 410.598 | 5 / 2 | 6.2 | No |
| 685 | 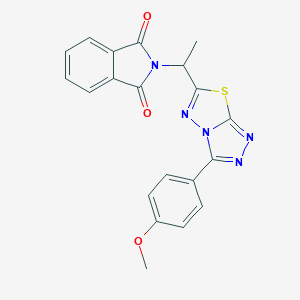 SMR000091760 SMR000091760 | C20H15N5O3S | 405.432 | 7 / 0 | 2.8 | Yes |
| 966 | 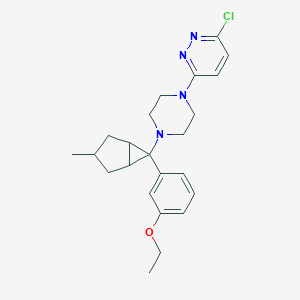 CHEMBL2397302 CHEMBL2397302 | C23H29ClN4O | 412.962 | 5 / 0 | 4.3 | Yes |
| 1561 | 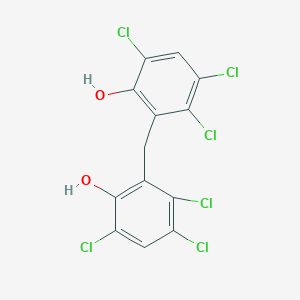 hexachlorophene hexachlorophene | C13H6Cl6O2 | 406.889 | 2 / 2 | 7.5 | No |
| 2019 | 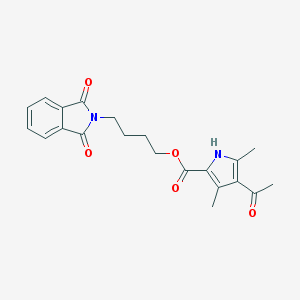 MLS000053810 MLS000053810 | C21H22N2O5 | 382.416 | 5 / 1 | 2.7 | Yes |
| 2345 | 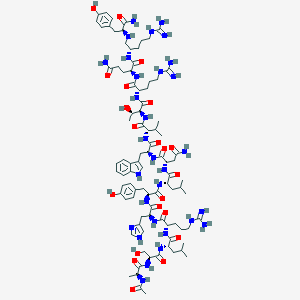 CID 16147801 CID 16147801 | C91H140N30O21 | 1990.31 | 26 / 33 | -3.7 | No |
| 3478 | 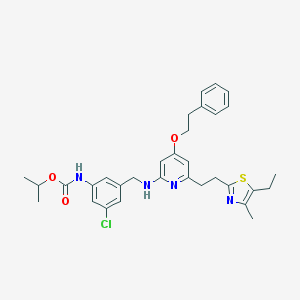 CHEMBL48897 CHEMBL48897 | C32H37ClN4O3S | 593.183 | 7 / 2 | 8.2 | No |
| 3873 |  MLS000041001 MLS000041001 | C17H19N3O | 281.359 | 4 / 0 | 4.2 | Yes |
| 3899 |  MLS000680598 MLS000680598 | C19H24BrNO | 362.311 | 2 / 0 | 5.2 | No |
| 4629 |  MLS000724783 MLS000724783 | C18H18ClF2N3O2S2 | 445.928 | 6 / 1 | 4.1 | Yes |
| 557422 |  MLS000060886 MLS000060886 | C20H16N2O2 | 316.36 | 3 / 2 | 4.0 | Yes |
| 5407 |  CHEMBL53876 CHEMBL53876 | C15H10ClN3O4S | 363.772 | 6 / 1 | 2.8 | Yes |
| 6484 |  CHEMBL557932 CHEMBL557932 | C33H45N3O3 | 531.741 | 4 / 1 | 4.8 | No |
| 6511 |  CHEMBL48456 CHEMBL48456 | C29H39ClN4O3S | 559.166 | 7 / 2 | 8.0 | No |
| 7864 |  CHEMBL562000 CHEMBL562000 | C32H45N3O3 | 519.73 | 4 / 1 | 4.7 | No |
| 7913 |  CHEMBL349328 CHEMBL349328 | C38H49IN4O2 | 720.74 | 5 / 0 | 8.2 | No |
| 7933 |  MLS000083032 MLS000083032 | C16H12Cl2N6S | 391.274 | 5 / 1 | 3.9 | Yes |
| 8486 |  CHEMBL1079731 CHEMBL1079731 | C18H18N4S2 | 354.49 | 5 / 2 | 4.5 | Yes |
| 8867 |  MLS000072668 MLS000072668 | C26H35ClN6O3 | 515.055 | 8 / 1 | N/A | No |
| 8873 |  CID 73351118 CID 73351118 | C184H287N53O53S2 | 4153.76 | 61 / 58 | -13.6 | No |
| 9241 |  CID 76335489 CID 76335489 | C177H272N52O55 | 4008.43 | 62 / 60 | -15.6 | No |
| 9272 |  2-[4-(2-methoxyphenoxy)-3-nitrophenyl]quinoxaline 2-[4-(2-methoxyphenoxy)-3-nitrophenyl]quinoxaline | C21H15N3O4 | 373.368 | 6 / 0 | 4.2 | Yes |
| 9309 |  MLS000778774 MLS000778774 | C21H18N2O4S2 | 426.505 | 7 / 1 | 4.2 | Yes |
| 9589 | 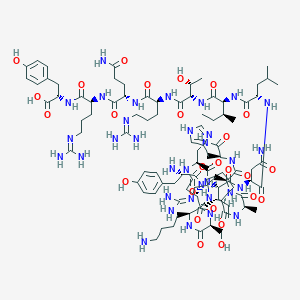 CHEMBL2370632 CHEMBL2370632 | C97H155N33O26 | 2199.51 | 32 / 33 | -10.1 | No |
| 9759 | 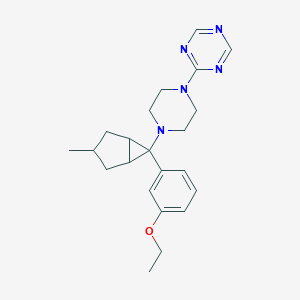 CHEMBL2397293 CHEMBL2397293 | C22H29N5O | 379.508 | 6 / 0 | 3.5 | Yes |
| 9882 | 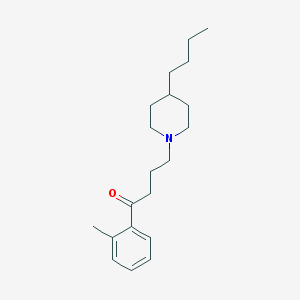 AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 9969 | 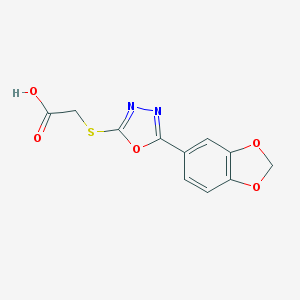 67572-43-4 67572-43-4 | C11H8N2O5S | 280.254 | 8 / 1 | 1.7 | Yes |
| 10005 | 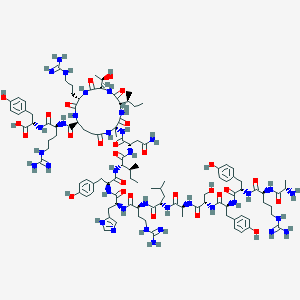 BDBM50003047 BDBM50003047 | C108H163N35O28 | 2399.71 | 34 / 41 | -6.2 | No |
| 10295 |  SMR000163162 SMR000163162 | C23H19F3N2O5 | 460.409 | 8 / 1 | 5.1 | No |
| 10608 | 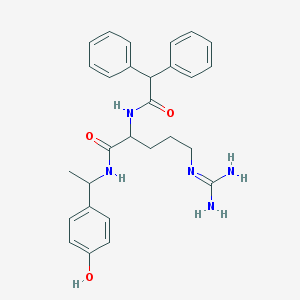 CHEMBL288355 CHEMBL288355 | C28H33N5O3 | 487.604 | 4 / 5 | 2.9 | Yes |
| 11431 | 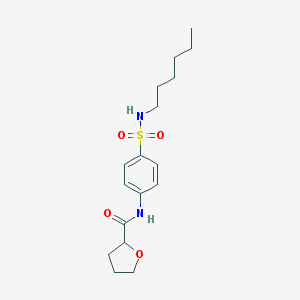 MLS000705653 MLS000705653 | C17H26N2O4S | 354.465 | 5 / 2 | 2.8 | Yes |
| 11435 | 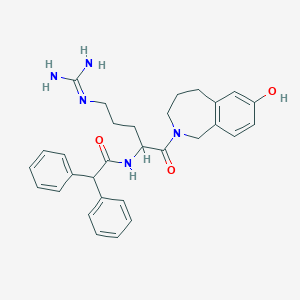 CHEMBL42747 CHEMBL42747 | C30H35N5O3 | 513.642 | 4 / 4 | 3.1 | No |
| 12144 |  Ac-Peptide YY(25-36) Ac-Peptide YY(25-36) | C74H118N26O18 | 1659.92 | 37 / 31 | 4.7 | No |
| 12162 |  CHEMBL52643 CHEMBL52643 | C15H10FN3O4S | 347.32 | 7 / 1 | 2.2 | Yes |
| 12294 |  CHEMBL566041 CHEMBL566041 | C22H27N5OS2 | 441.612 | 8 / 1 | 4.0 | Yes |
| 12341 |  CHEMBL585669 CHEMBL585669 | C22H27N5OS2 | 441.612 | 8 / 1 | 4.2 | Yes |
| 12650 | 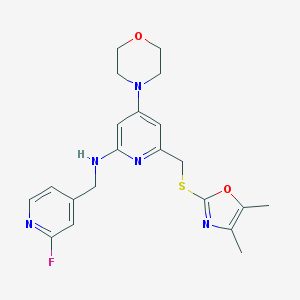 CHEMBL584908 CHEMBL584908 | C21H24FN5O2S | 429.514 | 9 / 1 | 3.4 | Yes |
| 12754 | 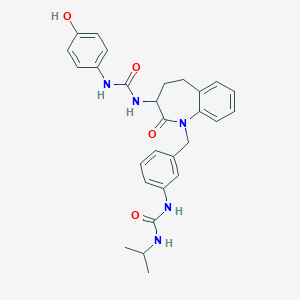 CHEMBL432188 CHEMBL432188 | C28H31N5O4 | 501.587 | 4 / 5 | 3.4 | No |
| 13868 | 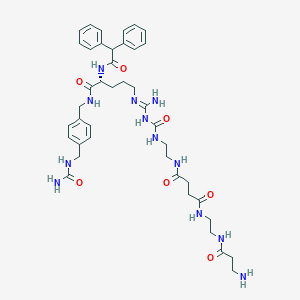 CHEMBL2440902 CHEMBL2440902 | C41H56N12O7 | 828.976 | 9 / 11 | -1.2 | No |
| 13873 | 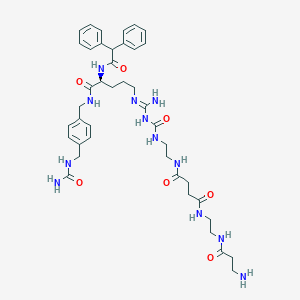 CHEMBL2440903 CHEMBL2440903 | C41H56N12O7 | 828.976 | 9 / 11 | -1.2 | No |
| 13891 |  5-Bromo-N,N-dimethyltryptamine 5-Bromo-N,N-dimethyltryptamine | C12H15BrN2 | 267.17 | 1 / 1 | 2.2 | Yes |
| 14041 |  MLS000685650 MLS000685650 | C16H12N2O3S2 | 344.403 | 6 / 3 | 4.1 | Yes |
| 14348 |  CHEMBL1771263 CHEMBL1771263 | C24H31Cl2N5O2 | 492.445 | 6 / 2 | 4.9 | Yes |
| 14632 |  D0P9KP D0P9KP | C180H279N53O54 | 4049.53 | 61 / 61 | -14.9 | No |
| 15150 |  MLS000570445 MLS000570445 | C25H20N2O3 | 396.446 | 4 / 1 | 4.8 | Yes |
| 15876 |  MLS000114342 MLS000114342 | C18H18ClNO3 | 331.796 | 3 / 0 | 2.2 | Yes |
| 16202 |  BU-E-76 BU-E-76 | C21H24F2N6 | 398.462 | 5 / 3 | 2.5 | Yes |
| 17121 |  CHEMBL2440659 CHEMBL2440659 | C15H11FN4O2S | 330.337 | 7 / 1 | 2.4 | Yes |
| 17341 | 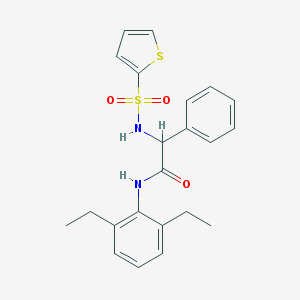 MLS000585997 MLS000585997 | C22H24N2O3S2 | 428.565 | 5 / 2 | 5.1 | No |
| 17744 | 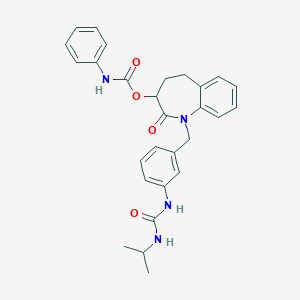 CHEMBL314637 CHEMBL314637 | C28H30N4O4 | 486.572 | 4 / 3 | 4.3 | Yes |
| 17776 | 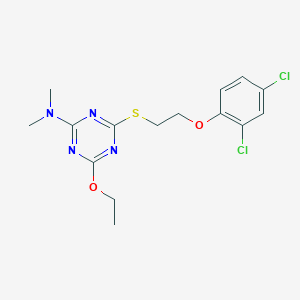 MLS000552453 MLS000552453 | C15H18Cl2N4O2S | 389.295 | 7 / 0 | 4.9 | Yes |
| 17853 | 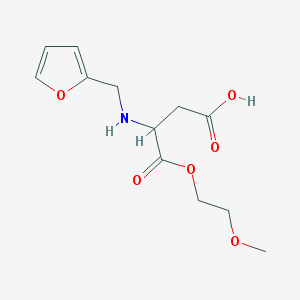 MLS000807054 MLS000807054 | C12H17NO6 | 271.269 | 7 / 2 | -2.6 | Yes |
| 18147 |  CHEMBL564536 CHEMBL564536 | C29H38ClN5O3S | 572.165 | 8 / 2 | 6.4 | No |
| 557823 |  SMR000047897 SMR000047897 | C17H16N2O4 | 312.325 | 5 / 3 | 3.1 | Yes |
| 18585 |  SMR000221562 SMR000221562 | C18H23N3S | 313.463 | 4 / 0 | 3.8 | Yes |
| 18659 |  MLS000771330 MLS000771330 | C20H17FN2O4 | 368.364 | 6 / 0 | 2.4 | Yes |
| 18716 | 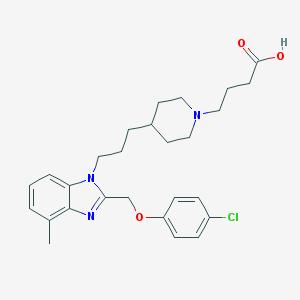 CHEMBL146116 CHEMBL146116 | C27H34ClN3O3 | 484.037 | 5 / 1 | 2.9 | Yes |
| 19832 | 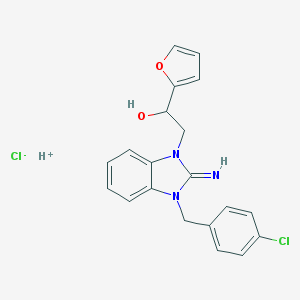 AC1LCZE6 AC1LCZE6 | C20H19Cl2N3O2 | 404.291 | 4 / 3 | N/A | N/A |
| 20082 | 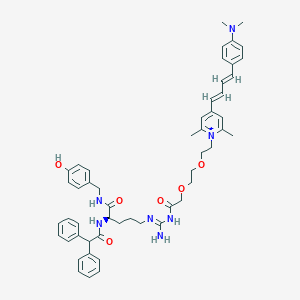 CHEMBL1774217 CHEMBL1774217 | C52H62N7O6+ | 881.111 | 8 / 5 | 7.5 | No |
| 20951 | 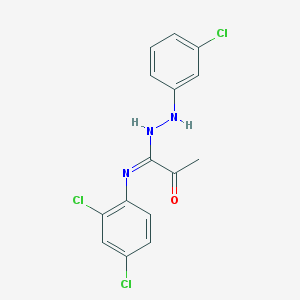 SMR000140824 SMR000140824 | C15H12Cl3N3O | 356.631 | 3 / 2 | 5.6 | No |
| 20991 | 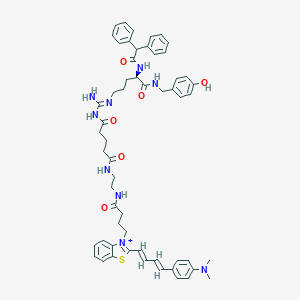 CHEMBL1774343 CHEMBL1774343 | C57H66N9O6S+ | 1005.27 | 9 / 7 | 7.7 | No |
| 21331 | 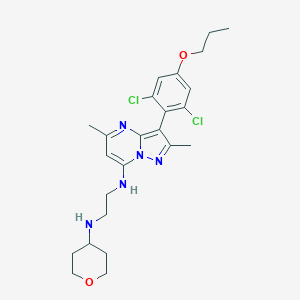 CHEMBL1771261 CHEMBL1771261 | C24H31Cl2N5O2 | 492.445 | 6 / 2 | 5.2 | No |
| 21385 | 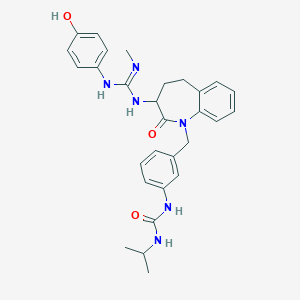 CHEMBL262904 CHEMBL262904 | C29H34N6O3 | 514.63 | 4 / 5 | 3.6 | No |
| 21632 | 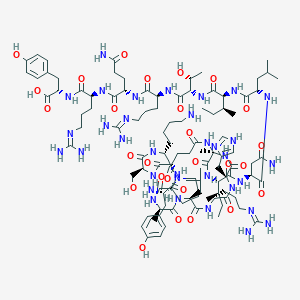 CID 44346875 CID 44346875 | C104H168N34O27 | 2326.7 | 33 / 34 | -8.4 | No |
| 22039 |  MLS000089771 MLS000089771 | C23H30N2O2 | 366.505 | 4 / 1 | 4.3 | Yes |
| 522108 |  CHEMBL3775436 CHEMBL3775436 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 522162 |  CHEMBL3775769 CHEMBL3775769 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 23372 |  CHEMBL311159 CHEMBL311159 | C23H23N3O5 | 421.453 | 5 / 2 | 3.7 | Yes |
| 23394 | 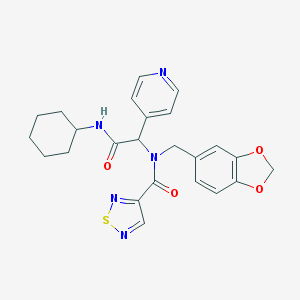 SMR000117962 SMR000117962 | C24H25N5O4S | 479.555 | 8 / 1 | 3.2 | Yes |
| 23746 | 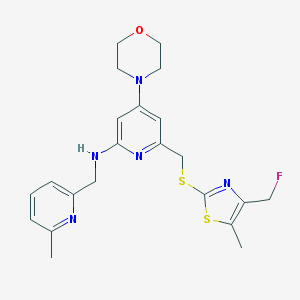 CHEMBL585711 CHEMBL585711 | C22H26FN5OS2 | 459.602 | 9 / 1 | 3.8 | Yes |
| 24258 | 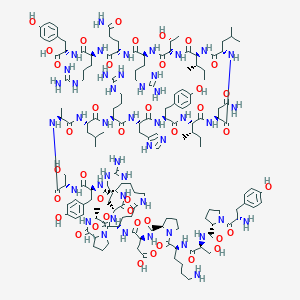 BDBM50046541 BDBM50046541 | C151H233N45O40 | 3318.8 | 47 / 50 | -11.0 | No |
| 24784 | 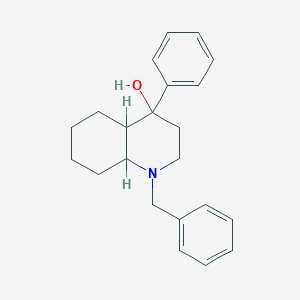 1-Benzyl-4-phenyl-decahydro-quinolin-4-ol 1-Benzyl-4-phenyl-decahydro-quinolin-4-ol | C22H27NO | 321.464 | 2 / 1 | 4.3 | Yes |
| 25225 |  Neuropeptide Y(NPY)(13-36) Neuropeptide Y(NPY)(13-36) | C135H208N40O37 | 2983.39 | 43 / 48 | -5.9 | No |
| 25400 |  MLS002589217 MLS002589217 | C22H21N3O5 | 407.426 | 6 / 2 | 3.9 | Yes |
| 25637 |  CHEMBL410502 CHEMBL410502 | C163H249N48O42+ | 3553.08 | 49 / 51 | -9.1 | No |
| 27183 |  CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27242 | 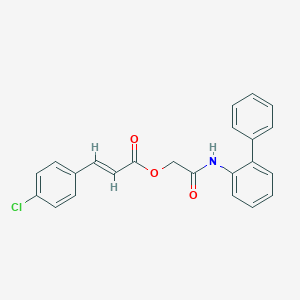 SMR000264080 SMR000264080 | C23H18ClNO3 | 391.851 | 3 / 1 | 5.4 | No |
| 27381 | 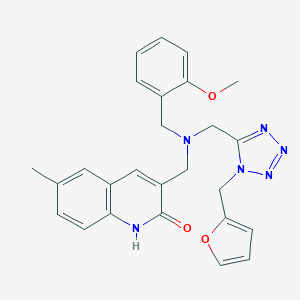 MLS000032393 MLS000032393 | C26H26N6O3 | 470.533 | 7 / 1 | 2.9 | Yes |
| 27747 | 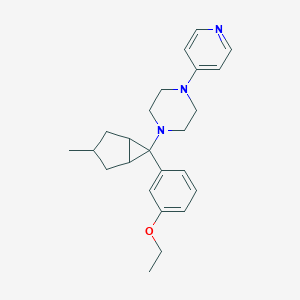 CHEMBL2397297 CHEMBL2397297 | C24H31N3O | 377.532 | 4 / 0 | 4.0 | Yes |
| 29077 | 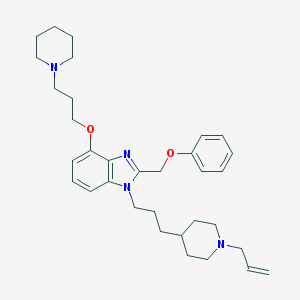 CHEMBL347804 CHEMBL347804 | C33H46N4O2 | 530.757 | 5 / 0 | 6.2 | No |
| 29551 | 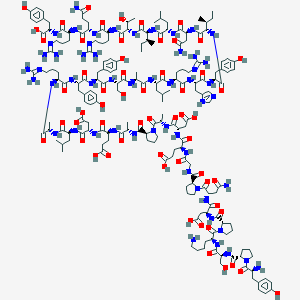 CID 77114776 CID 77114776 | C190H286N54O58 | 4254.7 | 65 / 59 | -16.4 | No |
| 29633 | 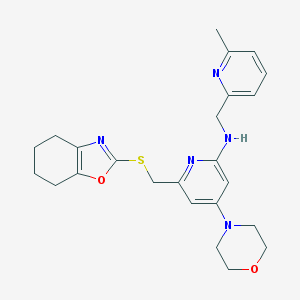 CHEMBL566040 CHEMBL566040 | C24H29N5O2S | 451.589 | 8 / 1 | 3.6 | Yes |
| 30142 | 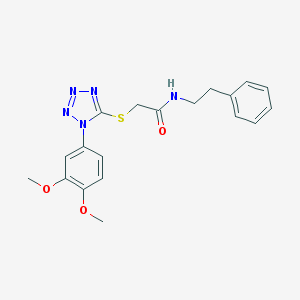 MLS000712137 MLS000712137 | C19H21N5O3S | 399.469 | 7 / 1 | 3.3 | Yes |
| 30516 | 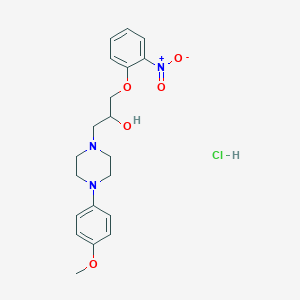 MLS000677133 MLS000677133 | C20H26ClN3O5 | 423.894 | 7 / 2 | N/A | N/A |
| 30877 |  CHEMBL90497 CHEMBL90497 | C28H30FN5O3 | 503.578 | 4 / 4 | 3.8 | No |
| 30977 |  MLS000569792 MLS000569792 | C19H19N3OS | 337.441 | 3 / 2 | 4.0 | Yes |
| 31096 |  CHEMBL315299 CHEMBL315299 | C29H31N7O3 | 525.613 | 4 / 5 | 3.4 | No |
| 31255 |  CHEMBL165506 CHEMBL165506 | C29H37N3O3S | 507.693 | 6 / 2 | 5.0 | No |
| 31375 |  CHEMBL1774210 CHEMBL1774210 | C58H68F3N9O6 | 1044.23 | 11 / 6 | N/A | No |
| 31605 |  CID 44439571 CID 44439571 | C81H123N27O18 | 1763.05 | 23 / 29 | -2.0 | No |
| 32747 |  CHEMBL144262 CHEMBL144262 | C24H29BrN2OS | 473.473 | 4 / 0 | 6.0 | No |
| 32872 |  CHEMBL440944 CHEMBL440944 | C24H26ClN3O2 | 423.941 | 4 / 0 | 4.0 | Yes |
| 558291 | 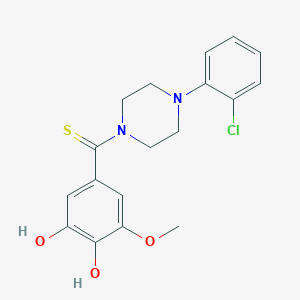 MLS000760186 MLS000760186 | C18H19ClN2O3S | 378.871 | 5 / 2 | 3.5 | Yes |
| 33417 | 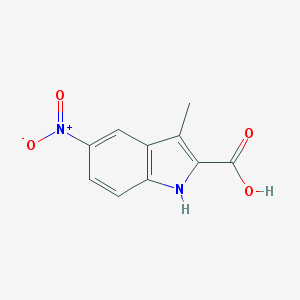 3-Methyl-5-nitro-1H-indole-2-carboxylic acid 3-Methyl-5-nitro-1H-indole-2-carboxylic acid | C10H8N2O4 | 220.184 | 4 / 2 | 2.1 | Yes |
| 33503 | 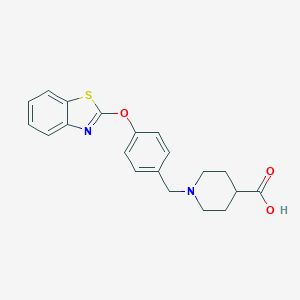 UNII-7W7511NPF8 UNII-7W7511NPF8 | C20H20N2O3S | 368.451 | 6 / 1 | 1.8 | Yes |
| 33639 | 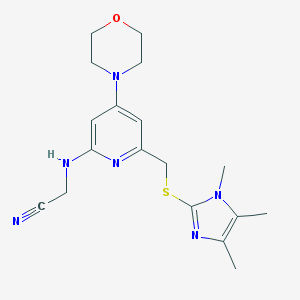 CHEMBL549699 CHEMBL549699 | C18H24N6OS | 372.491 | 7 / 1 | 2.0 | Yes |
| 33837 | 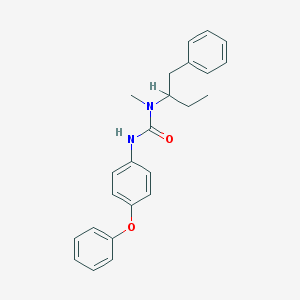 CHEMBL77589 CHEMBL77589 | C24H26N2O2 | 374.484 | 2 / 1 | 5.6 | No |
| 34392 | 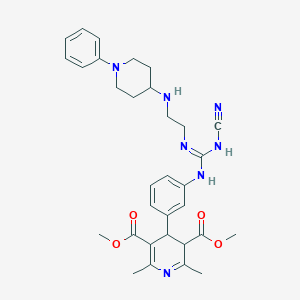 BDBM50156325 BDBM50156325 | C32H39N7O4 | 585.709 | 9 / 3 | 3.7 | No |
| 34549 | 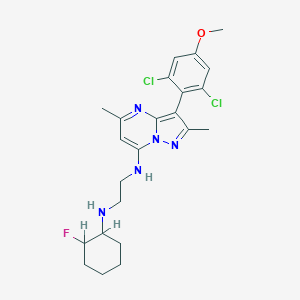 CHEMBL1771264 CHEMBL1771264 | C23H28Cl2FN5O | 480.409 | 6 / 2 | 5.5 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417